Abstract
Teneurins are type II transmembrane proteins expressed during pattern formation and neurogenesis with an intracellular domain that can be transported to the nucleus and an extracellular domain that can be shed into the extracellular milieu. In Drosophila melanogaster, Caenorhabditis elegans, and mouse the knockdown or knockout of teneurin expression can lead to abnormal patterning, defasciculation, and abnormal pathfinding of neurites, and the disruption of basement membranes. Here, we have identified and analyzed teneurins from a broad range of metazoan genomes for nuclear localization sequences, protein interaction domains, and furin cleavage sites and have cloned and sequenced the intracellular domains of human and avian teneurins to analyze alternative splicing. The basic organization of teneurins is highly conserved in Bilateria: all teneurins have epidermal growth factor (EGF) repeats, a cysteine-rich domain, and a large region identical in organization to the carboxy-half of prokaryotic YD-repeat proteins. Teneurins were not found in the genomes of sponges, cnidarians, or placozoa, but the choanoflagellate Monosiga brevicollis has a gene encoding a predicted teneurin with a transmembrane domain, EGF repeats, a cysteine-rich domain, and a region homologous to YD-repeat proteins. Further examination revealed that most of the extracellular domain of the M. brevicollis teneurin is encoded on a single huge 6,829-bp exon and that the cysteine-rich domain is similar to sequences found in an enzyme expressed by the diatom Phaeodactylum tricornutum. This leads us to suggest that teneurins are complex hybrid fusion proteins that evolved in a choanoflagellate via horizontal gene transfer from both a prokaryotic gene and a diatom or algal gene, perhaps to improve the capacity of the choanoflagellate to bind to its prokaryotic prey. As choanoflagellates are considered to be the closest living relatives of animals, the expression of a primitive teneurin by an ancestral choanoflagellate may have facilitated the evolution of multicellularity and complex histogenesis in metazoa.
Keywords: Teneurin, Odz, Ten-m, evolution, horizontal gene transfer, choanoflagellate, Monosiga brevicollis
Introduction
Teneurins are phylogenetically conserved transmembrane proteins (see reviews by Tucker and Chiquet-Ehrismann 2006; Young and Leamey 2009). The name “teneurin” honors their discovery in Drosophila melanogaster by combining the names of the two dipteran teneurin homologs, Ten-a (Baumgartner and Chiquet-Ehrismann 1993) and Ten-m (Baumgartner et al. 1994, also referred to as Odz [Levine et al. 1994]), with neurons, which are common sites of expression (e.g., Minet et al. 1999). In D. melanogaster, chicken, and mouse the teneurin homologs have the following conserved features: 1) teneurins are type II transmembrane proteins with an N-terminal intracellular domain (ICD) and a large extracellular domain (ECD); 2) teneurins have eight epidermal growth factor (EGF) repeats; 3) the third cysteine residue in the second and fifth EGF repeat is replaced with a tyrosine or phenylalanine residue, which results in the potential for teneurins to dimerize side-by-side through disulfide bonds (Oohashi et al. 1999); and 4) the C-terminal two-thirds of teneurins is similar to the YD-repeat proteins of prokaryotes, with characteristic NHL (from NCL-1, HT2A, and Lin-41) repeats, tyrosine and aspartate-rich YD repeats, and a region similar to the core-associated domain of retrotransposon hot spot (RHS) proteins. In addition, many teneurins can be proteolytically processed, freeing the ICD for transport to the nucleus (Bagutti et al. 2003; Nunes et al. 2005; Kenzelmann et al. 2008) and/or releasing the ECD for incorporation in the extracellular matrix (ECM; Rubin et al. 1999; Trzebiatowska et al. 2008). An additional cleavage site near the C-terminus can lead to the creation of a neuropeptide (reviewed by Lovejoy et al. 2009). Proline-rich Src homology 3 (SH3)–binding domains have been identified in the ICD of teneurins cloned from chordates and ecdysozoans, and ICD-interacting partners have been characterized that may mediate associations between teneurins and the cytoskeleton and methylated DNA (Nunes et al. 2005). Mutation analysis and RNAi-mediated knockdown of teneurin expression in D. melanogaster and Caenorhabditis elegans reveal fundamental roles for teneurins in pattern formation (Baumgartner et al. 1994; Rakovitsky et al. 2007), axonal fasciculation (Drabikowski et al. 2005), and the integrity of basement membranes (Trzebiatowska et al. 2008). In chordates, teneurins are best studied in mouse and chicken, where they are predominantly expressed in the developing nervous system in area-specific patterns mediated in part by EMX2 (Li et al. 2006; Beckmann et al. 2011). Knockout of the gene encoding mouse teneurin-3 by homologous recombination results in abnormal pathfinding in the visual system and a loss of binocular vision (Leamey et al. 2007).
In order to identify novel features and learn more about the potential evolutionary origin of teneurins, we searched for and compared sequences encoding teneurin-like proteins in opisthokont genomes and collections of expressed sequence tags (ESTs). We also cloned and sequenced cDNAs encoding the ICD of teneurins from human and chicken to study alternative splicing. By aligning and analyzing proteins for predicted nuclear localization sequences (NLSs), SH3-binding domains, and furin-type proteolytic cleavage sites, we have refined our knowledge of conserved teneurin structure and function. In addition, we identified a gene encoding a teneurin in the choanoflagellate Monosiga brevicollis, which suggests that teneurins may have played a role in the early evolution of metazoan tissues.
Materials and Methods
Sequence Analysis
Novel teneurin sequences were identified by sequence homology using tBLASTn (http://blast.ncbi.nlm.nih.gov/) and by domain architecture using Pfam (http://pfam.sanger.ac.uk/) with “view a family,” SMART (http://smart.embl-heidelberg.de/smart/) with “architecture queries,” and Superfamily (http://supfam.org/SUPERFAMILY/) with “domain combinations.” Boundaries of regions, domains, and repeats were determined using Pfam for EGF repeats, NHL repeats, RHS repeats (related to YD repeats), RHS protein, Ten_N domains, and PfamB PB025792 (the region between the transmembrane domain and the EGF repeats, which was identified as a phylogenetically conserved region by Pfam), and SMART for transmembrane domains and EGF repeats. Alignments and phylogenetic relationships were determined using ClustalW (http://www.genome.jp/tools/clustalw/) and the settings “pair alignment slow/accurate” (gap open penalty 10, gap extension penalty 0.1). Importin α/β pathway NLSs were identified using NLS Mapper (http://nls-mapper.iab.keio.ac.jp), and furin cleavage sites were predicted with ProP (http://www.cbs.dtu.dk/services/ProP/). SH3-binding domains were identified by hand from consensus sequences described by others (Kay et al. 2000; Mayer 2001; Kowanetz et al. 2003).
