Abstract
Background
Genetic variation may contribute to differential gene expression in the brain of individuals with psychiatric disorders. To test this hypothesis, we identified genes that were differentially expressed in individuals with bipolar disorder, along with nearby single nucleotide polymorphisms (SNPs) that were associated with expression of the same genes. We then tested these SNPs for association with bipolar disorder in large case-control samples.
Methods
We used the Stanley Genomics Database to extract gene expression and SNP microarray data from individuals with bipolar disorder (n = 40) and unaffected controls (n = 43). We identified 367 genes that were differentially expressed in the prefrontal cortex of cases vs. controls (fold change > 1.3 and FDR q-value < .05) and 45 nearby SNPs that were associated with expression of those same genes (FDR q-value < .05). We tested these SNPs for association with bipolar disorder in a meta-analysis of genome-wide association studies (GWAS) including 4,936 cases and 6,654 healthy controls.
Results
We identified 45 SNPs that were associated with expression of differentially expressed genes, including HBS1L (15 SNPs), HLA-DPB1 (15 SNPs), AMFR (8 SNPs), PCLO (2 SNPs) and WDR41 (2 SNPs). Of these, one SNP (rs13438494), in an intron of the piccolo (PCLO) gene, was significantly associated with bipolar disorder (FDR adjusted p < .05) in the meta-analysis of GWAS.
Conclusions
These results support the previous findings implicating PCLO in mood disorders and demonstrate the utility of combining gene expression and genetic variation data to improve our understanding of the genetic contribution to bipolar disorder.
Keywords: Allelic expression, expression quantitative trait loci (eQTL), genetic variants, functional genomics, risk factors
Bipolar disorder (BD) is a severe mental illness that affects approximately 1% of the population, with increasing morbidity and mortality. BD has a high heritability of at least 60% (1,2) which indicates a strong genetic contribution to this disorder. However, identification of specific genetic markers associated with BD has so far not been an easy task. Previous candidate gene studies implicated SLC6A4, TPH2, DRD4, SLC6A3, DAOA, DTNBP1, NRG1, DISC1, and BDNF (3) in BD, whereas genome-wide association studies (GWAS) with large case-control samples identified novel susceptibility loci (4 – 6) and genes such as CACNA1C (7), ANK3 (8) DFNBP31 (9), SORCS2 (10), and CDH7 (11) in BD. GWAS with large and phenotypically well-characterized samples may enhance our understanding of the genetic contribution to BD (12–14).
Genetic variation contributing to differential gene expression has provided insight into the genetic susceptibility of complex diseases (15). Studies have demonstrated the advantages of systematic mapping of single nucleotide polymorphisms (SNPs) that are associated with variations in gene expression in different tissue types and populations (16,17). These studies have analyzed gene expression values as expression quantitative trait loci (eQTL), and the eQTL were mapped to particular genomic loci by combining variations in their gene expression with genome-wide SNPs (15,18 –21). Emilsson et al. (22) found a marked association between gene expression and genetic variation in blood and adipose tissue samples. Using lymphoblastoid cell lines derived from individuals of European and African ancestry, others also reported that many local and distant SNPs are associated with the genes differentially expressed between these populations (23,24). These studies demonstrate the utility of combining genomic and transcriptomic data to identify potential genetic variants that contribute to differential gene expression in various phenotypes.
Although most eQTL studies have used peripheral tissue and blood cells (22,25), a few studies performed an eQTL analysis with postmortem brain tissue (26,27). Myers et al. (26) reported that, among the transcripts expressed in cortex (58%), 21% had expression profiles that are associated with SNP genotypes in normal human cortex. Here, we used a relatively narrow window size (100 kb up- and downstream of each gene) to map local SNPs adjacent to each gene, similar to the recent studies (15,28,29). The aim of the present study was to identify association between the genes differentially expressed in the prefrontal cortex (PFC) of individuals with BD and the local SNPs, and to test association between the local SNPs and BD using the results derived from a large scale meta-analysis of GWAS.
Materials and Methods
Postmortem Brains
Postmortem brain tissue from the two cohorts including the Neuropathology Consortium (n = 60) and the Array Collection (n = 105) of the Stanley Medical Research Institute were used in the study. The details of the sample collection have been described previously (30). Only BD subjects and unaffected controls from these cohorts were included in the current study. A summary of subject characteristics is shown in Table 1. The brain collection protocol was reviewed by the Uniformed Services University of the Health Sciences. Details on postmortem brain collection are available from the Stanley Medical Research Institute website (http://www.stanleyresearch.org).
Table 1.
