Abstract
Background and Aims
Since the early 1990s, research on genetic variation of phenotypic plasticity has expanded and empirical research has emphasized the role of the environment on the expression of inbreeding depression. An emerging question is how these two evolutionary ecology mechanisms interact in novel environments. Interest in this area has grown with the need to understand the establishment of populations in response to climate change, and to human-assisted transport to novel environments.
Methods
We compare performance in the field of outcrossed (O) and inbred lines (S1, S2) from 20 maternal families from each of two native populations of Mimulus guttatus. The experiment was planted in California in each population's home site, in the other populations's home site, in a novel site within the native range of M. guttatus, and in a novel site within the non-native range in North America. The experiment included nearly 6500 individuals. Survival, sexual reproduction and above-ground biomass were examined in order to evaluate inbreeding depression, and stem diameter and plant height were examined in order to evaluate phenotypic plasticity.
Key Results
Across all field sites, approx. 36 % of plants survived to flowering. Inbreeding depression differed among sites and outcrossed offspring generally outperformed selfed offspring. However, in the native-novel site, self-progeny performed better or equally well as outcross progeny. Significant phenotypic plasticity and genetic variation in plasticity was detected in the two architectural traits measured. The absolute value of plasticity showed the most marked difference between home and non-native novel site or non-native-novel site. Evidence was detected for an interaction between inbreeding and plasticity for stem diameter.
Conclusions
The results demonstrate that during initial population establishment, both inbreeding depression and phenotypic plasticity vary among field sites, and may be an important response to environments outside a species' currently occupied range. However, the interaction between inbreeding and plasticity may be limited and environment-dependent.
Keywords: Inbreeding depression, non-native species, Mimulus guttatus, opportunistic plasticity, phenotypic plasticity
INTRODUCTION
Widespread variation exists across plant species in their ability to self-pollinate (ability to fertilize ovules with their own pollen) and outcross (Vogler and Kalisz, 2001). Inbred progeny often differ from outcross progeny in a variety of phenotypic traits. One of the well-studied phenotypic differences between inbred progeny in comparison to outbred progeny is the reduction in fitness-related traits, termed inbreeding depression (Darwin, 1859). The reduction in fitness through inbreeding depression is due, in part, to exposure of deleterious or partially deleterious alleles when plants are homozygous (Charlesworth and Charlesworth, 1987; Dudash et al., 1997; Cheptou and Donohue, 2011). In a population, these deleterious or partially deleterious alleles can be selectively purged (Dudash and Carr, 1998).
Recent research has demonstrated that a wide range of environmental factors influence inbreeding depression (e.g. Carr and Dudash, 1995; Hayes et al., 2005; Ivey and Carr, 2005; Steets et al., 2007; Botham et al., 2009; Bello-Bedoy and Nunez-Farfan, 2010; O'Halloran and Carr, 2010). Environmental sensitivity to inbreeding is evaluated through many components of fitness including early plant size, flowering time, flower production, biomass and seed production. Additionally, inbreeding depression has been implicated in influencing the probability of population persistence (reviewed by Cheptou and Donohue, 2011). Ronce et al. (2009) have predicted that maladapted populations, such as those going through transient environmental shifts during establishment in novel environments, exhibit low levels of inbreeding depression.
Colonization of new locations, in response to climate change, or transport to novel geographic locations, results in many environmental differences compared with the home environment. Mates may be scarce and successful populations probably self-fertilize (e.g. Sakai et al., 2001; Dluglosh and Parker, 2008; Dudash and Murren, 2008). There is a paradox that in newly established populations outside the native range, population sizes are small and often with limited genetic variation (see for example Allendorf and Lundquist, 2003). Additionally, inbreeding may expose recessive mutations; thus, harmful effects such as inbreeding depression may be expected. Yet novel habitats are frequently considered to have abundant resources in association with the escape from co-adapted herbivores, competitors, etc. (Maron et al., 2004). Extreme resource availability and novel environmental conditions may mask effects of inbreeding depression according to a recent quantitative genetics model proposed by Ronce et al. (2009). Their predictions remain an open area of inquiry. In this study we ask the questions: is inbreeding depression observed in newly established populations in a novel environment within the native range, as well as in a non-native location? Does inbreeding interact with other mechanisms that contribute to population establishment and persistence?
Another important aspect of performance of a genotype in response to variation in the environment is phenotypic plasticity (Schlichting, 1986; Pigliucci, 2001). Evidence for the importance of phenotypic plasticity in long-established exotic species is growing (e.g. Bossdorf et al., 2005; Richards et al., 2006; Muth and Pigliucci, 2007; Davidson et al., 2011). In exotic taxa, phenotypic plasticity of morphological traits may act as a buffer in a new environment and contribute to population establishment success (Dudash et al., 2005; Ghalambor et al., 2007). Godoy et al. (2011) demonstrated in an established non-native colonist, Prunella vulgaris, in South American Valdivian forest that expansion into the forest environment was possible via loss of plasticity. In a recent meta-analysis of published studies contrasting invasive and native species, Davidson et al. (2011) demonstrated that invasive species exhibited greater plasticity than related native species. At initial establishment in a novel habitat, selection may not have had time to shape the responses to this environment. The maintenance or loss of phenotypic plasticity may play an important role in the process of population establishment. We term the particular case of performance in response to the novel habitat ‘opportunistic plasticity’ (Dudash et al., 2005).
The simultaneous occurrence of inbreeding depression and phenotypic plasticity has rarely been studied, although both are cited as critical evolutionary mechanisms in initial population establishment (Schlichting and Levin, 1986; O'Halloran and Carr, 2010). Two hypotheses have been discussed regarding the interaction between plasticity and inbreeding (Schlichting 1986). First, the developmental instability hypothesis (Pederson, 1968) states that plants with low levels of heterozygosity are more sensitive to environmental variation; thus, if more inbred they express greater developmental instability, and as a result are more plastic than outcrossed, more heterozygous individuals. In two tests of this hypothesis, inbreeding was not found to influence patterns of plasticity in either cultivated Phlox across greenhouse environments (Schlichting and Levin, 1986) or Mimulus ringens in native field sites or greenhouse environments (O'Halloran and Carr, 2010). In a second related hypothesis, Lerner (1954) suggested that inbreeding may decrease developmental stability. In an animal system, Auld and Relyea (2010) found that inbred lines of snails had reduced patterns of phenotypic plasticity to natural predators in comparison to outcrossed lines. These results are contrary to Lerner's hypothesis. These studies explored the interaction between inbreeding and phenotypic plasticity in native field habitats or in manipulated experimental conditions; however, in a novel environment, the developmental instability hypothesis may suggest one component of how newly established populations (which are often small and highly inbred) may be successful – through the expression of phenotypic plasticity.
Here, we address gaps in our understanding of the independent and joint influences of inbreeding depression and phenotypic plasticity as contributing factors in establishment of new populations within and outside the native range, by employing the emerging ecological–genetic model system (Wu et al., 2008), Mimulus guttatus. The yellow monkeyflower, M. guttatus, is native to western North America (Grant, 1924), has an extensive non-native range in eastern North America, Europe and New Zealand (van Kleunen and Fischer 2008; Murren et al., 2009), and is considered invasive in some areas of Europe (Truscott et al., 2006, 2008). Native and non-native populations of M. guttatus go through cycles of local extinction and recolonization, with commensurate fluctuations in population sizes (e.g. Vickery, 1999; Truscott et al., 2006; 2008). Mating systems vary widely among populations, from highly selfing, to mixed mating, to largely outcrossing (e.g. Ritland and Ritland, 1989; Dudash and Ritland, 1991; Fishman and Willis, 2008). Previous studies have documented inbreeding depression variation among maternal families and populations of M. guttatus across a range of traits (e.g. Carr and Dudash, 1997; Dudash et al., 1997; Carr and Eubanks, 2002; Carr et al., 2004; Ivey et al., 2004). Phenotypic plasticity has been investigated in relation to aspects of variation within and between populations to biotic and abiotic environments (Galloway, 1995; van Kleunen and Fischer, 2008; Holeski and Kelly, 2006; Murren et al., 2006). The present study has the explicit goal of examining phenotypic plasticity and inbreeding depression across field habitats. We examine whether differences in reproductive and morphological traits occur among cross types (outcross, and two generations of selfing) and across field locations, including the native home site, a native site occupied by M. guttatus, one novel native site near the home sites in California, and a novel, non-native site in Maryland. We investigate three specific hypotheses. (1) We hypothesize that the expression of inbreeding depression will be greater in home environments than other habitats, as homozygous mutational effects may disrupt patterns of local adaptation (Ronce et al., 2009). (2) We expect phenotypic plasticity to be greatest in self generations in comparison to the outcross generation, consistent with the developmental instability hypothesis (Pederson, 1968; Schlichting, 1986). (3) We expect plastic responses to be greatest to a native-novel environment and a non-native-novel environment in comparison to the native-occupied environment, as these field sites differ in a wide range of characteristics in comparison to the home site (Maron et al., 2004; Dudash et al., 2005; Auld, 2010; Baird et al., 2011).
