Abstract
P63, a member of the P53 tumor suppressor family, is known to play important functions in cancer and development. Interestingly, previous studies have shown that p63 null mice are absent or have truncated limbs, while mutations in human P63 cause several skeletal syndromes that also show limb and digit abnormalities, suggesting its essential role in long bone development. Indeed, we detected increased level of p63 transcript in hypertrophic MCT cells (an established cell model of chondrocyte maturation) than in proliferative MCT cells. To investigate the in vivo role of P63 upon endochondral bone formation, we have established transgenic mouse lines in which HA- and Flag-tagged TAP63α (the longest P63 isoform) is driven by the hypertrophic chondrocyte-specific Col10a1 regulatory elements. Skeletal staining of Col10a1-TAP63α transgenic mice at either embryonic day 17.5 (E17.5) or postnatal day 1 (P1) observed accelerated ossification in long bone, digit and tail bones compared to their wild-type littermates, suggesting a putative function of P63 during skeletal development. We also detected decreased level of Sox9 and Bcl-2 transcripts, while Alp and Ank are slightly upregulated in Col10a1-TAP63α transgenic mouse limbs. Further immunohistochemical analysis confirmed the decreased Sox9 expression in the proliferative and hypertrophic zone of these mice. Von Kossa staining suggests increased mineralization in hypertrophic zone of transgenic mice compared to littermate controls. Together, our results suggest a role of TAP63α upon skeletal development. TAP63a may promote endochondral ossification through interaction with genes relevant to matrix mineralization and chondrocyte maturation or apoptosis
Keywords: Col10a1, P63, Hypertrophic chondrocytes, Transgenic mice, Endochondral bone formation
1. Introduction
The p53 family of transcription factors consists of three members: p53, p63 and p73 (Mills et al., 2006). These family members differentially regulate target gene transcription and programmed cell death and therefore play pivotal roles during multiple tumor formation and development (Dietz et al., 2002; Wu et al., 2003; Wu et al., 2005). P63 shows high structural similarity to its mouse homologue, the p53 tumor suppressor. Due to its different promoter usage and alternative splicing, P63 is divided into two subtypes (TAP63 and ΔNP63) and each consists of three isoforms (−α, −β or −γ). The TAP63 isoforms act as transcription factors. The ΔNP63 isoforms lack the main transcription activation domain and act as dominant-negative inhibitors of transactivation (TA) isoforms (Petitjean et al., 2007). Although these P63 isoforms are known to play distinct functions in cancer and development, P63 is generally recognized as a critical transcription factor for epithelial development and maintenance of tumorigenesis since its discovery more than a decade ago (Perez CA, Pietenpol. JA., 2007). Interestingly, P63 has also been suggested to play putative roles in skeletal, especially, limb development. It was previously reported that, besides the severe defects of epithelial development, mice lacking P63 are either absent or have truncated limbs (Mills et al., 1999; Yang et al., 1999). Meanwhile, mutations in human P63 have clearly been associated with EEC (ectrodactyly, ectodermal dysplasia, and cleft lip/palate) syndrome, LMS (limb-mammary) syndrome, and isolated SHSF (Split Hand–Split Foot) malformation (van Bokhoven et al., 2007). Despite the craniofacial involvement in these syndromes, the severe limb defects in p63 null mice and the limb and digit abnormalities in P63 associated diseases strongly suggest a putative role of P63 during endochondral bone formation.
Endochondral bone formation or ossification is a major skeletal developmental process that gives rise to long bones including appendicular skeleton, facial bones, vertebrae, and the lateral medial clavicles (Ornitz DM., Marie PJ., 2002). Formation of these bones requires a cartilage intermediate, in which mesenchymal cells condense and form chondrocytes. Chondrocytes then undergo differentiation, proliferation, hypertrophy, and apoptosis, and eventually replaced by bone. This is a well-coordinated process and is regulated by multiple transcription factors and signaling pathways (de Crombrugghe et al., 2001). The obvious skeletal abnormalities in P63 related mouse models and human syndromes suggest that P63 might be a candidate that plays a pivotal role during skeletal development and the progression of skeletal diseases. However, currently, there is not much data that has been reported regarding the effects of P63 upon bone formation. The putative function of P63 isoforms during different skeletal developmental stages, especially, during endochondral bone formation is, therefore, largely unknown.