A Summary of Subject Characteristics
| Unaffected Control | Bipolar Disorder | |
|---|---|---|
| Subjects, n | 43 | 40 |
| Age | 43.7 ± 1.2 | 43.7 ± 1.8 |
| Gender (male) | 70% | 50% |
| Brain pH | 6.5 ± .1 | 6.4 ± .1 |
| BMI | 30.1 ± .5 | 28.2 ± .5 |
| Heavy Drug Use | 0% | 28% |
| Heavy Alcohol Use | 5% | 35% |
| Psychosis | 0% | 58% |
| Medication at TOD | 0% | 74% |
| Suicide | 0% | 50% |
For each variable, mean ± SEM or percentage value is reported. BMI, body mass index; TOD, time of death.
RNA/DNA Preparation and Microarray Experiment
Total RNA was extracted from gray matter of the middle frontal gyrus (Brodmann area 46) with the Trizol method (Invitrogen, Carlsbad, California) and purified through a Qiagen RNA miniKit column (Qiagen, Valencia, California). Purified RNA was carried through the protocol of the manufacturer (http://www.affymetrix.com), and each sample was hybridized to the Affymetrix U133A GeneChip platform (22,283 transcripts) to determine genome-wide expression profiles. For DNA extraction, a Norgen DNA purification kit (Norgen Biotek, Thorold, Canada) was used to extract high molecular weight genomic DNA from the frozen cerebellum tissue as described previously (27). Only high-quality DNA samples were used for genotyping study using the Affymetrix Genome-Wide Human SNP array 5.0 (500,568 SNPs). All microarray datasets are publicly available from the Stanley Online Genomics database (http://www.stanleygenomics.org). The gene expression microarray data were generated by Dr. Sabine Bahn at the University of Cambridge, Cambridge, UK (https://www.stanleygenomics.org/stanley/standard/studyDetail.jsp?study_id=3) and Dr. Anthony Altar at the Psychiatric Genomics, Gaithersburg, Maryland (https://www.stanleygenomics.org/stanley/standard/studyDetail.jsp?study_id=2). The SNP microarray data were generated by Dr. Chunyu Liu at the University of Chicago, Chicago, Illinois (https://www.stanleygenomics.org/stanley/standard/studyDetail.jsp?study_id=20).
Quality Control of Microarrays
Raw microarray data were processed and analyzed with the R statistical language (http://www.r-project.org) and the Bioconductor packages (31). The Affymetrix microarray Suite (MAS 5.0) was used for image processing, data acquisition, and normalization of expression values (log base 2) for each probeset (32). Normalized data were subjected to rigorous quality control procedures as described previously (33). Two analyses including principal components analysis and correlation coefficient heat map with hierarchical clustering of samples are shown in Figure S1 in Supplement 1. For gene filtering, probesets with absent/present call rate < 33% across all subjects were filtered out with the R package affy (http://www.bioconductor.org/packages/release/bioc/html/affy.html). The Affymetrix HGU133A array contains 22,283 probesets, of which 11,109 (approximately 50%) were retained after filtering.
For the SNP microarrays, raw data were normalized with the Bayesian Robust Linear Modeling with Mahalanobis Distance Genotyping Algorithm for genotype calling of 500,568 SNPs. SNPs were filtered based on the criteria of genotype missing rate > 10%, minor allele frequency < 5%, and Hardy–Weinberg Equilibrium p > .01 (34). After the frequency and genotype filtering, 335,074 SNPs were retained. For the subject filtering, we removed 1 subject based on missing genotype rate > 30%. We also computed multidimensional scaling with 335,074 SNPs and removed four subjects (non-Caucasian samples) that have aberrant genotype calls as compared to the rest of the subjects (Figure S2 in Supplement 1). For the gene and SNP association analysis, only subjects common to the gene expression and SNP microarray data were used (40 BD cases and 43 unaffected controls).
Gene Expression Analysis
Pre- and postmortem variables affecting the expression of a significant number of transcripts were identified as described previously (35,36). These variables include age, sex, body mass index, smoking at time of death (TOD), heavy alcohol use, heavy drug use, disease severity, suicide status, psychosis, lifetime antidepressant medication and medication at TOD, postmortem interval, brain pH, and refrigerator interval. We also analyzed the effects of microarray scan date because a recent study reported the confounding effects of this variable (e.g., batch effects) on gene expression (27). We identified four potential confounding variables, including brain pH, microarray scan date, psychotic features, and medication at TOD, that affect expression of more than 5% of the transcripts (p < .001), as shown in Figure S3 in Supplement 1. We then performed BD analysis using a multiple regression model to obtain an adjusted fold change (FC), standard error, and p value for each gene, including the four confounding variables as covariates. Based on the raw p values obtained from this analysis, the false discovery rate (FDR)-adjusted q-values (FDR 5%) were calculated using the q-value package (http://bioconductor.org/packages/release/bioc/html/qvalue.html). The FDR approach has been well-documented in the literature as a means to control type I error while maximizing power (37–39).