METHODS
Study species
Mimulus guttatus is a herbaceous plant species that is annual in habitats with seasonal moisture (as in this study), and perennial in areas that are wet year-round (Dole, 1992; Hall and Willis, 2006). Plants initially establish a rosette of leaves and, after internode elongation, flowering is initiated. Individual plants are upright and produce one to multiple perfect, often paired, yellow flowers that vary in size (Fenster and Ritland, 1994; Martin, 2004; Murren et al., 2009). Flowers are visited by small invertebrates (Ivey and Carr, 2005). Self-pollination occurs through geitonogamous pollination, or through within-flower corolla dragging (Dole, 1992). Numerous wind- and water-dispersed seeds are produced per flower (Waser et al., 1982; Vickery, 1999).
Field methods
We obtained seed from two M. guttatus populations located in Napa County, California (CA). The population that we name ‘LT’ is from Napa County Land Trust property on Snell Valley Road (38°41′N, 122°24′W). The population named ‘M13’ (38°33′N, 122°22′W) was initially studied by Carr and colleagues (e.g. Carr and Eubanks, 2002; Ivey and Carr, 2005), and we retain the name that they used. From 2002–2004, we observed M13 to be smaller in population size than LT (approx. 1000 vs. approx. 10 000 individuals).
Crossing design
We germinated seed collected from 20 maternal plants in the field from each population to create outcross, S1, S2 and S3 lines of the same maternal heritage in the University of Maryland greenhouse. All pollinations were conducted by researchers to ensure a known pollen source was transferred to the stigma (following methods described in Carr and Dudash, 1997). At each paired node, one flower received pollen from a randomly chosen mate from within the population, and one flower received pollen from the same individual to create outcross and S1 lines. Seed from these S1 selfed seed were then planted, grown up, and pollinated following the same method to create a second generation of selfed and outcross lines. This process was repeated to create S3 lines. From the original maternal plants, three replicate lines (called sublines) were created from single-seed descent for all self-generations. We included sublines to account for maternal effects. Plants were continuously bottom-watered and day length was extended to 18 h using sodium vapour lights to encourage flowering. Analyses (described below) initially included maternal effects; however, as maternal effects were not found to be significant in any of our statistical models, we omitted this as a factor in subsequent statistical models (Quinn and Keough, 2004). Ultimately, we created 20 maternal families per population, each represented by O, S1 and S2 generations, and occasionally an S3 generation.
Native California sites: native-home, native-occupied and native-novel
In the first week of January 2004, seeds of both populations and all generations were sown into seedling plug trays outside at the Wantrup Wildlife Sanctuary field station (Napa County Land Trust, Pope Valley, CA, 38°36′N, 122°22′W). Plug trays were dug into the ground and kept moist through natural rainfall and additional misting. After 3 weeks, seedlings were hand-transplanted into a randomized, incomplete design at three CA field sites. The first native site was the LT home site at the Snell Valley Wildflower Reserve managed by the Napa County Land Trust. The second native site was the M13 home site in a seep along the Pope Valley Road (see also Carr and Eubanks, 2002; Ivey and Carr, 2005). We reciprocally transplanted plants into both their native-home and the native site of the other population (termed native-occupied site, which is unique in that it is a suitable M. guttatus habitat, but one population is not locally adapted there). The third CA site was in a large seep on the Wantrup Wildlife Sanctuary and represented our native-novel site, as no M. guttatus had been reported from that location in the previous decade, during which censuses had been conducted (J. Callizo, Napa Land Trust, pers. comm.). We consider this site as novel habitat, as a population of M. guttatus was not known from the site.
We sowed seed at the field station from two populations (LT and M13), from 20 maternal families per population, up to three sublines per family, and up to four cross-treatments (O, S1, S2 and, when available, S3). Of those that germinated, individual seedlings were hand-transplanted at each of three field sites in an incomplete block design. Each site had 12 blocks with 160 planting locations marked with coloured swizzles (Soodhalter Plastics, Los Angeles, CA). Not all planting locations were filled in each block; sublines were often represented in a fraction of the blocks. Summing across three field sites in the native range, we planted 4742 plants (of the 20 maternal families from each of the two populations described above) with a total of 1382 outcross progeny, 1360 S1 progeny, 1295 S2 progeny, and 698 S3 progeny. Plants from the third generation of selfing were under-represented in the planting due to both lower seed production during the generation of seed in the greenhouse, and low germination in the field (Supplementary Data Table S1). Plants were harvested at the beginning of natural senescence in May after flowering was completed and prior to seed release (in accordance with an agreement with the land managers).
Non-native-novel site (Maryland)
We employed the same germination and planting methods that were used in CA in the Maryland (MD) site at USDA Beltsville Agricultural Research Center (BARC). The MD site was an area adjacent to a small stream that was naturally wet year-round and represented the non-native-novel site with similar moisture levels as the native range, and type of setting in other non-native populations of M. guttatus in eastern North America (Murren et al., 2009; USDA/NRCS, 2011). However, planting date differed due to climatic variation between the two coasts. Seeds were from both the LT and M13 populations from the same families (20 per population), sublines, and cross-treatments (O, S1, S2, and occasional S3) as planted in CA, but were sown in early March 2004, and transplanted 3 weeks later into 12 blocks. Two additional blocks (numbers 13 and 14) were planted 2 weeks after these initial blocks, because two of the original blocks were lost following flooding of the site. This planting effort resulted in a total of 1752 plants representing 20 maternal families from the two populations and composed of 558 outcross progeny, 545 S1 progeny, 464 S2 progeny, and 185 S3 progeny. We again harvested at the beginning of natural senescence when flowering was completed, since property managers had requested that we not allow seed to disperse into the sites. When all four field sites were summed, a total of 6497 plants were scored for survival and phenotypic traits.
Phenotypic traits
Survival was scored as either alive at harvest or died sometime prior to harvest, and corresponded with a census done prior to flowering (data not shown). Stem diameter and plant height were the morphological measures used in the plasticity analyses, and sexual reproduction and above-ground biomass were fitness traits used in the inbreeding depression analyses. After harvest, all plants from all planting sites were dried in a drying oven to constant weight. We measured the total height (length) of the main stem, and stem diameter just above the rosette leaves. Height of the main stem of the plant and stem diameter were previously shown to be phenotypically plastic architectural traits (e.g. Murren et al., 2006), and function as structural support and pollinator attraction. The hollow stems also contribute to air movement through the plant. Given the request of land managers that we limit seed release into the field site, when plants completed flowering they were harvested. This management request influenced our measure of fitness: thus, we employed total flower production per plant as our measure of lifetime sexual reproduction. We also obtained the biomass of the entire above-ground portion of the plant. These measures of fitness have previously been employed to evaluate inbreeding depression in this species (e.g. Dudash et al., 1997).