Reverse Transcriptase PCR and Sequencing
Human adult brain cDNA was generated out of total human brain RNA (AMS Biotechnology, Oxon, UK) using Superscript III (Invitrogen, Carlsbad, CA) polymerase and random hexamer primers (Invitrogen). Sequences corresponding to the ICDs of teneurins-1 through -4 were amplified with PfuTurbo polymerase (Stratagene/Agilent Technologies, Santa Clara, CA) using specific primers (teneurin-1: 5′-ACTAGCGGCCGCACCATGGAGCAAACTGACTGC-3′/5′-ACTACTCGAG GCAGCACCTGTAAGGTTTG-3′; teneurin-2: 5′-ACTAGCGGCCGCACCATGGATGTAAAGGACCGG-3′/5′-ACTACTCGAGGCAGTATTTGGAGGGCTTC-3′; teneurin-3: 5′-ACTAGCGGCCGCACCATGGATGTGAAAGAACGC-3′/5′-ACTACTCGAGACAGTACTTTGAAGACTTC-3′; teneurin-4: 5′-ACTAGCGGCCGCACCATGGACGTGAAGGAGAGG-3′/5′-ACTACTCGAGACAGTACTTGGAGGGCTTC-3′) including restriction sites NotI and XhoI. Amplified products were separated on a 0.8% agarose gel, and fragments were excised, gel extracted, and cloned into pcDNA3. Positive clones were sequenced using forward primer T7 and reverse primer Sp6.
The sequence of avian teneurin-3 including alternatively spliced variants was assembled from overlapping fragments cloned by polymerase chain reaction (PCR). cDNA was prepared from total RNA extracted from embryonic day 16 chicken cerebellum using the RNeasy Mini kit (Qiagen, Germantown, MD). PCR was performed with the Platinum Pfx DNA Polymerase System (Invitrogen). Five sets of primers were used to divide avian teneurin-3 into five segments. Segment 1 used primer pair 5′-ATGGATGTGAAAGAACGTCG-3′/5′-CACGTGGAGGGTAAACGATAA-3′; segment 2 used primer pair 5′-ACTGTGAAGAAGCGGATTGC-3′/5′- GACCGCCAAAAGTCACTAGA-3′; segment 3 used primer pair 5′-TGATGGGACCATGATCAGAA-3′/5′-ACCAGACGG CAGACATGAAC-3′; segment 4 used primer pair 5′-AGCGAGGGACGACTAGTGAA-3′/5′-GGAGAAAGGATAGAGT GAAA-3′; and segment 5 used primer pair 5′-AGGCTGTGGACAGAAGGAGA-3′/5′-GGTCCTCTACTTGGATGACT-3′. Each segment was TOPO cloned into the pCR-II vector (Invitrogen) for sequencing.
Results
Overview: Identification of Teneurins and Analysis of Teneurins from Homo sapiens
Genes encoding teneurin-like proteins and predicted proteins with the characteristic domain organization of known teneurins were identified by sequence similarity (e.g., tBLASTn) and by the presence of combinations of domains (e.g., predicted proteins with both EGF repeats and NHL repeats using Pfam or SMART; for details, see Materials and Methods). Teneurins identified in this way from chordates are summarized in supplementary table S1, Supplementary Material online; teneurins from nonchordates are summarized in supplementary table S2, Supplementary Material online.
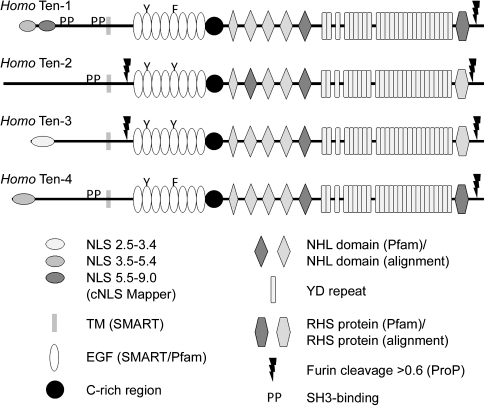
To illustrate the features of these teneurins identified through proteomic analysis, the four teneurins from H. sapiens are shown in figure 1. The variant of teneurin-1 shown in figure 1 is a type II transmembrane protein with a 317aa N-terminal ICD, a 23aa transmembrane domain, and a 2385aa C-terminal ECD. Within the ICD, there are two predicted importin α/β pathway NLSs. The first NLS (aa11-40) has an NLS Mapper score of 4.8, and the second NLS (aa60-69) has an NLS Mapper score of 6.0. Higher scores represent a greater likelihood of nuclear localization (Kosugi et al. 2009) and are indicated on the figure with progressively darker ovals. Two proline-rich SH3-binding domains (indicated by “PP” on fig. 1) are also found in the ICD of human teneurin-1. The first (aa193-199; RPLPPPP) is consistent with the consensus sequence for Class I SH3 ligands (+xφPxφP); the second (aa292-297; PRPLPR) is consistent with the atypical SH3-binding motif (PxxxPR) of cbl proteins (Kowanetz et al. 2003). In the ECD of human teneurin-1, there are eight EGF repeats (aa531-796). The third cysteine residue of the second EGF repeat has been replaced with a tyrosine residue, and the third cysteine residue of the fifth EGF repeat has been replaced with a phenylalanine (indicated by the “Y” and “F” in fig. 1). This substitution results in the potential for dimerization of teneurin monomers through disulfide bonds between cysteines that lack an intramodular partner (Oohashi et al. 1999). A cysteine-rich domain is found adjacent to the eighth EGF repeat (aa797-836). The carboxy two-thirds of human teneurin-1 shares the same domain organization as the YD-repeat proteins of some prokaryotes (e.g., Myxococcus xanthus, where it is required for gliding motility [Youderian and Hartzell 2007]): five NHL domains, YD-repeats (similar to “RHS repeats”), and a region near the C-terminal tail identified by Pfam as RHS protein (similar to “RHS-associated core domain”). The NHL domains of human teneurin-1 were identified by Pfam (dark gray, fig. 1) or by alignment using ClustalW (light gray, fig. 1). Similarly, the RHS protein domain identified by Pfam in human teneurin-1 is indicated in dark gray, and those identified in other teneurins by alignment are indicated by lighter shades. Finally, human teneurin-1 has a single predicted furin cleavage site with a ProP score at or above 0.55 (threshold = 0.50) at aa2618 (LNGRTRR/FA). This would create a 107aa C-terminal peptide similar to teneurin C-terminal-associated peptide-1 (Trubiani et al. 2007).
FIG. 1.
The human teneurins. Human teneurins share a basic domain organization but they differ in the presence of NLSs, SH3-binding motifs, and furin cleavage sites. Following processing the ICD of teneurin-1, for example, is predicted to be located in the nucleus but not the ICD of teneurin-2. Similarly, the ECD of teneurins-2 and -3 are predicted to be shed into the ECM but not the ECD of teneurins-1 and -4. Accession numbers, the positions of domains as well as the NLS and ProP scores of each teneurin are found in supplementary table S3, Supplementary Material online.
There are four teneurin genes in H. sapiens. The basic organization of teneurins-1, -2, -3, and -4 is the same, most notably in the ECD: each teneurin has eight EGF repeats with aromatic residues substituting for cysteines in the second and fifth repeat, each has a cysteine-rich domain and a C-terminal region similar to the YD-repeat proteins of prokaryotes, and each has a predicted furin cleavage site near the C-terminus (fig. 1; details can be found in supplementary table S3, Supplementary Material online). One difference is the presence of a second predicted cleavage site between the transmembrane domain and EGF repeats of teneurins-2 and -3 that would permit shedding of the ECD into the ECM; these cleavage sites are not found in teneurins-1 and -4. Additional differences are seen in the ICD. Teneurins-1, -2, and -4 have proline-rich motifs that match consensus SH3-binding domains but teneurin-3 does not. However, the proline-rich sequence PPTRPLPR is found in the ICD of human teneurin-3, which resembles, but does not exactly match, known SH3-binding motifs. Teneurin-2 does not have a predicted NLS, and the NLS of teneurin-3 has a lower NLS Mapper score than the NLSs of teneurin-1 and teneurin-4.