Gene Expression and SNP Association
For mapping local SNPs adjacent to the genes differentially expressed in the PFC of BD, we used a window size of 100 kb up- and downstream of each gene (cis SNPs). This cis SNP mapping retained 10,838 gene and SNP pairs. Probesets in the expression microarray (HGU-133A) that had any SNP within their sequence were identified using the UCSC Genome Browser (http://genome.ucsc.edu/) and AffyMAPSDetector software (40). These probesets were removed from the dataset, since those SNPs within the sequence may interfere with hybridization of the probesets (41). We implemented a genotypic model of the form AA, Aa, aa and regressed gene expression on the genotype calls, with the appropriate covariates. In this multiple regression analyses, we included sex and age as covariates, consistent with a previous study (28). To adjust for multiple testing of SNPs and genes, we implemented two steps of p value correction. First, we used a permutation test (10,000 iterations) to adjust the p values calculated from the association between gene expression and cis SNPs, similar to the method implemented in the eQTL study (22). Briefly, for each iteration, the disease classifications were randomly shuffled in the gene vector to remove class memberships, SNP genotypes predicting the gene expression were calculated, and the p value for the SNP coefficient was retained. Following the permutation tests for all gene and SNP pairs, permuted p values from all cis regions tested were further adjusted with the q-value method to control for multiple testing of the genes using a 5% FDR threshold.
Bipolar Disorder and SNP Association
To test disease association with the cis SNPs, we used the results derived from the BD subset of a meta-analysis of GWAS results (4), including 4,936 BD cases and 6,654 healthy controls as previously described (7,10,42,43). Genotype data were obtained from the db-GaP, the Wellcome Trust Case Control Consortium, the STEP-BD (7), and through collaborators at the National Institute of Mental Health (10). The data from all samples except the STEP-BD were used to impute genotypes for 2 million HapMap Phase two markers (http://hapmap.ncbi.nlm.nih.gov/) with the program Markov Chain Haplo-typing, version 1.0 (44). Allele-wise association results were meta-analyzed with a method that weights results by sample size (http://www.sph.umich.edu/csg/abecasis/Metal). The p values were corrected for population stratification by Genomic Control (45). In the disease association test, multiple testing of the cis SNPs was adjusted using a Bonferroni method adjusted for linkage disequilibrium (LD) among the SNPs. Each SNP that showed minimal LD with other SNPs in the set (at an r2 value > .3) was counted in the adjustment (9 independent SNPs).
Quantitative Polymerase Chain Reaction
Total RNA was re-extracted from the PFC of the same subjects that were used in the microarray experiments, and the quality of RNA was assessed with the Bioanalyzer 2100 (Agilent, Foster City, California). RNA was purified using the PureLink Total RNA Purification System (Invitrogen, Carlsbad, California), and complementary DNA was synthesized using reverse-transcriptase polymerase chain reaction (PCR) with oligo dT primers as previously described (46). Predesigned and validated QuantiTect SYBR primers (Qiagen) were used for the quantitative polymerase chain reaction (qPCR): HBS1L (QT00052626, NM_006620), HLA-DPB1 (QT00079338, NM_002121), PCLO (QT01843730, NM_014510), and POLR1D (QT01339856, NM_152705). Three reference genes, B2M (QT00088935, NM_004048), PP1A (QT00046046, NM_021130), and ACTB (QT00095431, NM_001101), were selected for the experiment. Using the Prism7900HT real-time detector (Applied Biosystems, Foster City, California), 1 μL aliquots of QuantiTect primer, 10 μL qPCR Master mix (Applied Biosystems), and 10 μL complementary DNA were mixed together and pipetted into each well of the 384-well qPCR plate. Thermocycle conditions were: 1) 1 cycle for 2 min at 50°C, 2) 1 cycle for 15 min at 95°C, and 3) 40 cycles for 15 sec at 95°C and 1 min at 60°C. Fluorescence was measured during the 60°C step for each cycle. Reactions were quantified by the cycle threshold method using the SDS2.2 software (Applied Biosystems). An average quantity value (Qty mean) for each sample from the triplicates of that sample was calculated for each gene. The data for each gene were expressed as Qty mean for the gene of interest/geometric mean of Qty mean for the three reference genes. Normalized values were expressed as a FC between BD subjects and unaffected controls.
Results
Demographic and Clinical Information
A summary of subject characteristics is shown in Table 1. Average age (p = .99), body mass index (p = .38), and brain pH (p = .05) were not significantly different between BD and control groups. Other clinical variables such as heavy drug use, psychosis, medication at TOD, and suicide were specific to BD subjects.
Differential Gene Expression in the PFC of BD Subjects
Using the gene expression microarray data from the Stanley Genomics Database, we analyzed various antemortem and postmortem variables. We found that brain pH (19.1%), microarray scan date (10.2%), psychosis (6.2%), and medication at TOD (5.3%) affected expression of more than 5% of the transcripts (p < .001), as shown in Figure S3 in Supplement 1. Other variables affected a relatively small number of transcripts (< 5%). Thus, we adjusted for the four confounding variables in BD analysis, and identified 400 transcripts (367 genes) as being differentially expressed in the PFC of BD subjects (FC > 1.3 and q-value < .05). Among those transcripts, 175 transcripts were upregulated and 225 transcripts were downregulated in BD (Table S1 in Supplement 1).