Analyses
All analyses were conducted in SAS Version 9·2 (SAS Institute, 2010).
Survival
Survival was calculated as a percentage of living plants out of the total seedlings planted at each site, population and generation. In a logistic regression model approach in the genmod procedure, we evaluated whether the fixed effects of populations, generations and sites differed in survival via likelihood ratio statistics (a rationale provided below for viewing these as fixed effects).
Population-level analysis of sexual reproduction and biomass among cross-types and field sites
Analysis of variance utilizing restricted maximum likelihood procedure (REML) in mixed or glimmix was employed to compare measures of reproductive success between populations, across cross-types (outcross, O, one generation of hand-selfing, S1, two generations of hand-selfing, S2), and across four planting sites (in CA: native-home site at the Napa Valley Land Trust, LT, native-home site at M13, native-novel site at Wantrup Wildlife Refuge; and in MD: non-native-novel site at USDA). Owing to limited germination of S3 seed and survival of those planted across the four sites (see Supplementary Data Table S1) we omitted the S3 generation from any formal statistical analyses. ‘Population’ examined an effect of overall genetic differentiation between the LT and the M13 populations in reproductive traits. ‘Population’ was considered as a fixed effect as we specifically chose to study a population (M13) previously examined in the native range in response to herbivory and inbreeding (e.g. Carr and Eubanks, 2002). The cross-type effect evaluated differences in phenotypes across outcross and generations of selfed progeny, and was considered a fixed effect, and a first step at evaluating inbreeding depression: a significant cross-type effect suggested an influence of inbreeding on performance of reproductive characters. ‘Site’ referred to the four planting locations, and was considered a fixed effect. Variation across planting sites suggested that aspects of the ecology associated with geographic location influenced differences in trait expression related to fitness, and was a measure of phenotypic plasticity of the fitness traits. Block effects nested within sites were considered random effects, and their inclusion resulted in an over-parameterized model. Therefore, they were omitted from this analysis (Quinn and Keough, 2004).
We also investigated pairwise and three-way interactions. Interaction terms with planting site suggested that populations or cross-types varied in their response to environmental variation and indicated genetic variation for phenotypic plasticity in fitness-related traits. Population by cross-type interactions suggested that populations varied in their response to inbreeding. We examined residuals of these models to assess assumptions of analysis of variance. Residuals were split when the homogeneity of variances assumption was not met (also employed in Murren et al., 2009; recommended by Dhulgosh and Parker, 2008), and log-transformation was required to meet ANOVA assumptions for sexual reproduction, but not biomass. Tukey post hoc tests were employed to further examine possible differences among levels of significant effects.
Family-level analysis of sexual reproduction and biomass among cross-types and field sites
Given our limited overall survivorship, we could not run a full model that included all the above effects as well as family effects and their interactions. Therefore, to specifically examine variation among maternal lines within populations for sexual reproduction and above-ground biomass, we ran a pair of mixed-model analyses of variance separately for the LT and M13 populations. These models allowed us to evaluate genetic variation within populations for reproductive traits and ask if maternal lines varied in their response to inbreeding and field sites within native and non-native habitats. The interaction term of cross-type by site was evaluated as an effect of inbreeding on phenotypic plasticity of the fitness-related traits. Maternal lines were considered random effects as were the pairwise interactions. The model included the fixed effects of cross-type, planting site and their interactions, and the random effects of family, block nested within site, planting site × family, and cross-type × family. Significances of random effects were evaluated via a χ2 test of the differential log-likelihoods.
Relative performance
As a companion to the analyses of variance described above for graphical interpretation, we chose to calculate the inbreeding depression coefficient for each maternal genotype. Relative performance was calculated as
RP = (outcross – self)/maximum
where maximum equals outcross when outcross > self, and maximum equals self when self > outcross. This approach restricts values of inbreeding depression to between 1 and –1, such that when self progeny outperform outcross progeny values are negative, and when outcross progeny outperform self progeny values are positive. This method allows comparison across studies because it is a standardized measure (e.g. Ågren and Schemske, 1993; Dudash et al., 1997; see recent meta-analysis Angeloni et al., 2011). Relative performance estimates were calculated separately for progeny resulting from both one and two generations of selfing compared to outcrossed progeny.
Phenotypic plasticity and genetic variation for plasticity in architectural traits among cross types and field sites
We assessed phenotypic plasticity with the specific aims of examining whether phenotypic responses were greatest at the two novel field sites, and if inbreeding influenced patterns of plasticity. We employed two approaches using REML analyses of variance in the mixed procedure. The first model included the fixed effects of population, planting location (‘site’), cross-type, and the site × cross-type interaction. Random effects included family nested within population, block within site, and the interactions family(population) × site, and family(population) × cross-type. Plasticity was evaluated by the planting site effect. A significant cross-type effect revealed an influence of inbreeding on trait expression. Evidence for influence of inbreeding on plasticity was evaluated by the site × cross-type interaction. Genetic variation for the traits was evaluated by the population, family nested within population, and family(population) × cross-type interactions. Genetic variation for plasticity was evaluated by the family nested within population × site interaction.
Differential plasticity analysis
In a second approach to examine how genotypes respond across environments in their phenotypic expression, we evaluated differential phenotypic expression between the original home site of the population in comparison to each of the three other planting locations. We employ this approach as our study sites are environmental categories rather a continuous environmental gradient. Reaction norms along continuous gradients are often evaluated by a slope, whereas two environment comparisons are often evaluated as a performance difference (see Via et al., 1995; and more recently Auld, 2010). Our goal was to compare the variation in phenotype between the home, locally adapted, environment and the other three environments. To examine how performance varied across pairs of environments, we followed the methods employed recently by Baird et al. (2011) and Auld (2010) to evaluate how a family (up to 20 families per population) and per-generation of inbreeding (O, S1, S2) differed in performance between sites. Our calculation was, for example, mean stem diameter of O progeny of family x from population LT at the LT home site minus mean stem diameter of O progeny of family x from population LT at the native-novel site. Maternal line was the replicate unit in this analysis. We conducted all differences in comparison to the native-home site specific to the M13 and LT population. These home vs. other planting site differences in phenotypes were our measures of plasticity, as it was a genotype-specific response across environments. As we cannot evaluate changes both in mean and slope in two-environment investigations (Pigliucci, 2001), we are unable to investigate changes in the shape of the reaction norm in this analysis.
We conducted a fixed effects analysis of variance model to examine if populations differed in this measure of cross-environment performance (population main effect, a measure of genetic variation for plasticity), if there was an effect of inbreeding on this measure of plasticity (cross-type main effect), if there was a difference in plasticity in response to novel vs. native planting sites (native-novel main effect), and if cross types differed between populations in plastic responses (population × cross-type interaction).
RESULTS
Survival
Overall, across all four planting sites survival of planted individuals to harvest was 36 % (2330 individuals). We found no significant difference in overall survival to harvest between the Land Trust (LT) population (37 %) and the Carr and Eubanks' ‘M13’ population (35 %; χ2 = 1·26, d.f. = 1, P = 0·26). Survival differed significantly across cross-types (χ2 = 12·85, d.f. = 2, P < 0·002) with significantly greater survival for the outcross group (39 %) than the first generation of selfing (35 %), the second generation of selfing (35 %) or the third generation of selfing (31 %, although the S3 only represented 13 % of all plants in the experiment – see Supplementary Data Table S1 – and are not included in analyses below). Survival differed significantly across planting sites (χ2 = 644·22, d.f. = 3, P < 0·0001) with greatest survival at the LT site (57 %), which was significantly greater than the M13 site (27 %), the native-novel site at Wantrup Wildlife Sanctuary (35 %) and the non-native-novel site in MD (25%, including the two blocks that were flooded just after planting).