Representative teneurins from major taxonomic groups were analyzed in this manner and are described below.
Identification and Analysis of Chordate Teneurins
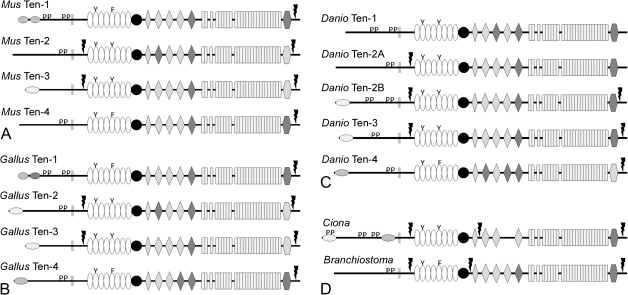
The teneurins of mouse (Mus musculus), chicken (Gallus gallus), zebrafish (Danio rerio), and the protochordates Ciona intestinalis and Branchiostoma floridae were identified and analyzed. There are four teneurins in M. musculus and G. gallus and they share many of the features described above for human teneurins. The few differences include the absence of a predicted NLS in murine teneurin-4, and the observation that chicken teneurin-2 has a predicted NLS, albeit a weak one (fig. 2A and B; supplementary table S3, Supplementary Material online). In D. rerio, there are five teneurins. ClustalW alignment and basic phylogenomic analysis identify two teneurin-2 paralogs that we have named teneurin-2A and teneurin-2B (figs. 2C and 3; supplementary table S3, Supplementary Material online). The predicted sequences of teneurin-1, teneurin-2B, and teneurin-4 appear to be complete, but the predicted N-termini of teneurin-2A and teneurin-3 were completed by translating potential open reading frames and aligning them with known teneurin sequences and by piecing together ESTs (supplementary table S3, Supplementary Material online; FASTA files can be found in supplementary table S4, Supplementary Material online). As with the chicken teneurins, the basic features of the zebrafish teneurins are conserved. Differences include the absence of a potential furin cleavage site near the C-terminus of teneurin-1, an additional potential furin cleavage site between the NHL repeats and YD repeats of teneurin-2A and the absence of predicted NLSs in the ICDs of teneurin-1 and teneurin-2A. In addition, the ICD of D. rerio teneurin-3 has a predicted proline-rich SH3-binding domain, unlike the ICDs of teneurin-3 in chicken and man.
FIG. 2.
The domain organization and possible relationships of chordate teneurins. There are four teneurins in mouse (Mus musculus) and chicken (Gallus gallus). Basic features are highly conserved between mouse (A), chicken (B), and human teneurins (fig. 1). There are five teneurin genes in the zebrafish, Danio rerio, including two teneurin-2 paralogs. The ICD of the predicted teneurin-1 does not have an NLS, and an additional furin cleavage site is found between the NHL domains and the YD repeats (C). The genomes of the tunicate Ciona intestinalis and the cephalochordate Branchiostoma floridae each contain a single teneurin gene (D).
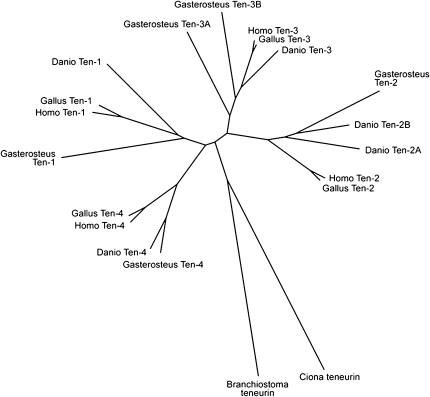
FIG. 3.
An unrooted phylogenetic tree based on ClustalW alignment of real and predicted teneurin amino acid sequences suggests that teneurins-1 and -4 share a common ancestor, as do teneurins-2 and -3. The stickleback Gasterosteus aculeatus has five teneurins, but unlike Danio rerio it has retained two teneurin-3 paralogs.
To determine if the duplication of teneurin-2 is a common feature in bony fish, the teneurins of the stickleback Gasterosteus aculeatus were also identified (supplementary table S1, Supplementary Material online) and aligned with the teneurins of other chordates. Like D. rerio, G. aculeatus has five teneurin genes. However, there are two teneurin-3 paralogs (teneurin-3A and teneurin-3B) and only one teneurin-2 (fig. 3). The teneurin-1 of G. aculeatus has a potential furin cleavage site near the C-terminus (aa2642; ProP score = 0.74), indicating that the absence of this site in D. rerio may not be typical of teneurin-1 in actinopterygians. The G. aculeatus teneurin-1 also has a predicted NLS in the ICD (aa11-41, NLS Mapper score = 4.0).
The genomes of C. intestinalis and B. floridae each encode a single teneurin (fig. 2D, supplementary table S3, Supplementary Material online). The basic organization of these teneurins resembles those of the craniate teneurins. The ICD of the predicted teneurin from C. intestinalis has two NLSs: one near the N-terminus and the other near the transmembrane domain. A predicted NLS near the transmembrane domain is commonly observed in the ICD of teneurins from protostomes (see below). The ICDs from both of the protochordate teneurins have predicted SH3-binding domains, but the ICD from B. floridae does not have an NLS that is recognized by NLS Mapper. Both of the predicted protochordate teneurins have potential furin cleavage sites that would shed the ECD and process the C-terminus like those of teneurin-2 and teneurin-3 in fish, birds, and man, but like D. rerio teneurin-2A (and unlike other craniate teneurins examined) they also have a third predicted furin cleavage site near the center of the ECD. A second teneurin-like sequence is found in the B. floridae genome when two adjacent predicted proteins (XP_002592160 and XP_002592161) are combined. However, the C-terminal two-thirds of the second predicted protein also has lysozyme and keratin-related sequences, and some teneurin and lysozyme-like sequences are encoded on the same large predicted exon, which leads us to suggest that this is a pseudogene. This is supported by the total absence of ESTs.
