Abstract
Emerging data from the coelacanth genome are beginning to shed light on the origin and evolution of tetrapod genes and noncoding elements. Of particular relevance is the realization that coelacanth retains active copies of transposable elements that once served as raw material for the evolution of new functional sequences in the vertebrate lineage. Recognizing the evolutionary significance of coelacanth genome in this regard, we employed an ab initio search strategy to further classify its repetitive complement. This analysis uncovered a class of interspersed elements (Latimeria Harbinger 1—LatiHarb1) that is a major contributor to coelacanth genome structure and gene content (∼1% to 4% or the genome). Sequence analyses indicate that 1) each ∼8.7 kb LatiHarb1 element contains two coding regions, a transposase gene and a gene whose function is as yet unknown (MYB-like) and 2) copies of LatiHarb1 retain biological activity in the coelacanth genome. Functional analyses verify transcriptional and enhancer activities of LatiHarb1 in vivo and reveal transcriptional decoupling that could permit MYB-like genes to play functional roles not directly linked to transposition. Thus, LatiHarb1 represents the first known instance of a harbinger-superfamily transposon with contemporary activity in a vertebrate genome. Analyses of LatiHarb1 further corroborate the notion that exaptation of anciently active harbinger elements gave rise to at least two vertebrate genes (harbi1 and naif1) and indicate that the vertebrate gene tsnare1 also traces its ancestry to this transposon superfamily. Based on our analyses of LatiHarb1, we speculate that several functional features of harbinger elements may predispose the transposon superfamily toward recurrent exaptive evolution of cellular coding genes. In addition, these analyses further reinforce the broad utility of the coelacanth genome and other “outgroup” genomes in understanding the ancestry and evolution of vertebrate genes and genomes.
Keywords: genome evolution, coelacanth, harbinger, exaptation, Latimeria
Introduction
The coelacanth, Latimeria, is known as a “living fossil” because it is the only vestige of a deep evolutionary lineage that predates the vertebrate invasion of the terrestrial environment and the evolution of associated anatomical features. As a consequence of its shared ancestry with the tetrapod lineage and long-independent evolutionary history (∼400 My) (Carroll 1988), the coelacanth can be used to provide critical perspectives on vertebrate evolution. Recent studies have used emerging sequence data from the coelacanth genome to illuminate the evolution of this key vertebrate lineage (Koh et al. 2003; Noonan et al. 2004; Shashikant et al. 2004; Bejerano et al. 2006; Nishihara et al. 2006; Xie et al. 2006; Gwee et al. 2008; Amemiya et al. 2010). These studies have revealed that 1) the coelacanth genome has experienced relatively slow rates of molecular change over the last 400 My (Noonan et al. 2004; Amemiya et al. 2010) and 2) the coelacanth genome retains large families of short interspersed repetitive elements (SINEs), which have acquired functionality as both regulatory and coding sequences in the “higher” vertebrates (Bejerano et al. 2006; Nishihara et al. 2006; Xie et al. 2006; Gwee et al. 2008). Thus, studies of the coelacanth genome are providing unique insights into the ancestry and origin of functional sequences in tetrapod genomes.
In mammalian genomes, dozens of gene families and microRNAs and thousands of presumptive regulatory elements have been identified that may trace their origin to transposable elements (Volff 2006; Lowe et al. 2007; Mikkelsen et al. 2007; Piriyapongsa et al. 2007). An example of particular relevance to the current study is the evolution of harbinger-derived genes. Harbinger transposable elements are suggested to have given rise to at least two genes during vertebrate ancestry (harbi1 and naif1) (Kapitonov and Jurka 2004; Sinzelle et al. 2008). The gene harbi1 is thought to have originated in the ancestor of bony vertebrates (class Osteichthyes) and evolved from a harbinger transposase. Harbinger elements are fairly unique among vertebrate transposons in that they also carry, in addition to a canonical transposase gene, a MYB-like gene. A harbinger-derived MYB-like gene is inferred to have served as the raw material for the evolution of naif1 in the vertebrate lineage (Sinzelle et al. 2008). Transposable elements therefore comprise an important reservoir of sequences that can be co-opted in the evolution of new gene functions.
The characterization of genomes from diverse evolutionary lineages is paramount to understanding the origins of transposon-derived genes (Lowe et al. 2010; Sela et al. 2010). Representatives of deep evolutionary lineages may retain transposable elements that are no longer active in well-sequenced lineages (e.g., mammals, ray-finned fish) or have long since become extinct. Some of these under-represented lineages provide a further advantage in identifying preserved transposable elements because their evolutionary history has been characterized by persistently low population sizes and (partially as a consequence) low rates of molecular evolution (Lynch and Conery 2003; Noonan et al. 2004; Lowe et al. 2010). Latimeria provides both these advantages, but at the disadvantage of being nearly intractable as a biological model, necessitating the use of surrogate model organisms for any functional analyses (Amemiya et al. 2010). Nonetheless, this issue does not outweigh the evolutionary perspective that can be gained through study of its genome. Moreover, as the sole vestige of a 400 My old lineage, the living coelacanths can provide key insight into the complement of repetitive elements that were present in, and contributed to, the evolution of the ancestral tetrapod lineage.