Population-level analysis of sexual reproduction and biomass among cross-types and field sites
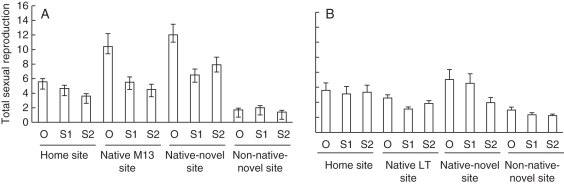
We detected significant variation among cross-types (generations of selfing and outcrossing), populations and sites, in both total sexual reproduction and above-ground biomass (Table 1) suggesting that inbreeding and population of origin as well as ecology influence the expression of these traits. The LT population outperformed the M13 population at the LT native-home planting site and in the M13 home planting site for both traits (post hoc tests, Table 1; Fig. 1; Supplementary Data Table S2). Performance (measured as sexual reproduction or biomass) overall was greatest in the native-novel site (Tukey post hoc tests, Table 1; Figure 1; Supplementary Data Table S2). Outcross progeny generally outperformed the two generations of selfing for both fitness traits (Tukey post hoc tests, Table 1; Fig. 1; Supplementary Data Table S2). Additionally, we found a significant interaction between population and site in sexual reproduction, and significant site × population × cross-type interactions in above-ground biomass (Table 1).
(b) Results of Tukey post hoc text
| Cross-type |
Planting site |
||||||
|---|---|---|---|---|---|---|---|
| O | S1 | S2 | Native-novel | Native-occupied | Home | Non-native-novel | |
| Sexual reproduction | 1·49a | 1·35b | 1·26b | 1·75a | 1·46b | 1·42b | 0·84c |
| Biomass | 0·11a | 0·07b | 0·05b | 0·16a | 0·07b | 0·07b | 0·003c |
d.d.f. (denominator degrees of freedom) sexual reproduction =1659, d.d.f. biomass = 1724. Lifetime sexual reproduction was log-transformed to meet assumptions for ANOVA. All effects in this model are fixed. The cross-types are outcross (O), first generation of selfing (S1) and second generation of selfing (S2). See Methods for description of sites and populations. Values are LS means and different letters indicate statistically significant differences.
Table 1.
(a) Analysis of variation to examine effects of inbreeding and planting site on sexual reproduction and above-ground biomass (g) for two populations of Mimulus guttatus. Performance was evaluated across four planting sites, and three cross-types (see text)
| Sexual reproduction |
Above-ground biomass |
|||||
|---|---|---|---|---|---|---|
| Source of variation | d.f. | F | P | d.f. | F | P |
| Site | 3 | 83·07 | < 0·0001 | 3 | 26·05 | < 0·0001 |
| Cross-type | 2 | 10·88 | < 0·0001 | 2 | 9·09 | 0·0001 |
| Population | 1 | 11·51 | 0·0007 | 1 | 0·08 | 0·78 |
| Site × Cross-type | 6 | 1·48 | 0·18 | 6 | 1·66 | 0·13 |
| Site × Population | 3 | 4·35 | 0·005 | 3 | 2·15 | 0·09 |
| Population × Cross-type | 2 | 1·58 | 0·21 | 2 | 2·85 | 0·06 |
| Site × Population × Cross-type | 6 | 1·95 | 0·07 | 6 | 2·25 | 0·04 |
Fig. 1.
Mean lifetime sexual reproduction across maternal lines for (A) population LT and (B) M13 across field sites and cross-types (O, outcross generation; S1, one generation of hand self-pollination; S2, two generations of hand self-pollination). Home site is the LT or M13 site of origin, native site is the reciprocally transplanted site, native-novel site is the CA native site with no M. guttatus population, and non-native-novel site is the non-native range in MD. Bars represent the standard error of the mean. See Supplementary Data Table S1 for survivorship.
Across both populations, sexual reproduction was reduced at the non-native-novel site in comparison to native range sites (Fig. 1). The LT population had increased sexual reproduction in both the M13 site (the non-native-occupied site) and the native-novel site, whereas the M13 population had reduced performance in the LT site compared to its home site. Above-ground biomass was greatest at the native-novel site (Supplementary Data Table S2), and outcross progeny were generally larger than S1 and S2 progeny (Supplementary Data Table S2). However, in the M13 population, biomass was greater in the S1 progeny than the outcross and S2 progeny in the native-novel site.
Family-level analyses among cross-types and field sites
In a separate pair of analyses for each population, performance of total sexual reproduction differed significantly among families in the LT population. However, for both populations, families differed in their response to planting location (family × site interaction), and we detected genetic variation among families to inbreeding (family × cross-type interaction; Table 2). Overall, sexual reproduction differed among cross-types and field sites, as seen in the previous analysis (Table 2, Fig. 1).
(b) Fixed effects
| d.f. | F | P | d.f. | F | P | |
|---|---|---|---|---|---|---|
| Site | 3 | 7·41 | 0·0005 | 3 | 3·03 | 0·04 |
| Cross-type | 2 | 8·66 | 0·0009 | 2 | 2·95 | 0·05 |
| Site × Cross-type | 6 | 1·65 | 0·13 | 6 | 0·56 | 0·77 |
For details of cross-types see text. Sites are the four planting sites in native-home sites of LT and M13, native-novel site in CA, and non-native-novel site in MD. Block effects nested within site were accounted for as a random effect in both models. – indicates no variance associated with the factor in the model. d.f. for family was 19 per population. Sexual reproduction was log-transformed to meet the assumptions of the analysis. Test for the random effects was based on Chi-square, and for the fixed effects, F-test.
Table 2.
Family-level mixed-model analysis of variance (REML) for sexual reproduction among cross types (see text) and four field planting sites for populations LT and M13 (a) Random effects
| LT |
M13 |
|||
|---|---|---|---|---|
| Source of Variation | Estimate | P | Estimate | P |
| Family | 0·02 | 0·002 | – | – |
| Family × Site | 0·008 | 0·007 | 0·02 | 0·004 |
| Family × Cross-type | 0·02 | 0·008 | 0·01 | 0·001 |
Relative performance
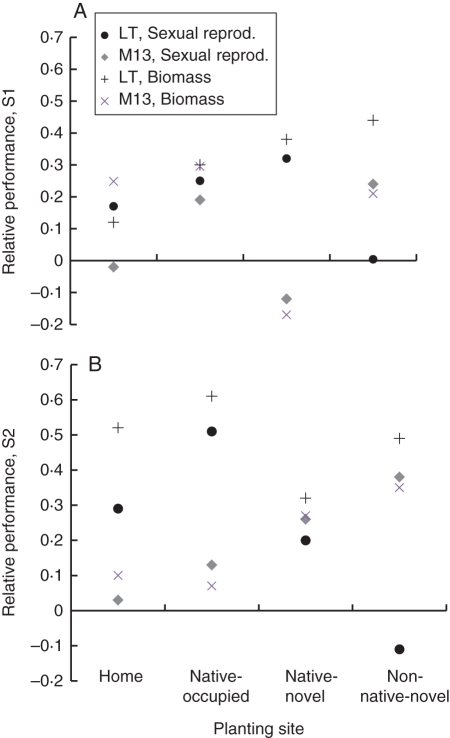
In an examination of relative performance of S1 and outcross progeny by population and site, outcross progeny outperformed selfed progeny at most sites for both sexual reproduction and above-ground biomass (Fig. 2A). However, S1 progeny outperformed outcross progeny in the M13 population at the home site in sexual reproduction, and in the native-novel environment for both sexual reproduction and biomass (Fig. 2A). Measures of relative performance were low at the home site for LT biomass, and increased with increasing novelty of the environment. Performance as measured by sexual reproduction of outcross to selfed progeny was nearly zero for LT at the non-native-novel site, and for M13 at home. In the S2 generation, most measures of relative performance across the four sites were positive, indicating outcross progeny consistently outperformed S2 progeny (Fig. 2B). Only for the LT population in the non-native-novel site did S2 progeny outperform outcross progeny. As in the S1 generation, at home, the relative performances of the S2 generation of the M13 population as measured by both sexual reproduction and above-ground biomass were nearly zero (Fig. 2B), indicating equal performance between the selfed and outcross cross-types.
Fig. 2.