In this manuscript, we report investigation of the putative role of P63 upon endochondral bone formation. We have detected an increased level of p63 transcript in hypertrophic MCT cells, a cell model known to express hypertrophic chondrocyte-specific type X collagen gene (Col10a1) abundantly upon growth arrest (Lefebvre et al., 1995; Zheng et al., 2003). We have also performed transgenic studies using the cell-specific Col10a1 control elements to selectively target TAP63α expression in hypertrophic chondrocytes. Skeletal phenotypic analysis revealed accelerated ossification in long bone, digit and tail bones of Col10a1-TAP63α transgenic mice at both E17.5 and the P1 stages, suggesting a putative function of TAP63α, the longest P63 isoform, upon late embryonic skeletal development.
2. Materials and Methods
2.1. Cell Culture, total RNA extraction, RT-PCR and qRT-PCR
Previously established mouse chondrocytes (MCT cells) were grown until sub-confluence at 32°C in standard DMEM with 8% FBS (Gibco BRL) and 8% CO2 as per published protocol (Lefebvre et al., 1995; Zheng et al., 2003). These MCT cells were further cultured at either 32°C or 37°C for additional 1–3 days before harvest. Total RNAs from MCT cells were isolated and reversely transcribed using Trizol reagent and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) to synthesize the first strand cDNA. The RT product was subjected to regular or real-time polymerase chain reaction (RT-PCR or qRT-PCR) to examine the mRNA level of relevant genes. These genes include p53 family members: p53, p63 and p73, as well as the type X collagen gene (Col10a1) and the endogenous control glyceraldehyde 3-phosphate dehydrogenase (Gapdh) for normalization. The primer sequence and the amplicon size are listed in Table 1. Regular RT-PCR was performed using standard protocol to amplify the cDNA templates from proliferative (32°C) and hypertrophic (37°C) MCT cells with designated primes. Following 30 cycles of amplification, the PCR products were subjected to ethidium bromide (EB) staining and agarose gel running. For qRT-PCR, same amount of cDNA templates were amplified with the designated primers using the Bio-Rad iQ™ SYBR Green supermix and the MyiQ Real-Time PCR Detection System (Bio-Rad Hercules, CA). Relative gene-transcript levels were analyzed by the manufacturer provided MyiQ Optical System Software. The mean CT (threshold cycle number) values of target genes were normalized to endogenous Gapdh using 2−ΔΔ Ct and student t-test (Zheng et al., 2003, Livak KJ, Schmittgen TD, 2001; Pfaffl MW, 2001). Data is collected from multiple runs of real-time PCR with duplicate templates. P<0.05 indicate significant fold-changes of mRNA level of genes of interest in different population of MCT cells.
Table 1.
Primers designed for real-time PCR
| Name | RefSeqID | Sense Primer (5′-3′) | Antisense Primer (5′-3′) | Amplicon (bp) |
|---|---|---|---|---|
| Gapdh | NM_008084 | ACCCAGAAGACTGTGGATGG | CACATTGGGGGTAGGAACAC | 171 |
| Col10α1 | NM_009925 | AAAGCTTACCCAGCAGTAGG | ACGTACTCAGAGGAGTAGAG | 331 |
| mTrp53 | NM_001127233 | TGGAAGACTCCAGTGGGAAC | CTGTAGCATGGGCATCCTTT | 319 |
| mTrp63 | NM_001127259 | GTCAGCCACCTGGACGTATT | CTCATTGAACTCACGGCTCA | 222 |