Gene Expression and SNP Association
Using those 400 transcripts, we mapped 10,838 cis SNPs (window size of 100 kb upstream and downstream of each gene) that are adjacent to each gene. After adjusting for multiple testing of SNPs and genes, we identified 45 gene and SNP pairs showing association between gene expression and SNP genotypes (FDR q-value < .05), as shown in Table 2.
Table 2.
A Summary of Significant Association Between Gene Expression and cis SNPs
| Probeset | Gene Symbol | Gene Title | Cytoband | FC | q-Value | SNPs |
|---|---|---|---|---|---|---|
| 202203-s-at | AMFR | autocrine motility factor receptor | 16q21 | −.866 | .016 | 8 |
| 209316-s-at | HBS1L | HBS1-like (S. cerevisiae) | 6q23-q24 | −.766 | .015 | 15 |
| 211990-at | HLA-DPA1 | MHC, class II, DPα1 | 6p21.3 | −.892 | .014 | 1 |
| 201137-s-at | HLA-DPB1 | MHC, class II, DPβ1 | 6p21.3 | −.423 | .039 | 15 |
| 210650-s-at | PCLO | piccolo (presynaptic cytomatrix protein) | 7q11.23-q21.3 | .774 | .013 | 2 |
| 218258-at | POLR1 | Polymerase (RNA) I polypeptide D, 16 kDa | 13q12.2 | −.397 | .017 | 2 |
| 218055-s-at | WDR41 | WD repeat domain 41 | 5q14.1 | −.419 | .013 | 2 |
FC, gene expression fold change between bipolar disorder and unaffected controls (log2); q-value, FDR-adjusted q-value (5% FDR); AMFR, autocrine motility factor receptor; MHC, major histocompatibility complex.
Expression levels of the major histocompatibility complex, class II, DP β 1 (HLA-DPB1) gene were associated with 15 cis SNPs, and 9 SNPs were in LD (Figure S4 in Supplement 1). Expression levels of the autocrine motility factor receptor gene were also associated with eight cis SNPs. Among those SNPs, four SNPs (rs2440467, rs2432540, rs6499837, and rs9937444) had the same association with autocrine motility factor receptor expression in the CEU adult population (HapMap Project). Details on the 45 gene and SNP pairs are shown in Table S2 in Supplement 1.
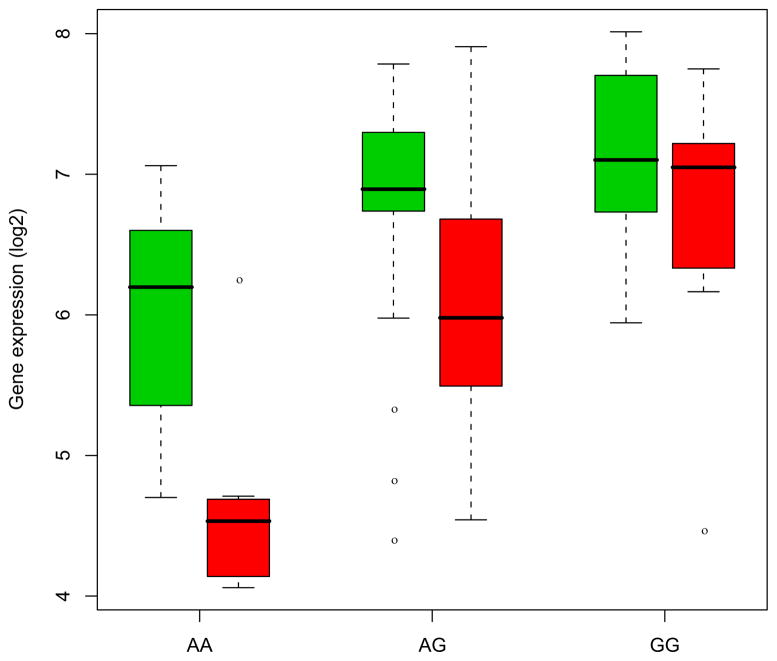
Figure 1 shows a significant association between HBS1L expression and an SNP (rs2150681) in the brain (FDR q-value < .05). Expression levels of HBS1L were decreased (FC −1.7, and q-value .02) in BD (red) as compared to the controls (green). Levels of the gene expression in the minor allele (A) were decreased in BD subjects as compared to the controls, indicating an allelic expression pattern. Other cis SNPs (rs1547247, rs4896128, rs6923765, rs7741515, rs1590975) that are in LD with the SNP rs2150681 also showed a similar pattern of association with HBS1L expression.
Figure 1.