Here, we describe the identification, analysis, and functional characterization of an 8.7 kilobase (kb) harbinger-like transposon in the Latimeria genome (LatiHarb1), which encodes a predicted harbinger transposase (tpase) and a second nontransposase gene (Latimeria harbinger-associated: lha). Recent activity appears to have precipitated the massive amplification of this transposon, such that this single element represents ∼1% to 4% of the coelacanth genome. Analysis of the MYB-like gene lha provides evidence that the vertebrate gene tsnare1 may also trace its ancestry to the harbinger transposons. Zebrafish reporter-enhancer assays and gene expression in a transgenic mouse harboring 162 kb of Latimeria genomic DNA, suggest that LatiHarb1 retains functionality in the coelacanth genome. We propose that similar functional features may have predisposed harbinger-type transposons toward exaptation in the vertebrate lineage in the past and perhaps presently in the coelacanth genome.
Materials and Methods
Identification of LatiHarb1
Menado coelacanth (Latimeria menandoensis, Abbreviated Lm) bacterial artificial chromosome (BAC) sequences that were used in this study included 11 sequences that were deposited in GenBank (accession numbers: AC140159, AC150283, AC150284, AC150308, AC150309, AC150310, EU284132, FJ497005, FJ497006, FJ497007, and FJ497008) and 3 additional sequences from the immunoglobulin heavy chain gene clusters (being submitted to NCBI). Ab initio identification of repetitive elements was performed using RepeatScout (Price et al. 2005). Prior to analysis with RepeatScout (Price et al. 2005), sequences corresponding to known repetitive elements were masked using RepeatMasker (Smit et al. 2004) and repeat library files from the Repbase database (Jurka et al. 2005). Based on the initial scan for repeated sequences, a 10 kb region surrounding three common and positionally associated repeats was realigned using AlignX (Invitrogen), and BLAT (Kent 2002) was used to search BAC sequences for additional instances of the repeat. Additional details are provided as supplementary materials and methods, Supplementary Material online.
Identification of Candidate Protein-Coding Regions and Similarity to Known Proteins
The programs GeneScan (Burge and Karlin 1998) and GeneMark.hmm-E (Besemer and Borodovsky 2005; Lomsadze et al. 2005) were used to search sequences of eight LatiHarb1 elements that were identified within the BAC sequence data set. The 8.7-kb repeat and predicted amino acids were used to query the GenBank NR database using the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Amino acid sequences for the two predicted coelacanth genes and their respective BLAST hits were aligned using the program CLUSTALX (Higgins and Sharp 1988; Thompson et al. 1997). We also screened the LatiHarb1 consensus sequence for microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs) using the programs miPred-II (Jiang et al. 2007) and snoSeeker (Yang et al. 2006), respectively. No candidate miRNAs or snoRNAs were identified in the LatiHarb1 consensus sequence.
Estimates of Repeat Frequencies, Gene Linkage, and Divergence in the Indonesian and African (L. chalumnae) Coelacanth Genomes
BACs were screened for LatiHarb1 by amplifying fragments of lha and tpase from 96 individual BAC clones using standard polymerase chain reaction (PCR) conditions (supplementary materials and methods, Supplementary Material online). In order to estimate the fraction of BACs that could be accounted for by LatiHarb1, it was necessary to take into account the probability that a positively screening BAC contained one or more instances of the LatiHarb1 element. To do this, we used the observed frequency of lha/tpase-positive BACs as an estimate of the probability of observing at least one BAC (34/96 = 0.354) and assumed that the probability of observing each additional instance was approximated by and contingent upon this value (e.g., the estimated number of BACs containing three elements was 96 × 0.354 × 0.354 × 0.354 = 4.2). This estimate assumes that the average BAC size is large (i.e., >150 kb) (Danke et al. 2004) and that all BACs contain inserts. Failure of PCRs and empty or small inserts will result in an underestimate of the true represented proportion. We tested for linkage (positional association) of lha and tpase using a G-test for independence (Sokal and Rohlf 1995), based on the results of PCR screens of these same 96 Lm BAC clones.
Shotgun sequence data were used to estimate the proportion of Lm and African coelacanth (L. chalumnae, abbreviated Lc) genomes represented by the LatiHarb1 element by aligning (Kent 2002) the LatiHarb1 to vector-trimmed shotgun reads, joining alignments that were separated by gaps of less than 50 bp using MapToGenome (Putta et al. 2007) and calculating the fraction of LatiHarb1-aligning bases relative to the full data set. These alignments were also used to determine the average depth of sequence (alignment) coverage for 100 bp windows along the length of LatiHarb1.