Alternatively Spliced Variants
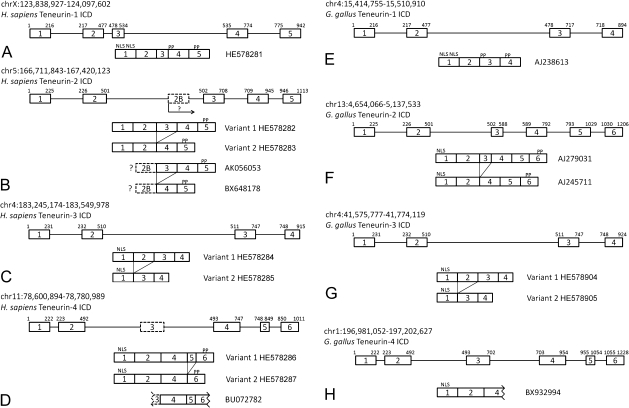
Previously we showed that there are a number of isoforms of avian teneurin-2 and that these variants are derived from alternative splicing of regions of transcripts encoding both the ICD and the ECD (Tucker et al. 2001). Here, we examined ESTs and used RT-PCR to determine if the ICD variants are specific for teneurin-2 and if they are conserved in mammals and birds. A single PCR product is amplified from adult human brain-derived cDNA using primers corresponding to the 3′ end of the first exon of the human teneurin-1 gene and the 5′ end of the fifth exon, which encodes the transmembrane domain (fig. 4A). When the same strategy is applied using primers based on human teneurin-2 sequences, two variants are observed: Variant 1 is encoded by all five previously identified exons and Variant 2 lacks the third exon (fig. 4B). These variants are supported by EST data, which also reveal the use of a sixth exon found between exon 2 and exon 3. EST sequences containing this alternative exon, which we have named exon 2B, do not contain sequences corresponding to either exon 1 or exon 2. As a putative start codon is found in exon 2B, this exon may be used as an alternative start site for teneurin-2 transcripts (and therefore would not have been amplified using our flanking primer pairs). The ICD of teneurin-3 is encoded on four exons and like teneurin-2 there are two ICD splicing variants: Variant 1 uses all four exons, whereas Variant 2 is encoded on exons 1, 3, and 4 (fig. 4C). Finally, RT-PCR reveals two alternatively spliced variants of the human teneurin-4 ICD. The larger is encoded by five exons, and a smaller by four exons. Interestingly, an EST (BU72782) shows the potential use of an additional exon that was not amplified by our primer pair (fig. 4D).
FIG. 4.
Comparison of alternative splicing in the ICD of human and chicken teneurins. The human teneurin-1 ICD is encoded on five exons. No splice variants were found using RT-PCR and primers corresponding to sequences in exon 1 and exon 5 (A). In contrast, RT-PCR reveals two variants amplified with primers based on sequences in the first and fifth exon of human teneurin-2. Variant 1 is encoded on 5 exons, whereas Variant 2 is composed of four. ESTs suggest an additional way to generate diversity in the ICD of human teneurin-2: An exon found between the second and third exon (exon 2B) is associated with long and short variants as well. This may represent an alternative start site for teneurin-2 (B). Human teneurin-3 has an ICD encoded on four exons, and exon 2 can be spliced out to generate a second variant (C). RT-PCR with primers based on sequences in exon 1 and exon 6 reveal long and short variants of human teneurin-4. Variant 1 is encoded on five exons, whereas Variant 2 is encoded on four. Use of an additional exon (exon 3) that does not contain a start codon is found in an EST (D). The ICDs of teneurins-1 and -2 from the chicken were described previously. As in human, there is a single teneurin-1 isoform, and there are long and short variants of teneurin-2. There is no evidence from chicken of an alternative start site in teneurin-2 (E, F). PCR using cDNA from embryonic chicken cerebellum and primers based on sequences in exons 1 and 4 reveal identical variants in chicken and human teneurin-3 (G). Sequence from a single EST from the chicken is consistent with the organization of the ICD of human teneurin-4 (H).
ESTs demonstrate that some of the ICD variants we observed in human teneurin-1 and teneurin-4 are conserved in G. gallus, just as our previous work with avian teneurin-2 showed the presence of two ICD variants (fig. 4E, F, and H). To study the alternative splicing of teneurin-3, we used RT-PCR to amplify products corresponding to the ICD and cDNA derived from embryonic chicken brain. Just as in human, the avian teneurin-3 ICD is encoded on four exons. A large variant contains sequences corresponding to all four exons, and exon 2 is spliced from a smaller variant (fig. 4G). Note that we could not identify an exon homologous to exon 2B of human teneurin-2 in the chicken genomic sequence, but there is a homologous potential exon 3 in chicken teneurin-4 DNA.
There is also evidence of teneurin variants derived from alternative splicing in the region encoding the ECD. For example, a short (8aa) stretch of amino acids can be present between the seventh and eighth EGF repeats of teneurin-2 from the chicken, and at least one variant of avian teneurin-2 is truncated between the seventh and eighth EGF repeats, resulting in an isoform lacking the cysteine-rich domain and the region homologous to the YD repeat proteins of prokaryotes (Tucker et al. 2001). Alternative splicing that results in additional sequence between the seventh and eighth EGF repeats may be common in teneurins, as similar variants are found in mRNA sequences in mouse teneurin-3 (NP_035987) and teneurin-4 (BAE28005). However, there is no evidence supporting grossly truncated isoforms of teneurins in other species.
Identification and Analysis of Teneurins from an Echinoderm and Protostomes
The same methods used to identify teneurins in chordates were applied to other metazoan sequences. In addition to the known teneurins of D. melanogaster and C. elegans, predicted complete or partial teneurins were found in the purple sea urchin (Strongylocentrotus purpuratus), a mollusk (Lottia gigantean), an annelid (Capitella teleta), a trematode (Schistosoma mansoni), and a wide variety of nematodes and arthropods (supplementary table S2, Supplementary Material online). Interestingly, no teneurin-like sequences were identified in the genomes or ESTs from cnidarians, ctenophores, sponges, or Trichoplax adhaerens.
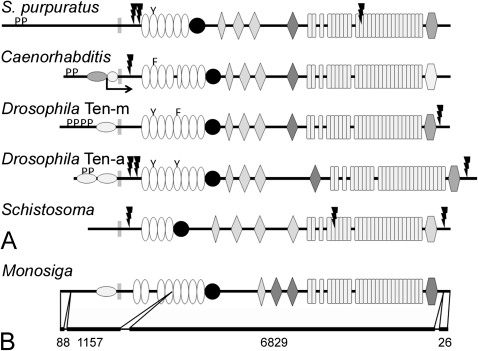
The single teneurin from S. purpuratus is remarkable for a few features not seen in teneurins from chordates: 1) it has only six EGF repeats and only the second EGF repeat has an aromatic residue substituting for a cysteine residue; 2) it lacks a predicted furin cleavage site near the C-terminus; and 3) it has two predicted furin cleavage sites between the transmembrane domain and the EGF repeats (fig. 5A). Like the teneurins from protochordates, it has a predicted furin cleavage site near the center of the ECD.
FIG. 5.
The domain organization of nonchordate teneurins. The ECDs of teneurins from the purple sea urchin (Strongylocentrotus purpuratus), Caenorhabditis elegans, Drosophila melanogaster, and the trematode Schistosoma mansoni have the same basic organization as chordate teneurins, but there is variation in the number of EGF repeats. Aromatic residues substitute for cysteines in at least one EGF repeats in all the species examined except the fluke, which suggests that the teneurin from S. mansoni does not dimerize (A). The genome of the choanoflagellate Monosiga brevicollis encodes a protein with a domain organization that is identical to metazoan teneurins. The predicted protein does not have furin cleavage sites or SH3-binding domains, and its EGF repeats contain a full complement of cysteines. The predicted M. brevicollis teneurin is encoded on just four exons, and most of the ECD is encoded on a single mega-exon of 6829 bp (B).