A significant association between expression levels of HBS1L and a single nucleotide polymorphism, rs2150681, in brain (false discovery rate q-value < .05). Overall expression levels of HBS1Lare decreased in the prefrontal cortex of bipolar disorder subjects (red) as compared to the unaffected controls (green). Levels of expression in the minor allele (A) were decreased in bipolar disorder subjects (red) as compared to the controls (green), indicating an allelic expression of HBS1L. X axis: genotype; Y axis: gene expression (log 2 scale).
BD and SNP Association
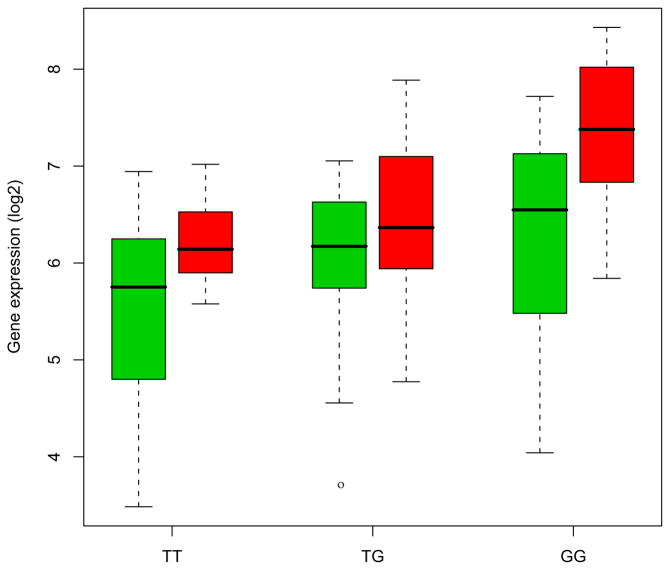
Using the 45 cis SNPs that are associated with differentially expressed in BD, we tested disease association using the results derived from a meta-analysis of GWAS. The LD (r2) was estimated from the HapMap Phase 2 genotypes and association p values were corrected by the number of independent SNPs using a Bonferroni method. We identified an SNP (rs13438494) that is significantly associated with BD (adjusted p < .05). Figure 2 shows that expression levels of PCLO are increased in the PFC of BD subjects (red) as compared to the controls (FC 1.71; q-value < .05). This also illustrates that the SNP rs13438494 is associated with expression levels of PCLO in the PFC of BD subjects (q-value < .05).
Figure 2.
A significant association between expression levels of PCLO and a single nucleotide polymorphism, rs13438494, in brain (false discovery rate q-value < .05). Overall expression levels of PCLO are increased in the prefrontal cortex of bipolar disorder subjects (red) as compared to the unaffected controls (green). X axis: genotype; Y axis: gene expression (log 2 scale).
Quantitative PCR
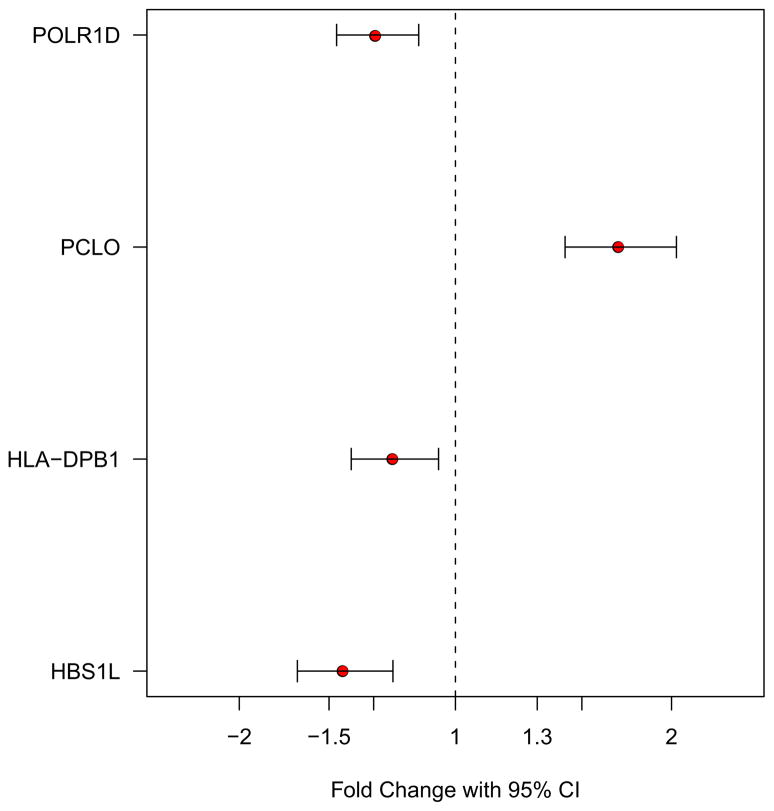
Following the microarray analysis, we performed a qPCR to validate gene expression changes that we observed in BD subjects. We confirmed that 4 genes (HBS1L, HLA-DPB1, PCLO and POLR1D) show the same directional changes in the PFC of BD subjects as compared to the controls. Figure 3 illustrates FCs and 95% confidence intervals of individual genes (p < .05). This suggests that the microarray and the qPCR data are consistent and the qPCR results further support the effectiveness of our significance criteria (FC > 1.3 and q-value < .05) in minimizing false positive discovery in the microarray data.