Amplification of LatiHarb1 Transcripts from Transgenic Mice
We assayed for transcription of tpase and lha genes in a system that approximates its native chromatin environment in the coelacanth genome: a mouse line that carries a 162 kb insert that was derived from the Lm BAC library. Details regarding the creation of this line are provided as supplementary materials and methods, Supplementary Material online. Assays for transcriptional activity used cDNAs that were reverse transcribed from adult germline (testes) RNA and RNA extracted from whole embryos and extraembryonic membranes. cDNAs were synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen) and manufacturer-specified reaction conditions. Fragments were then amplified using ExTaq (TaKaRa) under manufacturer recommended conditions (thermal cycling at 98 °C for 1 min; 30 cycles of 98 °C for 10 s, 62 °C for 30 s, and 72 °C for 1 min; and 72 °C for 10 min). Primers were designed to fall within adjacent exons (lha: Lha_ex2c.f.1—AAGGACACGTGGAGGAGGTA and Lha _ex1c.r.1—GAGCGTCAAGAGGAAGATGG; ltase: Ltase_ex4. f.2—TGTCACAAGCCTTGGATCAG and Ltase_ex5.r.2—CACAACACACGAGGAACACC).
Zebrafish Enhancer Assays
Enhancer assays employed the vector p339hsp70GFPrc (Mongin et al. 2011). This vector contains a multiple cloning site upstream of a copy of green fluorescent protein (GFP) that is driven by a promoter from the zebrafish gene hsp70. A copy of one LatiHarb1 from BAC clone VMRC47-217L16 was amplified using primers to the conserved inverted repeats that flank all copies of LatiHarb1 and contained a 5′ tail corresponding to the NotI restriction site (Lati_univ_NotI—ATG CGGCCGCGGGCTCTATCATGAACCAAC). This amplified fragment was inserted into the multiple cloning site of p339hsp70GFPrc via NotI restriction digestion and ligation (T4 ligase) of the two fragments. Ligation products were electrotransformed into DH10B-T1 cells (Invitrogen) and plated on LB agar plates containing ampicillin (100 μg/ml). A single transformant was selected and presence of LatiHarb1 fragment was authenticated by restriction digestion (BamHI and NotI) and end sequencing using vector primers. The construct was purified using Qiagen Plasmid Midi Kit prior to linearization and injection.
Transient transgenesis of the GFP construct was performed by microinjection of the animal pole of newly fertilized zebrafish eggs with approximately 200 pg of I-SceI linearized DNA. Linearized DNA was prepared by incubating constructs at a concentration of 40 ng/μl with 0.36 U/μl I-SceI in 2× I-SceI buffer on ice, until the time of injection. No further purification was performed prior to injection.
Images of live (shield stage) or fixed (26 h post-fertilization) embryos were captured using a motorized stereoscope (Leica M205FA). Z-stacks of bright field images were merged to produce a single deep focus image using the Leica Application Suite. Bright field images were then merged with GFP images (excitation filter 470 nm, barrier filter 525 nm) of the same embryo, using Adobe Photoshop CS5. Embryos were placed in small agar wells to prevent movement during capture of bright field and GFP images.