The teneurins from C. elegans and D. melanogaster are well known, and the sequences that were analyzed here came from cDNAs. There are two teneurins from C. elegans, Ten-1L and Ten-1S; they are encoded by the same gene, but two different promoters regulate the expression of “long” and “short” transcripts (Drabikowski et al. 2005). Ten-1L is illustrated in figure 4A; Ten-1S would be identical except the ICD is much smaller (the approximate location of the N-terminus of Ten-1S is indicated by the crooked arrow in fig. 5A). Ten-1L and the two D. melanogaster teneurins, Ten-m and Ten-a, have putative SH3-binding domains and one or more NLS. Unlike the NLSs of most chordate teneurins, the NLSs from the ecdysozoans tend to be found near the transmembrane domain and not at the N-terminus. The ECDs of these teneurins are similar to those found in chordate teneurins: note that both Ten-m and Ten-a (but not Ten-1L) have potential furin cleavage sites near the C-terminus, and both Ten-1L and Ten-a have potential furin cleavage sites that could shed the ECD into the ECM. The fifth EGF repeat of C. elegans Ten-1L is incomplete; the part of this repeat encoding both the second and third cysteine residues in other teneurins is missing. Also, the part of the ECD near the C-terminus that is predicted by Pfam to be homologous to the RHS core-associated protein domain is more unlike this domain in other teneurins, though some identity could be found by alignment.
Two teneurin genes that encode predicted proteins that align with either D. melanogaster’s Ten-a or Ten-m were identified in the genomes of a number of insect species, including Apis mellifera (honey bee), Tribolium castaneum (flour beetle), and the mosquitoes Aedes aegypti and Culex quinquefasciatus (supplementary table S2, Supplementary Material online). However, single teneurin genes were found in the genomes of the branchiopod crustacean Daphnia pulex and the arachnid Ixodes scapularis (deer tick). This suggests that the duplication of teneurins in the protostome lineage is limited to insects and is not a feature of all arthropods.
The trematode S. mansoni has a single predicted teneurin (fig. 5A; supplementary table S3, Supplementary Material online) that has a number of distinctive features. Its ICD is relatively short, and it does not contain predicted SH3-binding motifs or an NLS. Like many teneurins it has a putative furin cleavage site between the transmembrane domain and the EGF repeats, but it also has a second predicted furin cleavage site amidst the YD repeats. Finally, the fluke teneurin has only four EGF repeats, and all four EGF repeats have a full complement of cysteine residues, so unlike other teneurins studied to date it probably fails to dimerize.
A Teneurin Is Encoded in the Genome of the Choanoflagellate M. brevicollis
The absence of teneurin genes in the complete and assembled genomes of the cnidarian Nematostella vectensis and placozoan T. adhaerens (that could be identified using the search methods employed to find teneurins in other metazoans) initially suggested that teneurins may have evolved about the time of the Cambrian radiation. However, during a routine search of predicted protein domain architectures that included RHS core-associated protein domains using the Pfam program, a sequence encoding EGF repeats, NHL domains, and YD repeats (in addition to the RHS core-associated protein domain) was identified in the genome of the choanoflagellate Monosiga brevicollis. Further analysis revealed that this predicted protein has the basic features of a metazoan teneurin: it is a type II transmembrane protein with a putative NLS in the ICD, eight EGF repeats, a cysteine-rich domain, and a C-terminal two-thirds with the same domain architecture as a prokaryotic YD-repeat protein (i.e., NHL domains, YD repeats, and an RHS core-associated protein domain; fig. 5B; supplementary table S3, Supplementary Material online). The predicted sequence (XP_001749414) is shown in its entirety in supplementary figure S1, Supplementary Material online, together with relevant alignments generated with ClustalW. The expression of the M. brevicollis teneurin is supported by two nonoverlapping ESTs (FE890769 and FE895158), both of which correspond to regions encoding the YD repeats.
The ICD of the M. brevicollis teneurin does not align significantly with the ICDs from other teneurins, and it lacks SH3-binding motifs. In addition, ProP fails to identify any potential furin cleavage sites in this teneurin. There are eight EGF repeats, but there are no free cysteines to support dimerization. Adjacent to the EGF repeats is a cysteine-rich region that is highly conserved: the exact 23aa consensus sequence ExxCx(D/N)xxDx(D/E)xDxxxDCxxx(D/E)CCxxxxCxxxxxC is found in all the teneurins analyzed except for S. purpuratus teneurin (which has one additional “x” between the fourth and fifth cysteine) and S. mansoni teneurin (which is missing the sixth cysteine). In fact, using the M. brevicollis cysteine-rich domain sequence in a tBLASTn search of all nucleotide sequences uncovers all the teneurins identified above that are listed on GenBank. However, neither this method nor the other search methods we employed to identify teneurin sequences revealed teneurins in sequences from sponges, placozoans, ctenophores, cnidarians, fungi, ichthyospores or nucleariids. Interestingly, a similar cysteine-rich sequence is found in an endo-1,3-beta-glucosidase from the diatom Phaeodactylum tricornutum (XP_002181321). This sequence is 46% identical and 71% similar to the 35aa cysteine-rich domain of M. brevicollis and it includes a core stretch of 23aa that is 65% identical and 91% similar (table 1). This 23aa core domain aligns well with the Cu++-binding motif of MopE (Helland et al. 2008), and similar sequences are found in the metal-binding region of a predicted gametolysin from Volvox carteri (table 1 [XP_002958497]) and Chlamydomonas reinhardtii (XP_001695639). Alignment of the 23aa motif reveals that the core of the cysteine-rich region of M. brevicollis teneurin is most similar to the diatom sequence and the core of the cysteine-rich region of trematode teneurin; the same regions from other metazoan teneurins are conserved but not to the same extent (table 1).
Table 1.
Alignment of Representative Sequences with the Cysteine-Rich Domain Core Sequence of Monosiga brevicollis Teneurin.
| Species (common name) | Core Sequence | % Id | % Sim |
| Monosiga brevicollis (choanoflagellate) | CNDGIDNDNDRVTDCNDADCCSS | ||
| Teneurin | |||
| Phaeodactylum tricornutum (diatom) | CNDGIDNDNDGLFDCEDPDCAND | ||
| Endo-1,3-beta-glucosidase | ********** : **:*.**... | 65% | 91% |
| Schistosoma mansoni (fluke) | CDDGIDNDHDDLVDCLDPDCCTS | ||
| Teneurin | *:******:* :.** *.***:* | 65% | 91% |
| Caenorhabditis elegans (roundworm) | CDDGLDNDSDGLIDCDDPECCSS | ||
| > Ten-1L | *:**:***.* : **:*.:**** | 61% | 91% |
| Drosophila melanogaster (fruit fly) | CKDNIDNDGDGMTDCSDSECCSH | ||
| Ten-a | *:*.****.* :***.*::*** | 61% | 91% |
| Homo sapiens (human) | CGDNLDNDGDGLTDCVDPDCCQQ | ||
| Teneurin-1 | *.*.:***.* :*** *.***.. | 57% | 91% |
| Volvox carteri (volvox) | CDDGIDNDCDGLVDMDDPDCNTS | ||
| Gametolysin | *:****** * :.* :*.** :* | 57% | 83% |
| Strongylocentrotus purpuratus (purple sea urchin) | CTDEVDNDGDSLIDCEDPDCCLS | ||
| Teneurin | *.* :***.* : **:*.*** * | 57% | 83% |
| Pfam: Cu-binding_MopE | C.DGVDNNCDGQVD | ||
| PF11617* | * **:**: * .* | 54% | 77% |
NOTE.—*The Cu++-binding consensus domain of MopE and related proteins.