Figure 3.
A quantitative polymerase chain reaction validation of the genes (HBS1L, HLA-DPB1, PCLO, and POLR1D) that are differentially expressed in the prefrontal cortex of bipolar disorder subjects as compared to the unaffected controls. Each gene is shown with fold change and 95% confidence interval (CI) (p < .05).
Discussion
Previous eQTL studies suggested substantial heritable variation in gene expression in different cell types and populations. Genetic variants that influence gene expression levels can be mapped to local (cis) or distant (trans) SNPs (19). However, it is more difficult to interpret the biological mechanisms behind the trans SNPs as compared to the cis SNPs (18,20), due to a large number of comparisons presented between the gene and the trans SNPs. The cis SNPs are more proximal to their neighboring gene, and other studies demonstrated increased strength in association in the cis SNPs as compared to the trans SNPs (22,26). Another study, using postmortem brain tissue from psychiatric disorder subjects, also demonstrated a large number of cis associations with gene expression, while no trans associations were found to be significant after correcting for multiple comparisons (27). Thus, we used a relatively narrow window size (100 kb upstream and downstream of each gene) for mapping cis SNPs, similar to the studies that used the same window size for cis SNPs (15,28,29). Using the stringent criteria of significance, we identified 45 gene and SNP pairs that are associated with each other. Among those gene and SNP pairs, the same associations have been reported in 11 gene and SNP pairs in the CEU adult population (HapMap Project) and in the human frontal cortex (26,27). The remaining 34 gene and SNP pairs have not been reported elsewhere, and it is likely that novel associations may exist in the PFC of BD subjects.
We found that expression levels of HBS1L were associated with 15 cis SNPs, and those SNPs were in LD (Figure S4 in Supplement 1). Previous studies showed similar association between HBS1L expression and the SNPs. For example, five SNPs (rs1590975, rs2150681, rs4896128, rs6923765, and rs7741515) were associated with HBS1L expression in the human cortex (26). An SNP (rs1547247) was associated with HBS1L expression in the CEU adult population (HapMap Project). The HBS1L is located in chromosome 6q23-q24, a locus that appears to be associated with BD (47,48). By combining the genetic data from 11 different BD linkage studies (5,179 individuals from 1,067 families), the authors found a significant linkage to BD on chromosome 6q21-q25 (48). Another study using a linkage data-set (52 families of European descendent; 448 participants and 259 affected individuals) reported an interaction between BD genes on chromosomes 2q22-q24 and 6q23-q24, suggesting an epistasis in BD (47). Although more studies are needed, previous studies suggest that chromosome 6q23-q24 may be a risk locus associated with BD.
The major histocompatibility complex (MHC) region is located in chromosome 6p21.3-p22.1 and has been implicated in immune system function. A large-scale GWAS of European-ancestry subjects (3,322 cases and 3,587 controls) reported that the MHC region is associated with schizophrenia and BD (5). We found that 15 cis SNPs are associated with decreased expression of HLA-DPB1 (6p21.3) in the PFC of BD subjects. This strengthens the evidence that the MHC region (6p21.3-p22.1) may be associated with BD.
A GWAS reported that multiple SNPs located in PCLO are associated with major depression (49). Among the SNPs associated with major depression, a nonsynonymous SNP rs2522833 in PCLO yielded the lowest p value (6.4 × 10−8). Another study with reanalysis of 29 correlated SNPs supported the finding that the SNP rs2522833 is associated with major depression (50) and provided compelling evidence that PCLO may be a susceptibility gene for major depression. Here, we identified two cis SNPs associated with increased expression of PCLO in the PFC of BD subjects. Interestingly, PCLO encodes a protein that is localized to the presynaptic active zone and plays a significant role in monoaminergic neuro-transmission in the brain (51,52). Previous and current results suggest that PCLO is associated with both major depression and BD, and this gene may play a significant role in the development of major mood disorders (53–56).
Genetic variation that has been reported to contribute to gene expression in previous eQTL studies may not always reflect true changes in messenger RNA (mRNA) levels caused by SNPs. Hybridization differences caused by sequence polymorphisms in the 3′ end of the mRNA region that is targeted by the Affymetrix expression microarray probes may lead to false-positive eQTL findings (41,57). Previous eQTL studies did not always identify those SNPs interfering with gene expression and such putative cis SNPs should be considered with extra caution because differential gene expression could be due to sequence diversity in the microarray probe regions. Thus, we used the UCSC Genome Browser (http://www.genome.ucsc.edu) and the AffyMAPSDetector software (40) to exclude those SNPs located in the 3′ end of the mRNA region that are targeted by the Affymetrix microarray probes.