Relative performance across native and non-native planting sites sites of two populations of Mimulus guttatus for (A) first generation and (B) second generation of selfing progeny. Estimates are of the relative performance of outcross vs. the generation of selfing progeny in question standardized and bounded by 1 and –1, such that positive values indicate outcross > self and negative values indicate self > outcross. Data are for up to 20 maternal families for each of two populations, grown in four field locations including their home field site (either LT or M13), native-occupied site (reciprocal population site, either LT or M13), native-novel site, and non-native-novel site. Estimates of relative performance were calculated for sexual reproduction and above-ground biomass (see key).
Phenotypic plasticity and genetic variation for plasticity among cross-types
We found strong evidence for phenotypic plasticity of plant height and stem diameter (both architectural traits), assessed as variation in trait expression across the four planting sites (site effect in Table 3). Plants had the greatest height and widest stem diameter at the native-novel Wantrup site in comparison to trait expression at the other sites (Supplementary Data Table S2). We also detected a significant generation effect, indicating that inbreeding influenced architectural traits: outcross progeny generally had greater plant height and thicker stems than S1 and S2 progeny. A significant interaction was detected between planting site and cross-type for stem diameter, as either S1 outperformed outcross progeny (e.g. S1 of the M13 population outperformed outcross progeny) or S2 progeny outperformed S1 progeny (e.g. S1 from the M13 population at the LT and M13 planting sites; see also Supplementary Data Table S2). Phenotypes of families within populations differed across cross-types, a measure of genetic variation for plasticity (Table 3).
(b) Random effects
| Estimate | P | Estimate | P | |
|---|---|---|---|---|
| Family(Population) | 0·004 | 0·17 | 0·004 | 0·26 |
| Family(Population) × Cross Type | 0·01 | < 0·03 | 0·01 | 0·04 |
| Family(Population) × Site | 0·002 | 0·37 | 0·001 | 0·07 |
Height and stem diameter were log-transformed to meet assumptions of ANOVA. Model for height d.d.f. = 1404, stem diameter d.d.f. = 1410. For details of populations and cross-types see text. Random effects were evaluated by log-likelihoods. Block nested within site was also significant for both analyses.
Table 3.
Mixed model analysis of variance examining plasticity, genetic variation, and inbreeding effects on phenotypic plasticity of plant height (cm) and stem diameter (cm) in Mimulus guttatus grown in three native and one non-native field site. (a) Fixed effects
| Plant height |
Stem diameter |
|||||
|---|---|---|---|---|---|---|
| d.f. | F | P | d.f. | F | P | |
| Population | 1 | 3·53 | 0·06 | 1 | 0·40 | 0·53 |
| Site | 3 | 14·57 | < 0·0001 | 3 | 14·13 | < 0·0001 |
| Cross-type | 2 | 30·37 | < 0·0001 | 2 | 17·66 | < 0·0001 |
| Site × Cross-type | 6 | 0·92 | 0·47 | 6 | 2·18 | 0·04 |
Differential performance plasticity analysis
We assessed plasticity as the differential performance between the native-home site and the three other sites (Table 4), and examined if plasticity was greater in self or outcross progeny. Stem diameter plasticity and height plasticity were found to differ among sites, and between populations (Table 4). Greatest plasticity (highest absolute value of the differences in performance between home and another site) was observed by comparing performance in the home site to the native-novel site and the non-native-novel site (Supplementary Data Table S3). Note that the sign of the plasticity was negative for both the reciprocal site and the native-novel site, demonstrating that trait values in these sites were higher than at home. Plasticity between home and novel non-native site was positive, indicating that trait values were higher at home than in the distant novel site. Plasticity among populations was dependent on the trait examined (Supplementary Data Table S3), and we did not find cross-type to be a significant main effect in this analysis (Table 4, Supplementary Data Table S3). However, for stem diameter we did detect a population × generation interaction effect, suggesting that for this architectural trait plasticity varied by population and level of inbreeding.
Table 4.
Differential performance analysis. Plasticity is measured by differential family mean performance between home and other sites. Traits were investigated across populations (LT and M13) and cross-types (O, S1, S2; see text). Site-type is whether plasticity is calculated between home site and native-novel, reciprocal home sites, and home site and non-native site.
| d.f. | Stem diameter plasticity (F, P) | Height plasticity (F, P) | |
|---|---|---|---|
| Site-type | 2 | 79·6, <0·0001 | 89·6, <0·0001 |
| Population | 1 | 37·8, <0·0001 | 5·84, 0·01 |
| Cross-type | 2 | 2·25, 0·10 | 0·04, 0·95 |
| Population × Cross-type | 2 | 2·98, 0·05 | 0·70, 0·5 |
d.d.f. = 318, 317.
DISCUSSION
We investigated the impact of native-home site, native-occupied site, a novel site in the native range, and a novel site in the non-native range of M. guttatus on inbreeding depression for fitness-related traits and on phenotypically plastic responses for architectural traits. Our goal was to mimic conditions experienced during initial establishment of populations in the field in native-home environments, in a novel environment in the native range, and in an environment in the non-native range using the model ecological–genetic system M. guttatus. The yellow monkeyflower is known for its widespread native as well as extensive non-native ranges (Grant, 1924; Truscott et al., 2006, 2008; van Kleunen and Fischer, 2008; Murren et al., 2009). We confirmed that outcross offspring generally outperformed selfed offspring, yet in our native-novel site self progeny performed better or equally well as outcross progeny. These results in part support our hypothesis that inbreeding depression would be greatest in native environments. We also identified that phenotypic plasticity and genetic variation in plasticity are detectable through two generations of selfing. Plasticity was greater between home and our native-novel environment and our non-native-novel environment, than between the home and the native-occupied site. We observed limited survivorship overall, and while some replicates of each cross-type survived at each site, survival differed significantly among cross-types and sites. Differential survivorship may be an important aspect to pursue in future studies. Despite the observed patterns of survivorship, we still detected variation in inbreeding depression and phenotypic plasticity across sites. We found an interaction between inbreeding and plasticity for stem diameter. Future studies that expand the investigation of whether mating system and plasticity act independently, or interact, will be valuable in building predictions of native species responses to climate change and invasive species' establishment.
Inbreeding depression
Cheptou and Donohue (2011) advocate that understanding environmentally determined inbreeding depression is critical for predicting the dynamics of populations. Our focus was to investigate whether inbreeding depression was reduced in our two novel sites, as predicted by Ronce et al. (2009). We found that the expression of inbreeding depression was dependent upon location of the planting site. We detected inbreeding depression in the novel site inside of the native range whereas inbreeding depression was reduced in the novel site outside of the native range. These results support the idea of reduced inbreeding depression in a novel environment, as local adaptation probably plays an initially smaller role in such environments (Ronce et al., 2009). Responses to novel environments contiguous with the native range may be quite different than responses to novel environments in non-native habitats (such as occurs with introduction of exotic species) where environmental conditions could be dramatically different, and warrant further investigation.
Both greenhouse and field studies report variation in environmentally determined inbreeding depression (Dudash, 1990; Armbruster and Reed, 2005; Cheptou and Donohue, 2011). Inbreeding in the M. guttatus M13 population reduced tolerance to herbivory (Carr and Eubanks, 2002; Ivey et al., 2004) and to the cucumber mosaic virus (Eubanks et al., 2005), and decreased pollinator visitation (Ivey and Carr, 2005). These researchers have repeatedly observed both direct and indirect effects of inbreeding, and greater magnitudes of inbreeding depression in the field compared to greenhouse studies (Carr and Eubanks, 2002; Ivey et al., 2004; Ivey and Carr, 2005). Our results support Carr and colleagues' earlier findings of environmentally dependent inbreeding depression in the M13 population, and expands on their findings by demonstrating the occurence of increased expression of inbreeding depression at a novel site inside the native range, and decreased expression of inbreeding depression at a novel site in the non-native range. A next important set of studies would be to assess inbreeding depression across a large range of sites in the native and non-native range, allowing further characterization of the reaction norm (Schlichting, 2008). In the native range, Schmitt and Gamble (1990) identified that as distance from the parental site increased, inbreeding depression also increased. Within the native range this is also the pattern that we detected in the LT population but not in the M13 population. This suggests that populations may differ in response to new environments. Ecological differences between the home site and a set of newly dispersed sites may also vary. The home sites of the LT and M13 populations differ in multiple ecological attributes, with M13 being a disturbed roadside seep, with greater shading from trees, silty and rocky soil, and less water throughout the season. Its population size is considerably smaller and more variable than LT across years. The protected LT site is open and expansive, the seep stayed moist longer, the soil had more clay, and population size was large and stable across years. These habitat differences probably contribute to the differences in local adaptation (van Kleunen and Fischer, 2008; Murren et al., 2006), population size and variation in inbreeding depression observed. Had we chosen populations from distinct portions of the range, or from distinct habitat types that are native to M. guttatus, these differences would probaby be exaggerated.