The NHL repeats and YD repeats of M. brevicollis align better with the YD-repeat proteins of some prokaryotes than with the YD repeats found in metazoan teneurins. The NHL domains are most similar (31% identical) to the YD-repeat protein of Herpetosiphon aurantiacus (ABX04679), a predatory filamentous chloroflex bacterium that lives in soil and freshwater. A stretch of 103aa corresponding to YD repeats 17-21 of M. brevicollus teneurin was analyzed further using tBLASTn and the entire NCBI nucleotide collection. This stretch is most similar to the YD-repeat proteins of Syntrophobacter fumaroxidans (a freshwater bacterium) and Desulfococcus oleovorans (which lives in coastal waters) and then to the YD repeats found in human teneurin-4 (table 2).
Table 2.
Alignment of Most Similar Sequences with YD Repeats from Monosiga brevicollis Teneurina
|
M. brevicollis Syntrophobacter fumaroxidansb |
YDVDGQLTQVLEDGAEVESYSYDVNGNRVAWNVRG---AAHSATYGADDA YDSLGRLLAVRLDGVPAEEYRYDVNGNRVEETNTPRGIIGRTSTYSEEDH ** *:* * **. .*.* ******** . .:::**. :* |
|
|
M. brevicollis S. fumaroxidans |
VFTVDGQSYAVDVDGFLTSVRG----MSLAYSGRGELLSATLPSGAGTVR LLTSGGTVYRYDADGFLTTRTEGSAVTRYVYSSRGELLSVALPDGK-RIE ::* .* * *.*****: .**.******.:**.* :. |
|
|
M. brevicollis S. fumaroxidans |
YRYDGFGRRI YVNDPLGRRI 46% identical/69% similar * * :**** |
|
|
M. brevicollis Desulfococcus oleovoransc |
YDVDGQLTQVLEDGAEVESYSYDVNGN-----RVAWNVRGAAHSATYGAD YDEMGRLETVTKDGTLVESYSYDSTPYGTCTYQMNTLRGIAGRVLDYDAE ** *:* * :**: *******. :: *.: *.*: |
|
|
M. brevicollis D. oleovorans |
DAVFTVDGQSYAVDVDGFLTSVRG----MSLAYSGRGELLSATLPSGAGT DHLLSAGGTDYQYDLDGFLTSKTSGAETTYYDYSSRGELLSVDLPDGT-D * :::..* .* *:******. **.******. * *.*: |
|
|
M. brevicollis D. oleovorans |
VRYRYDGFGRRI ITYVHDPLGRRI 43% identical/67% similar: * :* :**** |
|
|
M. brevicollis Homo sapiens Ten-4 |
YDVDGQLTQVLEDGAEVESYSYDVNGNRVAWNVRGAAHS--ATYGADDAV YDADGQLQTVSINDKPLWRYSYDLNGNLHLLSPGNSARLTPLRYDIRDRI **.**** * :. : ****:***. .:*: *. * : |
|
|
M. brevicollis H. sapiens Ten-4 |
FTVDGQSYAVDVDGFLTSVRGMSLAYSGRGELLSATLPSGAGTVRYRYDG TRLGDVQYKMDEDGFLRQRGGDIFEYNSAGLLIKAYNRAGSWSVRYRYDG: .. .* :* **** . * : *.. * *:.* :*: :******* |
|
| M. brevicollis | FGRRI | |
| LGRRV 38% identical/63% similar :***: |
The M. brevicollis teneurin is predicted from sequences encoded on just four exons that are separated by introns of 129, 206 and 105 bp (in contrast, human teneurin-1 is encoded on 29 exons and the average intron is 8 kb). Remarkably, the region of the predicted protein corresponding to the four C-terminal EGF repeats, the cysteine-rich domain, the NHL repeats, the YD repeats, and the RHS core-associated protein domain is encoded on a single giant exon of 6829 bp (fig. 5B). For comparison, the corresponding regions of human teneurin-1, S. mansoni teneurin, and C. elegans Ten-1L are encoded on 20, 21, and 7 exons, respectively (see also Minet and Chiquet-Ehrismann 2000).
Discussion
Here, we have used predictions based on proteomics to determine which teneurins may be processed such that the ECD becomes incorporated into the ECM and which teneurins may be processed such that the ICD is transported to the nucleus. Our predictions are validated by our previous experimental studies with avian teneurins. For example, we showed that a recombinant fusion protein with the ECD of chicken teneurin-2 was cleaved in vitro at a furin site between the transmembrane domain and the EGF repeats (Rubin et al. 1999). Consistent with this observation, we also showed that antibodies to the ECD of chicken teneurin-2 not only labeled the cell surface but also the ECM of chicken embryos (Tucker et al. 2001). When tagged chicken teneurin-2 ICD is overexpressed in HT1080 cells the recombinant ICD localizes to the nucleus (Bagutti et al. 2003), but there is no evidence published to date that the teneurin-2 ICD is processed and transported to the cell nucleus in vivo. In contrast, antibodies to the ECD of chicken teneurin-1 failed to stain the ECM, but antibodies to the ICD of teneurin-1 routinely stained the nuclei of cells in vitro and in histological sections of embryos (Kenzelmann et al. 2008). Moreover, when the sequence RKRK in the avian teneurin-1 ICD is mutated to AAAA it no longer localizes to the nucleus in vitro (Kenzelmann et al. 2008). Here, we show that the ICD of chicken teneurin-1 (specifically, the RKRK and flanking sequences) is predicted with a high likelihood to be located in the nucleus (NLS Mapper score = 9.0) and that the ICD of chicken teneurin-2 is much less likely to be nuclear (NLS Mapper score = 2.7). Similarly, the chicken teneurin-2 furin-cleavage site that we previously demonstrated to be functional is predicted by ProP to be active (score = 0.65), but no such site is found in chicken teneurin-1. Thus, teneurin-1 and/or teneurin-4 are most likely to be processed (by a yet unknown mechanism) so that the ICD can move to the nucleus, and teneurin-2 and/or teneurin-3 are more likely to have the ECD shed into the ECM. The shared features of these pairs of teneurins are also reflected in their predicted origins: teneurins-1 and -4 appear to have evolved from a gene duplication, as do teneurins-2 and -3 (fig. 3; see also Minet and Chiquet-Ehrismann 2000). All the chordate teneurins examined here except D. rerio teneurin-1 are likely to be cleaved near the C-terminus. This may be a step that precedes the formation of the teneurin-derived C-terminal neuropeptides characterized by others (reviewed by Rotzinger et al. 2010).
Using a yeast two-hybrid screen, Nunes et al. (2005) found that the SH3 domains of CAP/ponsin interact with the second proline-rich SH3-binding motif of chicken teneurin-1; the identical motif is present in teneurin-4. CAP/ponsin in turn binds to vinculin, which could anchor the ICD of teneurins to the actin cytoskeleton. A predicted SH3-binding motif at the same location in teneurin-2 does not bind CAP/ponsin even though it varies from the motif in teneurins-1 and -4 by only a single amino acid (Nunes et al. 2005). This led us to analyze teneurins from a broad range of taxa for SH3-binding motifs. The teneurin ICDs from each species examined, except for S. mansoni and M. brevicollis, contained one or more consensus SH3-binding motif. Interestingly, S. mansoni and M. brevicollis are the only species examined with teneurins lacking the capacity to dimerize. Perhaps dimerization is necessary for the ICD-interacting proteins to link teneurins to the cytoskeleton or to regulate the processes necessary for ICD nuclear localization.