Results
Identification of an 8.7-kb Repeat in the Coelacanth Genome
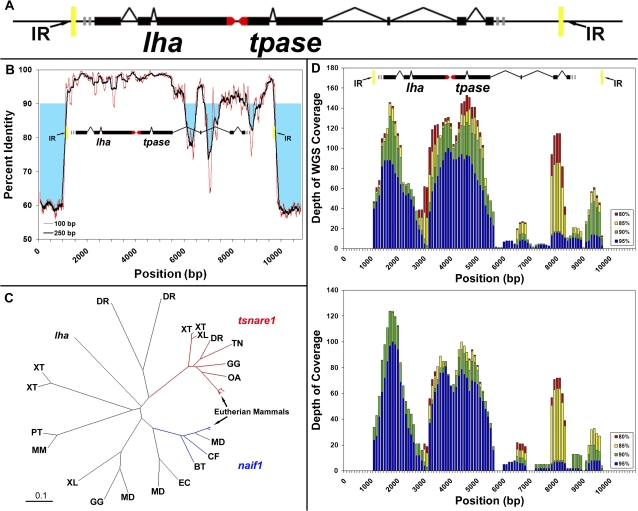
We surveyed 14 BAC assemblies that spanned more than 3.3 megabases (Mb), using ab initio repeat finding strategies that do not rely upon comparative information from other genomes (Price et al. 2005). These BACs were derived from specific genic regions of the Lm (Menado coelacanth) genome, including the HOX and protocadherin clusters, and immunoglobulin loci. Our analysis of these sequences found several known repeats that were previously identified among a subset of these BACs, such as the lmSINEs (Bejerano et al. 2006; Nishihara et al. 2006; Xie et al. 2006) and other previously described classes of repetitive elements (Smit et al. 2004; Jurka et al. 2005) (supplementary table 1, Supplementary Material online). Our survey also identified several repetitive sequences that did not correspond to these known elements. Most striking among these uncharacterized repetitive regions were three highly conserved sequences that always co-occurred with one-another, and invariably found, in the same relative position and orientation. Alignment of broader regions that contained these respective sequences revealed that all three were contained within a single larger repeated sequence that was approximately 8.7 kb in length (fig. 1A). Alignment to Lm BAC sequences revealed 7 nearly complete copies of the repeat (and 11 incomplete or disrupted copies larger than 1 kb in aligned length) among the 3.3 Mb of combined sequence. This repeat therefore accounted for approximately 2.3% of known Lm BAC sequences (ungapped alignment or 3.9% in gapped alignment). By comparison, the entire protein coding complement comprises roughly 1.5–2.0% of the human genome (Lander et al. 2001). This single 8.7-kb repeat was found to be dispersed among numerous genic regions and accounts for a substantial fraction of the coelacanth genome. Copies of LatiHarb1 occupy a fraction of the coelacanth genome that is similar to other highly abundant retrotransposable elements (supplementary table 1, Supplementary Material online).
FIG. 1.
Structure, evolution, and genomic distribution of LatiHarb1. (A) Colored boxes represent the consensus position and direction of predicted gene structures and within LatiHarb1 (black = coding, red = untranslated regions). The two predicted genes tpase and lha are nonoverlapping and are oriented in opposite directions (transcribed toward one-another). These are bounded by 35-bp inverted repeats (yellow boxes). (B) Percent sequence identity among five independent instances of the LatiHarb1 element. Percent identity is averaged over sliding windows of 100 or 250 bp. Regions of high percent identity (>90%) overlap with predicted exons, whereas regions of low identity correspond to predicted intronic sequences. (C) Several genes from different vertebrate gene families align to a common region of the lha but none is strikingly similar to the lha gene. Branch termini designate individual lha homologs and labels denote the species from which these sequences were identified. Scale bar = 0.1 amino acid substitutions per site. Abbreviations are defined below. (D) Histogram showing the alignment depth of randomly sampled reads along the length of the LatiHarb1 element (100-bp intervals). The number of aligning reads is shown for several thresholds of percent identity. Regions of high sequence coverage correspond to predicted exons. Upper panel: reads are from a shotgun sequencing data set generated from heterospecific (Latimeria chalumnae) whole genome (0.02× genome coverage). Lower panel: reads are from a shotgun data set generated from a homospecific (Latimeria menadoensis) BAC library (<0.01× genome coverage) (Thomson et al. 1973; Cimino and Bahr 1974). Abbreviations: DR (Danio rerio), XT (Xenopus tropicalis), XL (X. laevis), TN (Tetraodon nigroviridis), GG (Gallus gallus), OA (Ornithorhynchus anatinus), MD (Monodelphis domestica), CF (Canis familiaris), BT (Bos taurus), EC (Equus caballus), MM (Mus musculus), PT (Pan troglodytes).
Structure and Function of the LatiHarb1 Transposon
The LatiHarb1 transposon consists of two open reading frames flanked by a 35-bp inverted repeat. LatiHarb1 is structurally similar to other harbinger-superfamily transposons in that 1) it carries a second gene along with its transposase (lha) and 2) the transposase reading frame is disrupted by introns (fig. 1A). Besides harbinger, which has not been observed as an endogenously active copy within the vertebrates (Kapitonov and Jurka 2004; Sinzelle et al. 2008) only a few superfamilies of transposons (e.g., IS4EU in vertebrates [http://www.girinst.org/2007/vol7/issue4/IS4EU-1_DR.html] and En/Spm [Kunze and Weil 2002] and MuDR [Walbot and Rudenko 2002] in plants) are known to carry a second gene along with their canonical transposase gene (Kapitonov and Jurka 1999).