Expanding this idea geographically, Pujol et al. (2009) examined populations of Mercurialis annua along the distribution of a historic migration path after the Pleistocene glaciation from North Africa through the Iberian Peninsula. Their data demonstrated that marginal populations had reduced inbreeding depression in comparison to central populations. These data are a partial explanation for the common finding across plant species of increased selfing at range margins. The authors suggested that a history of range expansion can contribute to the evolution of self-fertilization. In contrast, in more recent ecological time, invasive populations of Spartina alternaflora exhibited substantial and varied levels of inbreeding depression when assessed in the greenhouse (Daehler, 1999). In both the Mercurialis and Spartina studies, populations had been established for long periods of time, and demonstrate that inbreeding depression exists in populations at the limits of their range or in invasive populations.
Our data examined somewhat different questions: was inbreeding depression observed during initial establishment at a novel site in the native range and a novel site in the non-native range? As we did not find substantial inbreeding depression in our non-native-novel site in an experiment aimed to mimic the establishment phase, this opens a new question as to whether inbreeding depression changes between the initial establishment phase and the fully established invasive phase. Our expectation is that inbreeding depression may be influenced by whether populations are a result of a single or multiple introductions and by subsequent selection on lines that successfully establish. Our findings here complement our previous work (Murren et al., 2009) suggesting that evolutionary change may occur in breeding and mating system through time (across generations) outside of the native range.
An intriguing result is that performance of both populations increased in our native-novel site. Perhaps escape from co-evolved pathogens or herbivores is a factor, (Carr and Eubanks, 2002) although what distance from a maternal plant is required to escape from co-evolved antagonists remains an important open question in species' range expansion (Wolfe, 2002). In addition, abiotic site quality may play a role in performance in the native-novel site. In our study, this site remained moist throughout the season, and water availability had been shown to influence performance in this species (Galloway, 1995; van Kleunen and Fischer, 2008; Murren et al., 2006). We conclude that inbreeding depression may be important in local population persistence in a novel habitat inside the native range. Outside the native range, reduction in inbreeding depression may contribute to initial population establishment and perhaps population persistence. This may be a partial answer to the paradox described by Allendorf and Lundquist (2003) of how small populations with limited genetic variation are able to establish.
Phenotypic plasticity
We observed phenotypic plasticity as well as genetic variation for plasticity across geographic locations in M. guttatus, as has been observed for other architectural and morphological traits of invasive species (e.g. Sakai et al., 2001; Richards et al., 2006; Davidson et al., 2011). Our approach was to examine plasticity of architectural traits by calculating the differences in trait expression between the home sites and all other native and non-native sites (see Via et al., 1995; Auld, 2010; Baird et al., 2011, for similar approach). Both lines of evidence suggest the occurrence of variation in trait performance across sites for architectural traits. Our results build upon previous investigations in the native range and in greenhouse experiments (e.g. Galloway, 1995; Carr and Eubanks, 2002). We found that plasticity was generally greatest to either of the novel field locations than the native field site, as predicted. In addition, the LT population exhibited greater plasticity than M13, and we detected no difference in the magnitude of plasticity among self and outcross cross-types.
Phenotypic plasticity has been heralded as an important mechanism in the establishment success of new populations and in range expansion (Dudash et al., 2005; Ghalambor et al., 2007). van Kleunen and Fischer (2008) investigated the importance of plasticity to water for a suite of native and non-native populations that varied in life histories. By contrasting allozyme data to quantitative genetic variation, the authors argued that adaptive evolutionary processes were at work in the non-native range for these long-established populations. We also find that plasticity may be important during initial population establishment. In addition, other ecological research in M. guttatus has found that particular aspects of both the abiotic (e.g. moisture) and biotic environment (e.g. herbivores) serve as important ecological determinants of success of the species within its native range (Carr and Eubanks, 2002; Hall and Willis, 2006; Murren et al., 2006) and outside its native range (Truscott et al., 2006; Murren et al., 2009). Here, we focused on large-scale differences and found plasticity to be greatest in response to our non-native site and our native-novel site in comparison to the reciprocal home site. Opportunistic plasticity may play a role in the establishment of populations both when performance is higher or lower than the home site. Further field and greenhouse studies are warranted across subsequent generations, micro-environments within habitats, and across a broader range of novel habitat types, in order to understand the mechanisms behind the differences in plasticity between native and non-native sites (Davidson et al., 2011).
Inbreeding and plasticity interaction
We found limited evidence for a relationship between inbreeding and expression of plasticity, which agrees with the few other published studies on this topic in plants. For one trait, stem diameter, we detected an interaction between inbreeding and plasticity in the field. In the greenhouse, Schlichting and Levin (1986) examined ornamental lines of Phlox drummondii for which the self-incompatibilty was broken: across manipulated environments and for 12 traits, no differences in plasticity were observed across generations. More recently, in M. ringens, O'Halloran and Carr (2010) failed to detect inbreeding depression (contrary to our study here). These authors did not find a relationship between inbreeding and phenotypic plasticity for a wide variety of floral and size traits in relation to water manipulations in both greenhouse and field.
Taken together, these studies suggest that inbreeding depression and phenotypic plasticity may in fact function independently (Cheptou and Donohue, 2011). However, studies on the interaction between inbreeding and plasticity in plants have largely examined morphological or architectural traits. In a mixed-mating animal system, induced plastic responses in response to predator cues were found to be greater in outcross progeny compared to self progeny (Ivey and Carr, 2005; Auld and Relyea, 2010). Thus the nature of the plastic response (whether an induced plastic response, transgenerational, etc.) or type of trait (e.g. physiological or morphological) may be an important indicator of the influence of inbreeding, and warrants further study in plants.
New populations, outside the native range, suffer from small size, potentially reduced genetic variation and risk of inbreeding depression, yet a paradox exists as many of these populations are successful (Allendorf and Lundquist, 2003). Environmentally determined inbreeding depression and its relationship to phenotypic plasticity may in fact offer a partial solution to this conundrum. Those environment–population combinations that fail to expose inbreeding depression, yet result in expression of phenotypic plasticity, may be the ones where establishment and spread are successful. This scenario does not require purging of the genetic load: populations may maintain genetic variation in the face of small population size as outcross and selfed offspring may both contribute to the next generation. Differential purging of genetic load via bottlenecks may become important in future generations. Thus environmentally determined inbreeding depression and plasticity may both contribute to invasive species' initial success, even if seed production is not initially high.
Genetic source populations may respond differently to the challenges of a new environment. For example, in our data, survivorship at the M13 site was low across the board. The low water availability in the M13 site may have played an important role in reduction of survivorship and contributed to the observed patterns of inbreeding depression and plasticity. At the LT home site, water was present throughout the season (see also Murren et al., 2006), resulting in greater survivorship and consistently better performance of outcross progeny compared to self progeny (Fig. 1). Contribution to these responses may in part be explained by differential history of purging, as has been found in other work on M. guttatus (Dudash and Carr, 1998), although the two populations in the present study were indistinguishable in population outcrossing rates for the two flowering seasons observed (Murren and Dudash, unpubl. res.). Therefore, further careful examination of the multivariate environmental conditions among populations and their genetic composition is needed in order to uncover the ecological and genetic origins of inbreeding responses (Armbruster and Reed, 2005; Cheptou and Donohue, 2011). Regional differences in populations to stress may also shed light on patterns of invasion success.