Recent studies demonstrated the confounding effects of microarray scan date (batch effects) on gene expression profiles (27,58). Liu et al. (27) reported that brain pH and batch effects were the two most influential factors affecting gene expression profiles in postmortem brain tissue. We confirmed that brain pH (19%) and microarray scan date (10%) affected expression of a significant number of transcripts as compared to other variables. In order to adjust for the confounding variables, we included them as covariates in the multiple regression analysis. Thus, we were able to obtain a set of genes as being differentially expressed in the PFC of BD subjects after correcting for the confounding factors including brain pH, microarray scan date, psychosis and medication at TOD.
We obtained a relatively small number of genes (7 of 367) that show a significant association with cis SNPs as compared to the previous studies that reported a larger portion of gene expression and SNP associated pairs (22,23). There are several possibilities for this discrepancy. First, we mapped local SNPs with a narrow window size (100 kb upstream and downstream) so that the total number of local SNPs mapped to the genes was smaller than previous studies that used a larger window size. Second, we used a permutation test (10,000 iterations) followed by FDR-adjusted q-value method to ensure that we have a strict threshold to obtain association between gene expression and SNP genotypes. Third, we used postmortem brain tissue from individuals with psychiatric disorder with a relatively small sample size (n = 83) for eQTL analysis. It has been shown that gene expression changes are subtle in the PFC of individuals with psychiatric disorders (35) as compared to other studies that investigated expression profiles in different populations (25). Finally, there are many unknown factors such as disease comorbidity, complex substance abuse and medication history that may affect gene expression profiles in postmortem brain tissue. Thus, it is possible that those factors may confound some of the significant associations and may result in a relatively small number of genes associated with cis SNPs in the current study.
In conclusion, we identified a set of SNPs that show cis association with expression of the genes that are themselves differentially expressed in the PFC of individuals with BD. We showed that these cis associations were not attributable to common biases present in postmortem gene expression data, and that they met stringent thresholds of significance and multiple testing corrections. One of the identified SNPs, rs13438494 in an intron of PCLO, showed association with BD in a large meta-analysis of GWAS, consistent with prior evidence that PCLO is involved in mood disorders. We have demonstrated the utility of combining genomic and transcriptomic data to identify potential genetic markers that are associated with both gene expression and disease status in a complex genetic disorder.
Supplementary Material
Acknowledgments
We thank the Stanley Medical Research Institute for supporting this work, in particular Dr. E Fuller Torrey and Theodore and Vada Stanley. We thank Drs. Anthony Altar, Sabine Bahn, and Chunyu Liu for contributing the microarray data for the current study. We thank many researchers and collaborators for making GWAS data/results available for the meta-analysis. Complete acknowledgments for the data used in the meta-analysis are given in the primary publication (4).
Footnotes
All authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online.
References
- 1.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 2.Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36:585–594. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serretti A, Mandelli L. The genetics of bipolar disorder: Genome “hot regions,” genes, new potential candidates and future directions. Mol Psychiatry. 2008;13:742–771. doi: 10.1038/mp.2008.29. [DOI] [PubMed] [Google Scholar]
- 4.McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WTCCC . Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soronen P, Ollila HM, Antila M, Silander K, Palo OM, Kieseppa T, et al. Replication of GWAS of bipolar disorder: Association of SNPs near CDH7 with bipolar disorder and visual processing. Mol Psychiatry. 2010;15:4–6. doi: 10.1038/mp.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craddock N, Sklar P. Genetics of bipolar disorder: Successful start to a long journey. Trends Genet. 2009;25:99–105. doi: 10.1016/j.tig.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, et al. The bipolar disorder phenome database: A resource for genetic studies. Am J Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- 15.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heap GA, Trynka G, Jansen RC, Bruinenberg M, Swertz MA, Dinesen LC, et al. Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genomics. 2009;2:1. doi: 10.1186/1755-8794-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastinen T, Ge B, Hudson TJ. Influence of human genome polymorphism on gene expression. Hum Mol Genet. 2006;15:R9–16. doi: 10.1093/hmg/ddl044. [DOI] [PubMed] [Google Scholar]