Sequence analyses indicate that both predicted coding regions within LatiHarb1 have experienced purifying selection over recent evolutionary history, which might be considered further evidence indicative of contemporary functionality. Specifically, alignment of five full length and nonidentical copies of LatiHarb1 reveal high levels of sequence conservation that correspond to the location of predicted exons in both the lha and tpase regions (fig. 1B). Analysis of predicted lha protein revealed that it was similar to genes from several vertebrate species, which could be separated into several distinct classes, including: homologs of t-snare domain containing (tsnare1), nuclear apoptosis-inducing factor 1 (naif1), and several other genes that were not similar to either of these classes except within the region that aligned to our predicted protein. The topology of a neighbor joining tree of lha alignments revealed that orthologs of the human genes tsnare1 and naif1 are clustered in distinct regions of the tree that are also distinct from the location of lha sequence and other groups (fig. 1C). Functional studies have previously yielded strong evidence that naif1 is derived from a nontransposase coding (MYB-like) region within the harbinger superfamily of transposons (Kapitonov and Jurka 2004; Sinzelle et al. 2008). Notably, lha shares several amino acid residues with tsnare1 that are absent from naif1 homologs, possibly suggesting a closer relationship with tsnare1 (supplementary fig. S1, Supplementary Material online).
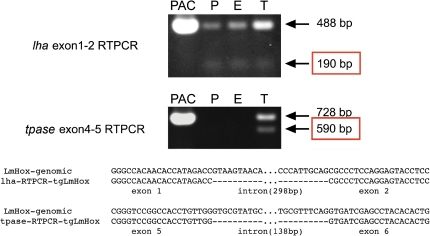
Given the complex structure of this transposon and circumstantial evidence that the transposon is currently, or recently, functional in the coelacanth genome (i.e., high copy number and high sequence identity over evolutionarily relevant regions), we sought to verify predicted transcription and splicing of the two predicted genes within LatiHarb1. However, tissue collection from coelacanths has proven notoriously difficult and to our knowledge, tissue samples sufficient for extraction of good-quality RNA have not been reported for the species. We therefore took advantage of an existing PAC (P1-derived artificial chromosome)-derived mouse transgenic line (Tg [pPAC-GFP-1054-RFP-iTol2-Kan] B6C3) that carries a 162 kb region from the 5′ end of the Lm HOXA cluster (Powers and Amemiya 2004) (see also supplementary materials and methods, Supplementary Material online). This line carries one complete copy of LatiHarb1 centered at 75.6 kb of the insert and a second incomplete copy of the element (supplementary fig. S2, Supplementary Material online). Reverse-transcriptase PCR from testes and embryonic tissues (placenta, whole embryo, and adult testes) revealed transcripts that correspond in size and sequence to predicted splicing products and unspliced pre-mRNAs (fig. 2). Expression of lha was observed in all three samples, whereas the expression of tpase was limited to the testes. The observation of testes-restricted expression of tpase is consistent with the inferred propagation of this element by cut-and-paste transposition, and the wider expression of lha suggests the possibility that the gene could influence coelacanth biology in a way that is not directly related to the propagation of LatiHarb1.
FIG. 2.
Transcripts derived from LatiHarb1. PCR was performed on cDNAs that derived from placenta (P), embryo (E), and adult testes (T) of transgenic mice carrying 162 kb of Latimeria genomic DNA, including one intact copy of the LatiHarb1 element (supplementary figure S3, Supplementary Material online). Fragments corresponding in size and sequence to spliced lha mRNA and unspliced lha pre-mRNAs are observed in all three transcriptomes, whereas fragments corresponding to spliced tpase and tpase pre-mRNAs are only observed in adult testes. The respective PCR fragments were sequenced in order to validate their identities and organization (bottom of figure).
Distribution of the LatiHarb1 Transposon in Coelacanth Genomes
We employed empirical and in silico approaches to better characterize the distribution of LatiHarb1 in the coelacanth genome. Our in silico approach took advantage of several thousand pilot shotgun sequences for Lm (N = 56,064 pooled BAC shotgun reads) and African coelacanths (L. chalumnae abbreviated Lc: N = 167,423 whole-genome shotgun reads) that are deposited in the GenBank Trace Archives. These databases correspond to ∼0.02× (Lm) and 0.06× (Lc) coverage of the coelacanth genome, given an estimated genome size between ∼3.6 and 1.8 pg/C (Thomson et al. 1973; Cimino and Bahr 1974; Danke et al. 2004). Searches for LatiHarb1 identified several hundred hits to shotgun sequences from Lm (N = 846) and Lc (N = 1740), which accounted for ∼1% of sequence from Lm and ∼0.6% of sequence from Lc. Notably, these hits are localized to regions of high sequence conservation and thus underestimate the total contribution of LatiHarb1 elements to these genomes (i.e., intronic and noncoding regions are underrepresented in the alignments; fig. 1D). Empirical estimates based on PCR amplification of tpase and lha and hybridizations of an lha probe to both BAC library high-density filters (Danke et al. 2004) and genomic Southern blot also revealed patterns that are entirely consistent with this repeated sequence being present at high copy number in the genome (supplementary figs. S3, S4, and supplementary materials and methods, Supplementary Material online).