Conclusions
We detected variation in both outcross and selfed lines in response to our two native-home sites and a novel habitat inside and a novel habitat outside the native range. Consistent with our predictions, we found greater plasticity to both our native-novel site and non-native-novel site, although in different directions. Our finding of reduced inbreeding depression in our novel site outside the native range is consistent with previous predictions by Ronce et al. (2009). The implications of these findings are that both selfed and outcross progeny can contribute to establishment of new populations within both the native and non-native range, and together with phenotypic plasticity may play an important role in the dynamics of establishing small populations. Invasive species and populations in novel environments offer fruitful grounds for the study of the genetic mechanisms of inbreeding depression (sensu Waller et al., 2008). The relationship between inbreeding depression and phenotypic plasticity warrants further examination as the genetic mechanisms behind the expression of these two important evolutionary mechanisms may turn out to be overlapping, or at least to be important to further elucidate for particular suites of phenotypic traits in novel environments.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the Napa County Land Trust for facilities support and site access for our research, USDA Beltsville for access to a field study site and UMD for greenhouse space and assistance. We are indebted to J. Callizo for his logistical and botanical support in California and to honour his memory dedicate this paper to him. We thank E. Byrnes for help with data collection, A. Matthews and M. Rutter for critical read of our manuscript. We thank J. Karron, A. Case, R. Peakall, C. Ivey and R. Mitchell for organizing this volume and for the invitation to contribute. We thank D. Levin, J. Karron and P. Heslop-Harrison for their editorial work at Annals of Botany, and for comments from anonymous reviewers. This work was supported by the United States Department of Agriculture USDA-CRES [#2001-35320-10899 MRD].
LITERATURE CITED
- Ågren J, Schemske DW. Outcrossing rate and inbreeding depression in two annual monoecious herbs. Begonia hirsuta and B. semiovata. Evolution. 1993;47:125–135. doi: 10.1111/j.1558-5646.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Lundquist LL. Introduction: population biology, evolution and control of invasive species. Conservation Biology. 2003;17:24–30. [Google Scholar]
- Angeloni F, Ouborg NJ, Leimu R. Meta-analysis on the association of population size and life history with inbreeding depression in plants. Biological Conservation. 2011;11:35–43. [Google Scholar]
- Armbruster P, Reed DH. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. [DOI] [PubMed] [Google Scholar]
- Auld JR. The effects of predation risk on mating system expression in a freshwater snail. Evolution. 2010;64:3476–3494. doi: 10.1111/j.1558-5646.2010.01079.x. [DOI] [PubMed] [Google Scholar]
- Auld JR, Relyea RA. Inbreeding depression in adaptive plasticity under predation risk in a freshwater snail. Biology Letters. 2010;6:222–224. doi: 10.1098/rsbl.2009.0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Henson CA, Daily NM, Baccari GV, Murren CJ. Differential patterns of plasticity to water availability along native and naturalized latitudinal gradients. Evolutionary Ecology Research. 2011;13:55–73. [Google Scholar]
- Bello-Bedoy R, Nunez Farfan J. Cost of inbreeding in resistance to herbivores in Datura stramonium. Annals of Botany. 2010;105:747–753. doi: 10.1093/aob/mcq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Botham R, Collin CL, Ashman T-L. Plant-mycorrhizal fungus interactions affect the expression of inbreeding depression in wild strawberry. International Journal of Plant Sciences. 2009;170:143–150. [Google Scholar]
- Carr DE, Dudash MR. Inbreeding under a competitive regime in Mimulus guttatus: consequences for potential male and female function. Heredity. 1995;75:437–445. [Google Scholar]
- Carr DE, Dudash MR. The effects of five generations of enforced selfing on potential male and female function in Mimulus guttatus. Evolution. 1997;51:1707–1807. doi: 10.1111/j.1558-5646.1997.tb05103.x. [DOI] [PubMed] [Google Scholar]
- Carr DE, Eubanks MD. Inbreeding alters resistance to insect herbivory and host plant quality in Mimulus guttatus (Scrophulariaceae) Evolution. 2002;56:22–30. doi: 10.1111/j.0014-3820.2002.tb00846.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual Review of Ecology and Systematics. 1987;18:237–268. [Google Scholar]
- Cheptou P-O, Donohue K. Environment-dependent inbreeding depression: its ecological and evolutionary significance. New Phytologist. 2011;189:395–407. doi: 10.1111/j.1469-8137.2010.03541.x. [DOI] [PubMed] [Google Scholar]
- Daehler C. Inbreeding depression in smooth cordgrass (Spartina alterniflora, Poaceae) invading San Francisco Bay. American Journal of Botany. 1999;86:131–139. [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Davidson AM, Jennions M, Nicotra AB. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecology Letters. 2011;14:419–431. doi: 10.1111/j.1461-0248.2011.01596.x. [DOI] [PubMed] [Google Scholar]
- Dluglosh KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Dole JA. Reproductive assurance mechanisms in three taxa of the Mimulus guttatus complex (Scrophulariaceae) American Journal of Botany. 1992;79:650–659. [Google Scholar]
- Dudash MR. Relative fitness of selfed and outcrossed progeny in a self-compatible, protandrous species, Sabatia angularis L. (Gentianaceae): a comparison in three environments. Evolution. 1990;44:1129–1139. doi: 10.1111/j.1558-5646.1990.tb05220.x. [DOI] [PubMed] [Google Scholar]
- Dudash MR, Carr DE. Genetics underlying inbreeding depression in Mimulus with contrasting mating systems. Nature. 1998;393:682–684. [Google Scholar]
- Dudash MR, Murren CJ. Evolution in action. Oxford, UK: Oxford University Press; 2008. The influence of breeding systems and mating systems on conservation genetics and conservation decisions In: Carroll SC, Fox CW; pp. 68–80. [Google Scholar]
- Dudash MR, Ritland K. Multiple paternity and self-fertilization in relation to floral age in Mimulus guttatus (Scrophulariaceae) American Journal of Botany. 1991;78:1746–1753. [Google Scholar]
- Dudash MR, Carr DE, Fenster CB. Five generations of enforced selfing and outcrossing in Mimulus guttatus inbreeding depression variation at the population and family level. Evolution. 1997;51:54–65. doi: 10.1111/j.1558-5646.1997.tb02388.x. [DOI] [PubMed] [Google Scholar]
- Dudash MR, Murren CJ, Carr DE. Using Mimulus as a model system to understand the role of inbreeding in conservation: genetic and ecological approaches. Annals of the Missouri Botanical Garden. 2005;92:36–51. [Google Scholar]
- Eubanks MD, Carr DE, Murphy JF. Effects of virus infection of Mimulus guttatus (Phyrmaceae) on host plant quality for meadow spittlebugs, Philaenus spumarius (Hemiptera: Cercopidae) Environmental Entomology. 2005;34:891–898. [Google Scholar]
- Fenster CB, Ritland K. Evidence for natural selection on mating system in Mimulus (Schrophulariaceae) International Journal of Plant Science. 1994;155:588–596. [Google Scholar]
- Fishman L, Willis J. Pollen limitation and natural selection on floral characters in the yellow monkeyflower. Mimulus guttatus. New Phytologist. 2008;177:802–810. doi: 10.1111/j.1469-8137.2007.02265.x. [DOI] [PubMed] [Google Scholar]