Databases (e.g., GenBank, Ensembl, JGI, UniProt) contain listings for numerous teneurin variants. Most of these variants are based on predicted sequences, but some are based on cDNAs and ESTs. Here, we chose to study the range of alternative splicing in the ICD of human and chicken teneurin by PCR. The ICDs of human and chicken teneurins tend to be encoded on two pairs of neighboring exons separated by a large intron. Additional exons, which often are not conserved between birds and man and which frequently are subjected to alternative splicing, are sometimes found between the two pairs of exons. Alternatively spliced exons do not contain recognizable SH3-binding domains or NLSs, so the significance of these variations is not clear. Interestingly, an alternatively spliced exon in human teneurin-2 may represent an alternative start site, as ESTs with this sequence do not contain sequence from exons 1 or 2, and sequences encoded on this exon are not found in the PCR products amplified using a primer based on sequences found in exon 1. A similar method for generating teneurin splice variants was shown previously for C. elegans (Drabikowski et al. 2005).
The extraordinary diversity of teleost fish is commonly attributed to the duplication of their genome followed by the selective retention of certain duplicated genes (see Jozefowicz et al. 2003; Postlethwait et al. 2004; Volff 2005). This has been supported by studies of Hox genes (Kurosawa et al. 2006; Zou et al. 2007). In contrast, comparisons of Sox genes in the zebrafish D. rerio and the stickleback G. aculeatus (Cresko et al. 2003) show the mutual retention of Sox9a and Sox9b, albeit with subtle differences in their patterns of expression. Here, we show that the zebrafish has retained genes encoding teneurin-2A and teneurin-2B, whereas the stickleback has retained genes encoding teneurin-3A and teneurin-3B. Current models of selective gene retention in teleosts predict that genes are preserved following degeneration of regulatory elements and the partitioning of function between the duplicated gene products (Force et al. 1999). It is likely that a large, multifunctional protein like a teneurin would be selected in this way, and differential retention and expression could contribute to speciation.
Previously we speculated that the RHS proteins of bacteria, which share significant sequence homologies with the C-terminal portion of teneurins, may have evolved from horizontal gene transfer from a metazoan teneurin to a symbiotic or pathogenic prokaryote (Minet and Chiquet-Ehrismann 2000). However, the presence of a teneurin gene in M. brevicollis, but not in the other nonmetazoan opisthokonts (e.g., fungi), suggests that the gene evolved in a choanoflagellate, and the horizontal gene transfer was from a prokaryote to a eukaryote instead of the other way around. Horizontal gene transfer between predatory M. brevicollis and their prokaryotic prey has been described previously. For example, Foerstner et al. (2008) reported that a nitrile hydratase is encoded in the M. brevicollis genome that is most closely related to enzymes from proteobacteria; the absence of this enzyme from other eukaryotic genomes strongly implies horizontal gene transfer from prokaryotic prey to eukaryotic predator. Over a hundred genes originating from haptophytes and diatoms have also been found in M. brevicollis (Nedelcu et al. 2008; Sun et al. 2010), indicating that gene transfer may be a relatively common occurrence in these organisms. In fact, this may explain the origin of the highly conserved cysteine-rich domain, which is nearly identical to part of an enzyme from the diatom P. tricornutum (and is also similar to an enzyme in V. carteri) but is only found in teneurins in metazoa. If this is the case, M. brevicollis teneurin originated as a fusion protein acquired by horizontal gene transfer from both a prokaryote and a diatom or algae.
Choanoflagellates are believed to be the closest living relatives of metazoansn (King and Carroll 2001; Philippe et al. 2004; King et al. 2008). The presence of teneurins (which have been shown to play roles in cell–cell and cell–ECM interactions in a variety of tissues) on the surface of an ancestral choanoflagellate may have facilitated the evolution of metazoan multicellularity and the development of complex tissues. Similar roles have been proposed for cadherins, which appear to have evolved in a choanoflagellate as well (Abedin and King 2008). The two cadherins of M. brevicollis are found in the microvilli that form the feeding collar that surrounds the base of the flagellum, which has led to the hypothesis that this family of proteins, which is indispensable in the formation of meaningful cell–cell contacts in animal tissues, evolved as a means of catching prey. Teneurins may have evolved to do something similar. YD-repeat proteins are found on the surface of aquatic bacteria, and in vitro studies with eukaryotic cells show that teneurin expression leads to increased cell–cell adhesion (Rubin et al. 2002). The acquisition of the carbohydrate-rich YD-repeat proteins from a prokaryote by a choanoflagellate may have improved “fishing” for bacterial prey in the feeding collar. It will be interesting to determine where M. brevicollis teneurin is expressed to test this hypothesis.
The lowest branches of the metazoan tree of life include the ancestors of sponges, ctenophores, and cnidarians. Therefore, it is puzzling that we were able to identify teneurins in a choanoflagellate and in all the available genomes of Bilateria (i.e., deuterostomes and protostomes) but not in modern sponges or cnidarians. It is possible that our search methods were insufficient to find them. More likely they are present in some of these organisms but not in the organisms with complete and well-assembled genomes like N. vectensis. It will be important to scrutinize newly sequenced and assembled sponge and cnidarian genomes for teneurin genes as they become available. Another possibility is that teneurins evolved in a relatively advanced common ancestor of protostomes and deuterostomes, after the evolution of sponges and cnidarians. In this scenario, the teneurin in M. brevicollis would have been acquired by horizontal gene transfer from metazoan-derived detritus and not a prokaryote. Evidence against this hypothesis includes the relative similarity of the core region of the cysteine-rich domain and a diatom enzyme and the YD repeats to YD-repeat proteins from bacteria, as well as the organization of the M. brevicollis teneurin gene: most of the ECD is encoded on a single huge exon, not unlike the YD-repeat proteins of prokaryotes, and unlike the ECD of metazoan teneurins. Others (King et al. 2008) have reported that the number of introns per gene in M. brevicollis is similar (6.6) to the number found in human genes (7.7), so the unusually large exon encoding the ECD of M. brevicollis teneurin argues for origins from a prokaryotic and not metazoan, horizontal gene transfer, and the subsequent loss of teneurins from the genomes of modern sponges and cnidarians.
Teneurins are phylogenetically conserved among Bilateria, where they have been demonstrated to play critical roles in pattern formation, the organization of the ECM, and the development of the nervous system. Genomic analysis reveals an ancient origin of teneurins in single-celled choanoflagellates that may have assembled teneurins via horizontal gene transfer from two of its prey: diatoms and prokaryotes. Thus, the talent for gene acquisition by an ancestral choanoflagellate, perhaps to diversify its metabolic pathways and improve its ability to capture prey, may have contributed to the development of multicellularity in metazoans.
Supplementary Material
Supplementary tables S1–S4 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We would like to thank John Hess for his comments on the manuscript and for assistance with the cloning and sequencing of avian teneurin-3.