Notably, the proportion of shotgun sequence data that corresponded to LatiHarb1 in Lm differs significantly from Lc, both in terms of the proportion of nucleotides (1.0% of 49,955,567 bases in Lm vs. 0.6% of 146,680,543 bases in Lc, Z = 274.2 P < 0.0001) and the proportion of transposon-containing reads (1.5% of 56,064 Lm reads vs. 1.0% of 167,423 Lc reads, Z = 9.00, P < 0.0001). The average nucleotide sequence identity between our prototype LatiHarb1 sequence and the aligned shotgun reads from Lm and Lc species was 95.0% and 92.4%, respectively. The expansion of this sequence therefore appears to predate and the divergence between the two coelacanth species, which has been estimated to be up to 30–40 Ma (Inoue et al. 2005) or less than 5 My (Holder et al. 1999). Although LatiHarb1 is highly abundant in both genomes, differences in its relative abundance within the Lc and Lm data sets may reflect differential expansion/loss of LatiHarb1 subsequent to the divergence of the two species or, alternatively, sampling differences in generating sequence reads for the two species. High sequence identity among multiple copies of LatiHarb1 in both coelacanth species suggests the evolution of exonic sequences has been strongly constrained in both lineages over recent evolutionary history.
Effect of LatiHarb1 on the Transcription of Neighboring Genes
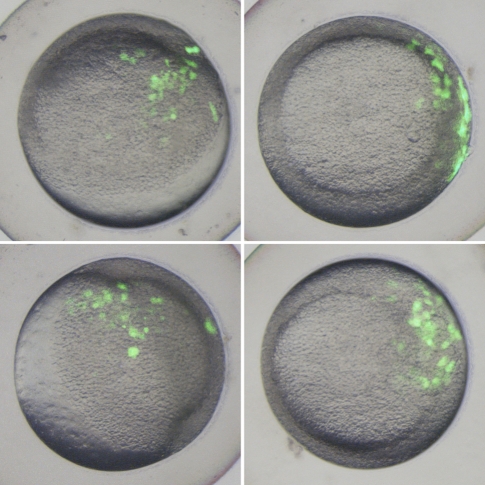
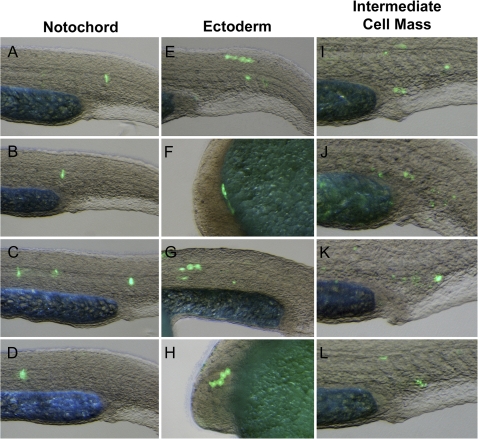
A fundamental outcome of transposition is the joining of sequences that previously existed in separate genomic locations. If a transposed sequence possesses transcriptional enhancer or repressor activity, then transposition may alter the gene regulatory environment in the vicinity of excision or insertion sites. To further investigate how transposition of LatiHarb1 might influence coelacanth biology, we used transient transfection of zebrafish embryos to assay the ability of the element to drive in vivo transcription of a GFP gene from a minimal promoter (Mongin et al. 2011). Zebrafish embryos exhibited a significantly higher incidence of gene expression (fluorescence) when injected with a reporter construct that included a copy of LatiHarb1 (LatiHarb1-GFP), as compared with control embryos that were injected with the minimal construct alone. During gastrulation (6 h post-fertilization, i.e., shield stage), GFP expression was significantly enhanced in LatiHarb1-GFP embryos, whereas only a small fraction of control embryos exhibited fluorescence (106/132 in test group, 7/154 in control group, Fisher’s Exact: P = 4.97 × 10−43) (fig. 3). Moreover, significantly enhanced fluorescence persisted through the first day of development (43/44 GFP positive in test group, 9/41 GFP positive in control group, Fisher’s Exact: P = 3.86 × 10−14). At 26 h post-fertilization, recurrent patterns of expression were observed in the developing epidermis (n = 16/44), intermediate cell mass (n = 11/44), and notochord (n = 8/44), while GFP expression was not observed in any of these three tissues in the control group (fig. 4). In the context of this experimental system, it may be worth noting that all embryos are expected to be (as observed) highly chimeric for GFP expression. Overall, these studies indicate that LatiHarb1 possesses substantial enhancer activity in vivo, which can promote expression in diverse cell types during zebrafish embryonic development.
FIG. 3.
LatiHarb1 enhancer activity in gastrulating embryos (6 h post-fertilization). Images show four examples of fluorescence patterns that were observed among embryos injected with a GFP reporter construct for LatiHarb1 enhancer activity. Bright field and GFP images were merged to show the location of fluorescent cells in relation to morphology.
FIG. 4.