- Galloway LF. Response to natural environmental heterogeneity: maternal effects and selection on life-history characters and plasticities in Mimulus guttatus. Evolution. 1995;49:1095–1107. doi: 10.1111/j.1558-5646.1995.tb04436.x. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll S, Reznick DN. Adaptive vs non-adaptive phenotypic plasticity and the potential for contemporary adaptation to new environments. Functional Ecology. 2007;21:394–407. [Google Scholar]
- Godoy O, Saldana A, Fuentes N, Valladares F, Gianoli E. Forests are not immune to plant invasions: phenotypic plasticity and local adaptation allow Prunella vulgaris to colonize a temperate evergreen rainforest. Biological Invasions. 2011;13:1615–1625. [Google Scholar]
- Grant AL. A monograph of the genus Mimulus. Annals of the Missouri Botanical Garden. 1924;11:99–388. [Google Scholar]
- Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- Hayes CN, Winsor JA, Stephenson AG. Environmental variation influences the magnitude of inbreeding depression in Cucurbita pepo ssp. texana (Curcurbitaceae) Journal of Evolutionary Biology. 2005;18:147–155. doi: 10.1111/j.1420-9101.2004.00785.x. [DOI] [PubMed] [Google Scholar]
- Holeski LM, Kelly JK. Mating system and the evolution of quantitative traits: an experimental study of Mimulus guttatus. Evolution. 2006;60:711–723. [PubMed] [Google Scholar]
- Ivey CT, Carr DE. Effects of herbivory and inbreeding on the pollinators and mating system of Mimulus guttatus (Phrymaceae) American Journal of Botany. 2005;92:1641–1649. doi: 10.3732/ajb.92.10.1641. [DOI] [PubMed] [Google Scholar]
- Ivey CT, Carr DE, Eubanks ME. Effects of inbreeding in Mimulus guttatus on tolerance to herbivory in natural environments. Ecology. 2004;85:567–574. [Google Scholar]
- van Kleunen M, Fischer M. Adaptive rather than non-adaptive evolution of Mimulus guttatus in its native range. Basic and Applied Ecology. 2008;9:213–223. [Google Scholar]
- Lerner IM. Genetic homeostasis. New York: Dover Publishing; 1954. [Google Scholar]
- Maron JL, Vila M, Bommarco R, Elmendorf S, Beardsley P. Rapid evolution of an invasive plant. Ecological Monographs. 2004;74:261–280. [Google Scholar]
- Martin NH. Flower size preferences of the honeybee Apis mellifera foraging on Mimulus guttatus (Scrophulariaceae) Evolutionary Ecology Research. 2004;6:777–782. [Google Scholar]
- Murren CJ, Douglass L, Gibson A, Dudash MR. Individual and combined effects of Ca/Mg ratio and water on trait expression in Mimulus guttatus. Ecology. 2006;87:2591–2602. doi: 10.1890/0012-9658(2006)87[2591:iaceom]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Murren CJ, Chang CC, Dudash MR. Patterns of selection of two North American native and nonnative populatons of monkeyflower (Phrymaceae) New Phytologist. 2009;183:691–701. doi: 10.1111/j.1469-8137.2009.02928.x. [DOI] [PubMed] [Google Scholar]
- Muth NZ, Pigliucci M. Implementation of a novel framework for assessing species plasticity in biological invasions: responses of Centaurea and Crepis to phosphorus and water availability. Journal of Ecology. 2007;95:1001–1013. [Google Scholar]
- O'Halloran LR, Carr DE. Phenotypic plasticity and inbreeding depression in Mimulus ringens (Phrymaceae) Evolutionary Ecology Research. 2010;12:617–632. [Google Scholar]
- Pederson DG. Environmental stress, heterogzygote advantage and genotype-environment interaction in Arabidopsis. Heredity. 1968;23:127–138. doi: 10.1038/hdy.1968.11. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Phenotypic plasticity: beyond nature vs. nurture. Baltimore, MD: Johns Hopkins University Press; 2001. [Google Scholar]
- Pujol B, Zhou SR, Vilas JS, Pannell JR. Reduced inbreeding depression after species range expansion. Proceedings of the National Academy of Sciences USA. 2009;106:15379–15383. doi: 10.1073/pnas.0902257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn G, Keough M. Experimental design and analysis for biologists. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- Richards CL, Bossdorf O, Gurevitch J, Muth NZ, Pigliucci M. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecology Letters. 2006;9:981–993. doi: 10.1111/j.1461-0248.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- Ritland C, Ritland K. Variation of sex allocation among eight species of the Mimulus guttatus complex. American Journal of Botany. 1989;76:1731–1739. [Google Scholar]
- Ronce O, Shaw RH, Rousset F, Shaw RG. Is inbreeding depression lower in maladapted populations? a quantitative genetics model. Evolution. 2009;63:1807–1819. doi: 10.1111/j.1558-5646.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- SAS Institute Inc. SAS 9·2 help and documentation. Cary, NC: SAS Institute; 2010. [Google Scholar]
- Schlichting CD. The evolution of phenotypic plasticity in plants. Annual Review of Ecology and Systematics. 1986;17:667–693. [Google Scholar]
- Schlichting CD. Hidden reaction norms, cryptic variation and evolvability. Annals of the New York Academy of Science. 2008;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- Schlichting CD, Levin DA. Effects of inbreeding on phenotypic plasticity in cultivated Phlox. Theoretical and Applied Genetics. 1986;72:114–119. doi: 10.1007/BF00261465. [DOI] [PubMed] [Google Scholar]
- Steets JA, Knight TM, Ashman T-L. The interactive effects of herbivory and mixed mating for the population dynamics of Impatiens capensis. American Naturalist. 2007;170:113–127. doi: 10.1086/518178. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Gamble SE. The effect of distance from parental site on offspring performance and inbreeding depression in Impatiens capensis: a test of the local adaptation hypothesis. Evolution. 1990;44:2022–2030. doi: 10.1111/j.1558-5646.1990.tb04308.x. [DOI] [PubMed] [Google Scholar]
- Truscott AM, Soulsby C, Palmer SC, Newell L, Hulme PE. The dispersal characteristics of the invasive plant Mimulus guttatus and the ecological significance of increased occurrence of high-flow events. Journal of Ecology. 2006;94:1080–1091. [Google Scholar]
- Truscott AM, SCF Palmer, Soulsby C, Hulme PE. Assessing the vulnerability of riparian vegetation to invasion by Mimulus guttatus: relative importance of biotic and abiotic variables in determining species occurrence and abundance. Diversity and Distributions. 2008;14:412–421. [Google Scholar]
- USDA/NCRS. Plants Database. Baton Rouge, LA: National Plant Data Center; 2011. http://plants.usda.gov. (accessed 16 January 2011) [Google Scholar]
- Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Schlichting CD, van Tienderen PH. Adaptive phenotypic plasticity: consensus and controversy. Trends in Ecology and Evolution. 1995;10:212–217. doi: 10.1016/s0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Vickery R. Remarkable waxing and waning and wandering of populations of Mimulus guttatus: an unexpected example of global warming. Great Basin Naturalist. 1999;59:112–126. [Google Scholar]
- Vogler DW, Kalisz S. Sex among flowers: the distribution of plant mating systems. Evolution. 2001;55:202–204. doi: 10.1111/j.0014-3820.2001.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Waller DM, Dole J, Bersch AJ. Effects of stress and phenotypic variation on inbreeding depression in Brassica rapa. Evolution. 2008;62:917–931. doi: 10.1111/j.1558-5646.2008.00325.x. [DOI] [PubMed] [Google Scholar]
- Waser NM, Vickery RK, Price MV. Patterns of seed dispersal and population differentiation in Mimulus guttatus. Evolution. 1982;36:753–761. doi: 10.1111/j.1558-5646.1982.tb05441.x. [DOI] [PubMed] [Google Scholar]
- Wolfe LM. Why alien invaders succeed: support for the escape-from-enemy hypothesis. The American Naturalist. 2002;160:705–711. doi: 10.1086/343872. [DOI] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW, Willis JH. Mimulus is an emerging model system for the integration of ecology and genetics. Heredity. 2008;100:220–230. doi: 10.1038/sj.hdy.6801018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.