References
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Bagutti C, Forro G, Ferralli J, Rubin B, Chiquet-Ehrismann R. The intracellular domain of teneurin-2 has a nuclear function and represses zic-1-mediated transcription. J Cell Sci. 2003;116:2957–2966. doi: 10.1242/jcs.00603. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Chiquet-Ehrismann R. Tena, a Drosophila gene related to tenascin, shows selective transcript localization. Mech Dev. 1993;40:165–176. doi: 10.1016/0925-4773(93)90074-8. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Martin D, Hagios C, Chiquet-Ehrismann R. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 1994;13:3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Vitobello A, Ferralli J, Kenzelmann Broz D, Rijli FM, Chiquet-Ehrismann R. Human teneurin-1 is a direct target of the homeobox transcription factor EMX2 at a novel alternate promoter. BMC Dev Biol. 2011;11:35. doi: 10.1186/1471-213X-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, Yan YL, Baltrus DA, Amores A, Singer A, Rodríguez-Marí A, Postlethwait JH. Genome duplication, subfunction partitioning, and lineage divergence: sox9 in stickleback and zebrafish. Dev Dyn. 2003;228(3):480–489. doi: 10.1002/dvdy.10424. [DOI] [PubMed] [Google Scholar]
- Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol. 2005;282:27–38. doi: 10.1016/j.ydbio.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Foerstner KU, Doerks T, Muller J, Raes J, Bork P. A nitrile hydratase in the eukaryote Monosiga brevicollis. PLoS One. 2008;3:e3976. doi: 10.1371/journal.pone.0003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J Biol Chem. 2008;283:13897–13904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- Jozefowicz C, McClintock J, Prince V. The fates of zebrafish Hox gene duplicates. J Struct Funct Genomics. 2003;3:185–194. [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Kenzelmann D, Chiquet-Ehrismann R, Leachman NT, Tucker RP. Teneurin-1 is expressed in interconnected regions of the developing brain and is processed in vivo. BMC Dev Biol. 2008;8:30. doi: 10.1186/1471-213X-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, et al. (36 co-authors) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz K, Szymkiewicz I, Haglund K, Kowanetz M, Husnjak K, Taylor JD, Soubeyran P, Engstrom U, Ladbury JE, Dikic I. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J Biol Chem. 2003;278:39735–39746. doi: 10.1074/jbc.M304541200. [DOI] [PubMed] [Google Scholar]
- Kurosawa G, Takamatsu N, Takahashi M, et al. (15 co-authors) Organization and structure of hox gene loci in medaka genome and comparison with those of pufferfish and zebrafish genomes. Gene. 2006;370:75–82. doi: 10.1016/j.gene.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Leamey CA, Merlin S, Lattouf P, Sawatari A, Zhou X, Demel N, Glendining KA, Oohashi T, Sur M, Fässler R. Ten_m3 regulates eye-specific patterning in the mammalian visual pathway and is required for binocular vision. PLoS Biol. 2007;5:e241. doi: 10.1371/journal.pbio.0050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R. Odd Oz: a novel Drosophila pair rule gene. Cell. 1994;77:587–598. doi: 10.1016/0092-8674(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Li H, Bishop KM, O’Leary DD. Potential target genes of EMX2 include Odz/Ten-M and other gene families with implications for cortical patterning. Mol Cell Neurosci. 2006;33:136–149. doi: 10.1016/j.mcn.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Lovejoy DA, Rotzinger S, Barsyte-Lovejoy D. Evolution of complementary peptide systems: teneurin C-terminal-associated peptides and corticotropin-releasing factor superfamilies. Ann N Y Acad Sci. 2009;1163:215–220. doi: 10.1111/j.1749-6632.2008.03629.x. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Minet AD, Rubin BP, Tucker RP, Baumgartner S, Chiquet-Ehrismann R. Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J Cell Sci. 1999;112:2019–2032. doi: 10.1242/jcs.112.12.2019. [DOI] [PubMed] [Google Scholar]
- Minet AD, Chiquet-Ehrismann R. Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene. 2000;257:87–97. doi: 10.1016/s0378-1119(00)00388-7. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Miles IH, Fagir AM, Karol K. Adaptive eukaryote-to-eukaryote lateral gene transfer: stress-related genes of algal origin in the closest unicellular relatives of animals. J Evol Biol. 2008;21:1852–1860. doi: 10.1111/j.1420-9101.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- Nunes SM, Ferralli J, Choi K, Brown-Luedi M, Minet AD, Chiquet-Ehrismann R. The intracellular domain of teneurin-1 interacts with MBD1 and CAP/ponsin resulting in subcellular codistribution and translocation to the nuclear matrix. Exp Cell Res. 2005;305:122–132. doi: 10.1016/j.yexcr.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Oohashi T, Zhou XH, Feng K, Richter B, Morgelin M, Perez MT, Su WD, Chiquet-Ehrismann R, Rauch U, Fassler R. Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J Cell Biol. 1999;145:563–577. doi: 10.1083/jcb.145.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Rakovitsky N, Buganim Y, Swissa T, Kinel-Tahan Y, Brenner S, Cohen MA, Levine A, Wides R. Drosophila Ten-a is a maternal pair-rule and patterning gene. Mech Dev. 2007;124:911–924. doi: 10.1016/j.mod.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Rotzinger S, Lovejoy DA, Tan LA. Behavioral effects of neuropeptides in rodent models of depression and anxiety. Peptides. 2010;31:736–756. doi: 10.1016/j.peptides.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Tucker RP, Martin D, Chiquet-Ehrismann R. Teneurins: a novel family of neuronal cell surface proteins in vertebrates, homologous to the Drosophila pair-rule gene product Ten-m. Dev Biol. 1999;216:195–209. doi: 10.1006/dbio.1999.9503. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Tucker RP, Brown-Luedi M, Martin D, Chiquet-Ehrismann R. Teneurin 2 is expressed by the neurons of the thalamofugal visual system in situ and promotes homophilic cell-cell adhesion in vitro. Development. 2002;129:4697–4705. doi: 10.1242/dev.129.20.4697. [DOI] [PubMed] [Google Scholar]
- Sun G, Yang Z, Ishwar A, Huang J. Algal genes in the closest relatives of animals. Mol Biol Evol. 2010;27:2879–2889. doi: 10.1093/molbev/msq175. [DOI] [PubMed] [Google Scholar]
- Trubiani G, Al Chawaf A, Belsham DD, Barsyte-Lovejoy D, Lovejoy DA. Teneurin carboxy (C)-terminal associated peptide-1 inhibits alkalosis-associated necrotic neuronal death by stimulating superoxide dismutase and catalase activity in immortalized mouse hypothalamic cells. Brain Res. 2007;1176:27–36. doi: 10.1016/j.brainres.2007.07.087. [DOI] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 2008;19:3898–3908. doi: 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol. 2006;290:237–245. doi: 10.1016/j.ydbio.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R, Chevron MP, Martin D, Hall RJ, Rubin BP. Teneurin-2 is expressed in tissues that regulate limb and somite pattern formation and is induced in vitro and in situ by FGF8. Dev Dyn. 2001;220:27–39. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1084>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Youderian P, Hartzell PL. Triple mutants uncover three new genes required for social motility in Myxococcus xanthus. Genetics. 2007;177:557–566. doi: 10.1534/genetics.107.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TR, Leamey CA. Teneurins: important regulators of neural circuitry. Int J Biochem Cell Biol. 2009;41:990–993. doi: 10.1016/j.biocel.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Zou SM, Jiang XY, He ZZ, Yuan J, Yuan XN, Li SF. Hox gene clusters in blunt snout bream, Megalobrama amblycephala and comparison with those of zebrafish, fugu and medaka genomes. Gene. 2007;400:60–70. doi: 10.1016/j.gene.2007.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.