LatiHarb1 enhancer activity in developing embryos (26 h post-fertilization). Examples of fluorescence patterns that were observed among embryos injected with a GFP reporter construct for LatiHarb1 enhancer activity. Recurrent patterns were observed in the notochord (A–D), ectoderm (E–H), and intermediate cell mass (I–L). Bright field and GFP images were merged to better show the location of fluorescent cells in relation to morphology.
Discussion
This study identifies a new group of long harbinger-superfamily transposons with high sequence identity across multiple members, conservation of predicted exonic sequences, and surmised biological activity. Functional assays indicate that copies of LatiHarb1 are actively transcribed in testes of transgenic mice harboring the transposon, suggest a role for the lha gene beyond its presumed role in transposition, and imply that the element can significantly alter transcriptional regulation in its local environment. Altogether, these analyses reveal the considerable biological potential of harbinger elements within the coelacanth genome and in vertebrate genomes in general.
The harbinger superfamily of transposons (harbinger/PIF) is distributed throughout the eukaryotes (Jurka and Kapitonov 2001; Zhang et al. 2001), although active members of the superfamily have not been observed in vertebrates (with the exception of an artificially resurrected copy from the zebrafish genome) (Sinzelle et al. 2008). Our experiments demonstrate that one member of the harbinger superfamily (LatiHarb1) is present at exceedingly high copy numbers in the coelacanth genome and that the element shows patterns of evolutionary conservation that are indicative of recent expansion and selection to maintain functionality. Moreover, direct experimental evidence demonstrates that at least one copy of the transposon is transcriptionally active in the context of its local chromatin environment (71.5 kb of Lm sequence 5′ and 82.4 kb 3′).
Observed patterns of transcription in testes, embryo, and extra-embryonic tissues parallel the evolution and expansion of LatiHarb1 and underscore the evolutionary potential of this element to serve as the raw material for exaptive evolution of cellular coding genes. Over an evolutionary timeframe, only mobilization in the germline can lead to an increase in copy number. Transcription and splicing of LatiHarb1 in testes are therefore consistent with the hypothesis that the element is undergoing expansion in the coelacanth genome over a contemporary timescale or at least retains many of the activities that are necessary for expansion. However, assays for transcription of individual LatiHarb1-encoded loci (lha and tpase) reveal important differences in the regulation of genes within the transposon. Specifically, transcripts from lha are observed in embryonic and extra-embryonic tissues, whereas tpase expression is not observed in either of these tissues. If lha is playing a functional role outside of the testes, then this role must be peripheral to the proposed role of harbinger-associated (MYB-like) genes in mediating target site specificity of harbinger transposase activity (Sinzelle et al. 2008).
We propose that regulatory uncoupling of lha and tpase (similar to that observed for one LatiHarb1 element) might promote the evolution of new gene functions for lha (or other harbinger-associated MYB-like genes), and similar regulatory differences may have contributed to the evolution of genes such as naif1 and tsnare1 from ancestral harbinger elements. Our analyses provide some indication that LatiHarb1 might not strictly adhere to the concept of a “selfish” DNA element (Doolittle and Sapienza 1980; Orgel and Crick 1980) in the context of the coelacanth genome. The lha gene shows evidence for strong purifying selection and patterns of expression (broader than its associated transposase and including non-germline tissues) that hint at a biological role beyond the transposition of LatiHarb1. It has been suggested that MYB-like proteins (presumptive homologs of lha, including naif1 and tsnare1) possess functions related to transcriptional regulation, nuclear import, and DNA binding (Kapitonov and Jurka 2004; Sinzelle et al. 2008). Ostensibly, one or more of these functions might prove beneficial to tissues in which lha genes are expressed, which may be promoting selection for retained functionality of lha and amplification of the gene along with its associated transposase. Thus, something akin to exaptation of lha genes might occur even in the context of contemporary transposition.
In summary, analysis of the repetitive fraction of the coelacanth genome permitted the identification of a class of harbinger-like elements that exhibit signs of contemporary activity. The identification of LatiHarb1 allows us to account for a substantial fraction of the genome, adds to our understanding of the evolutionary contribution of harbinger elements to modern day vertebrate genomes and illustrates how genes encoded within transposons might transition from selfish to more altruistic roles in the context of genome evolution. These findings further illustrate the utility of coelacanth and other evolutionarily and genomically underrepresented “outgroup” lineages in understanding the history, structure, and functionality of vertebrate genomes.
Supplementary Material
Supplementary materials and methods, figures S1–S4, and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Nil Ratan Saha and Evan Eichler for helpful discussions. This research was supported by grants from the National Science Foundation (grant number MCB 0719558 to C.T.A.), the National Institutes of Health (grant number R24 RR14085 to C.T.A. and grant number T32 HG00035 and F32 GM087919 to J.J.S.), and the Japanese Ministry of Education, Culture, Sports, Science, and Technology Grant-in-Aid for Scientific Research (grant numbers 18570007, 20657003 to K.S.). Andrew Stuart and Michael Rego assisted with zebrafish transgenesis and husbandry.