- 19.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: The promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sladek R, Hudson TJ. Elucidating cis- and trans-regulatory variation using genetical genomics. Trends Genet. 2006;22:245–250. doi: 10.1016/j.tig.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 22.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Duan S, Bleibel WK, Wisel SA, Huang RS, Wu X, et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum Genet. 2009;125:81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Cheng L, Badner JA, Zhang D, Craig DW, Redman M, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 29.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The Stanley Foundation Brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim WK, Wang K, Lefebvre C, Califano A. Comparative analysis of microarray normalization procedures: Effects on reverse engineering gene networks. Bioinformatics. 2007;23:i282–i288. doi: 10.1093/bioinformatics/btm201. [DOI] [PubMed] [Google Scholar]
- 33.Choi KH, Zepp ME, Higgs BW, Weickert CS, Webster MJ. Expression profiles of schizophrenia susceptibility genes during human pre-frontal cortical development. J Psychiatry Neurosci. 2009;34:450–458. [PMC free article] [PubMed] [Google Scholar]
- 34.Sindhi R, Higgs BW, Weeks DE, Ashokkumar C, Jaffe R, Kim C, et al. Genetic variants in major histocompatibility complex-linked genes associate with pediatric liver transplant rejection. Gastroenterology. 2008;135:830–839. 839, e831–e810. doi: 10.1053/j.gastro.2008.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi KH, Elashoff M, Higgs BW, Song J, Kim S, Sabunciyan S, et al. Putative psychosis genes in the prefrontal cortex: Combined analysis of gene expression microarrays. BMC Psychiatry. 2008;8:87. doi: 10.1186/1471-244X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ, et al. Meta-analysis of 12 genomic studies in bipolar disorder. J Mol Neurosci. 2007;31:221–243. doi: 10.1385/jmn:31:03:221. [DOI] [PubMed] [Google Scholar]
- 37.Storey JD. A direct approach to false discovery rates. J R Stat Soc B Stat Methodol. 2002;64:479–498. [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 39.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 40.Kumari S, Verma LK, Weller JW. AffyMAPSDetector: A software tool to characterize Affymetrix GeneChip expression arrays with respect to SNPs. BMC Bioinform. 2007;8:276. doi: 10.1186/1471-2105-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, Jansen RC. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Abecasis G. MACH. 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 45.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 46.Choi KH, Higgs BW, Weis S, Song J, Llenos IC, Dulay JR, et al. Effects of typical and atypical antipsychotic drugs on gene expression profiles in the liver of schizophrenia subjects. BMC Psychiatry. 2009;9:57. doi: 10.1186/1471-244X-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abou Jamra R, Fuerst R, Kaneva R, Orozco Diaz G, Rivas F, Mayoral F, et al. The first genomewide interaction and locus-heterogeneity linkage scan in bipolar affective disorder: Strong evidence of epistatic effects between loci on chromosomes 2q and 6q. Am J Hum Genet. 2007;81:974–986. doi: 10.1086/521690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bochdanovits Z, Verhage M, Smit AB, de Geus EJ, Posthuma D, Boomsma DI, et al. Joint reanalysis of 29 correlated SNPs supports the role of PCLO/Piccolo as a causal risk factor for major depressive disorder. Mol Psychiatry. 2009;14:650–652. doi: 10.1038/mp.2009.37. [DOI] [PubMed] [Google Scholar]
- 51.Cen X, Nitta A, Ibi D, Zhao Y, Niwa M, Taguchi K, et al. Identification of Piccolo as a regulator of behavioral plasticity and dopamine transporter internalization. Mol Psychiatry. 2008;13:451363. doi: 10.1038/sj.mp.4002132. [DOI] [PubMed] [Google Scholar]
- 52.Leal-Ortiz S, Waites CL, Terry-Lorenzo R, Zamorano P, Gundelfinger ED, Garner CC. Piccolo modulation of Synapsin1a dynamics regulates synaptic vesicle exocytosis. J Cell Biol. 2008;181:831–846. doi: 10.1083/jcb.200711167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeng S, Hunsberger JG, Pearson B, Yuan P, Wang Y, Wei Y, et al. BAG1 plays a critical role in regulating recovery from both manic-like and depression-like behavioral impairments. Proc Natl Acad Sci U S A. 2008;105:8766–8771. doi: 10.1073/pnas.0803736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sequeira A, Turecki G. Genome wide gene expression studies in mood disorders. OMICS. 2006;10:444–454. doi: 10.1089/omi.2006.10.444. [DOI] [PubMed] [Google Scholar]
- 55.Dorz S, Borgherini G, Conforti D, Scarso C, Magni G. Depression in inpatients: Bipolar vs unipolar. Psychol Rep. 2003;92:1031–1039. doi: 10.2466/pr0.2003.92.3.1031. [DOI] [PubMed] [Google Scholar]
- 56.Kupfer DJ, Carpenter LL, Frank E. Is bipolar II a unique disorder? Compr Psychiatry. 1988;29:228–236. doi: 10.1016/0010-440x(88)90046-6. [DOI] [PubMed] [Google Scholar]
- 57.Sliwerska E, Meng F, Speed TP, Jones EG, Bunney WE, Akil H, et al. SNPs on chips: The hidden genetic code in expression arrays. Biol Psychiatry. 2007;61:13–16. doi: 10.1016/j.biopsych.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 58.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.