References
- Amemiya CT, Powers TP, Prohaska SJ, et al. (11 co-authors) Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc Natl Acad Sci U S A. 2010;107:3622–3627. doi: 10.1073/pnas.0914312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Karlin S. Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Carroll RH. Vertebrate paleontology and evolution. New York: W. H. Freeman & Co; 1988. [Google Scholar]
- Cimino MC, Bahr GF. The nuclear DNA content and chromatin ultrastructure of the coelacanth Latimeria chalumnae. Exp Cell Res. 1974;88:263–272. doi: 10.1016/0014-4827(74)90240-7. [DOI] [PubMed] [Google Scholar]
- Danke J, Miyake T, Powers T, Schein J, Shin H, Bosdet I, Erdmann M, Caldwell R, Amemiya CT. Genome resource for the Indonesian coelacanth, Latimeria menadoensis. J Exp Zool A Comp Exp Biol. 2004;301:228–234. doi: 10.1002/jez.a.20024. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Gwee PC, Amemiya CT, Brenner S, Venkatesh B. Sequence and organization of coelacanth neurohypophysial hormone genes: evolutionary history of the vertebrate neurohypophysial hormone gene locus. BMC Evol Biol. 2008;8:93. doi: 10.1186/1471-2148-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Holder MT, Erdmann MV, Wilcox TP, Caldwell RL, Hillis DM. Two living species of coelacanths? Proc Natl Acad Sci U S A. 1999;96:12616–12620. doi: 10.1073/pnas.96.22.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Venkatesh B, Nishida M. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: Coelacanthiformes) and divergence time estimation between the two coelacanths. Gene. 2005;349:227–235. doi: 10.1016/j.gene.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV. PIFs meet Tourists and Harbingers: a superfamily reunion. Proc Natl Acad Sci U S A. 2001;98:12315–12316. doi: 10.1073/pnas.231490598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–324. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EG, Lam K, Christoffels A, Erdmann MV, Brenner S, Venkatesh B. Hox gene clusters in the Indonesian coelacanth, Latimeria menadoensis. Proc Natl Acad Sci U S A. 2003;100:1084–1088. doi: 10.1073/pnas.0237317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R, Weil C. The hAT and CACTA superfamilies of plant transposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 565–610. [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (255 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Salama SR, Haussler D. Endangered species hold clues to human evolution. J Hered. 2010;101:437–447. doi: 10.1093/jhered/esq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, et al. (62 co-authors) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mongin E, Auer TO, Bourrat F, Gruhl F, Dewar K, Blanchette M, Wittbrodt J, Ettwiller L. Combining computational prediction of cis-regulatory elements with a new enhancer assay to efficiently label neuronal structures in the Medaka fish. PLoS One. 2011;6:e19747. doi: 10.1371/journal.pone.0019747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, Amemiya CT, Myers RM. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers TP, Amemiya CT. Evidence for a Hox14 paralog group in vertebrates. Curr Biol. 2004;14:R183–R184. doi: 10.1016/j.cub.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Putta S, Smith JJ, Staben C, Voss SR. MapToGenome: a comparative genomic tool that aligns transcript maps to sequenced genomes. Evol Bioinform Online. 2007;3:15–25. [PMC free article] [PubMed] [Google Scholar]
- Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashikant C, Bolanowski SA, Danke J, Amemiya CT. Hoxc8 early enhancer of the Indonesian coelacanth, Latimeria menadoensis. J Exp Zool B Mol Dev Evol. 2004;302:557–563. doi: 10.1002/jez.b.21018. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Kapitonov VV, Grzela DP, Jursch T, Jurka J, Izsvak Z, Ivics Z. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc Natl Acad Sci U S A. 2008;105:4715–4720. doi: 10.1073/pnas.0707746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. [updated 2008 Aug 01] 2004. Available from: http://www.repeatmasker.org. [Google Scholar]
- Sokal FJ, Rohlf RR. Biometry: the principles and practice of statistics in biological research. NewYork: W. H. Freeman & Co; 1995. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson KS, Gall JG, Coggins LW. Nuclear DNA contents of coelacanth erythrocytes. Nature. 1973;241:126. doi: 10.1038/241126a0. [DOI] [PubMed] [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Walbot V, Rudenko G. MuDR/Mu transposable elements of maize. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington (DC): ASM Press; 2002. pp. 533–564. [Google Scholar]
- Xie X, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci U S A. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34:5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feschotte C, Zhang Q, Jiang N, Eggleston WB, Wessler SR. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci U S A. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.