| mTrp73 | NM_0011276331 | CAAAGTGTCCACACCACCAC | CATACGGCACAACCACACTC | 233 |
| Bax | NM_0007527 | TGCAGAGGATGATTGCTGAC | GATCAGCTCGGGCACTTTAG | 173 |
| Bcl-2 | NM_009741 | CTGGCATCTTCTCCTTCCAG | GACGGTAGCGACGAGAGAAG | 183 |
| Runx2 | NM_001145920 | ACCCAGC CACCTTTACCTAC | TATGGAGTGCTGCTGGTCTG | 150 |
| Sox9 | NM_011448 | TTCATGAAGATGACCGACGA | ATGCACACGGGGAACTTATC | 200 |
| Alp | NM_007431.2 | AGTGAGCGCAGCCACAGAGC | GTGTGGCGTGGTTCACCCGA | 134 |
| Ank | NM_020332.4 | CCTGGCCACGCAGCGAATCA | GACCCACGGGGTAGGTGGCT | 119 |
| Enpp1 | NM_008813.3 | CTCATGCCCCTCTGGGCGTC | CGGTGGCGTGAGGAACCCAT | 168 |
| Phospho1 | NM_153104.3 | GCTCTGGCTCCCCGGCATTTA | AGCCCCGAGGGACTGTGGATG | 105 |
Gapdh: glyceraldehydes-3-phosphate dehydrogenase; Col10α1: type X collagen, alpha 1; mTrp53/63/73: murine transformation related protein 53/63/73; Bax: BCL-2-associated X protein; Bcl-2: B-cell leukemia (lymphoma) 2; Runx2: runt-related transcription factor 2; Sox9: SRY-box containing gene 9; Alp: Alkaline phosphatase; Ank: progressive ankylosis gene; Enpp1: ectonucleotide pyrophosphatase/phosphodiesterase 1; Phospho1: phosphatase, orphan 1
2.2. Generation of Col10a1-TAP63α transgenic mice
Transgenic mice were generated in which HA and Flag-tagged human TAP63α cDNA was driven by the hypertrophic chondrocyte-specific Col10a1 regulatory elements (Fig. 2A, 2B) that we recently described (Zheng et al., 2009). Specifically, the Col10a1 regulatory elements contain four copies of the 288-bp Col10a1 distal promoter (4296 to −4209 bp) followed by a Col10a1 basal promoter (−220 to +45 bp) as illustrated (Fig. 2B). These combined Col10a1 promoter elements were released from plasmid pBluescript II by BamHI and SalI (blunted) digestion and cloned into the BamHI and Asp718 (blunted) sites of the pcDNA3.1(−) vector (Invitrogen). The full length human TAP63α cDNA in-frame with a 5′-HA- and a 3′-flag fragment was released from pcDNA 3.1(−) by BamHI (blunted) and XhoI (blunted) digestion and cloned into the HindIII (blunted) site of the pcDNA3.1 (−) downstream of the Col10a1 regulatory elements. After sequence confirmation, a 3.4 kb Bsrb1 fragment containing the whole transgenic cassette, which includes the cell-specific Col10a1 promoter elements, the HA- and flag-tagged TAP63α cDNA, and the bovine growth hormone polyadenylation signal sequence, was released and purified for DNA microinjection. Generation of transgenic mice was conducted at the University of Illinois at Chicago (UIC) Transgenic Production Service core facility. Purified DNA construct was injected into pronuclei of FVBN/J mouse zygotes and transplanted into pseudopregnant ICR mice. Transgenic founders were identified by PCR genotyping using following primer pairs: HA-Forward: 5′-GTA CCT GAC TAT GCA TAT CCG-3′ and human P63-Reverse: 5′-CAG TGG AAT ACG TCC AGG TG-3′; or human P63-Forward: 5′-TCC TGC GGA CCC CAA GCA GT-3′ and Flag-Reverse: 5′-AGA AGG CAC AGT CGA GGC TGA -3′. The animal studies were approved by the animal care and oversight committees at University of Illinois at Chicago and Rush University Medical Center.
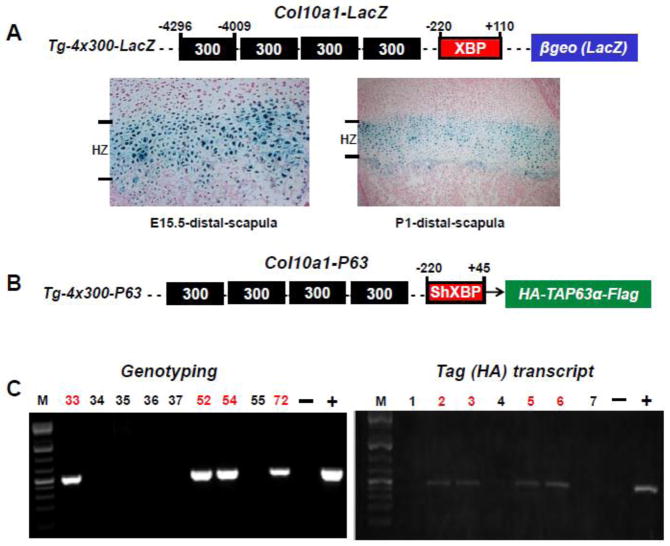
Fig 2. Generation of Col10α1-TAP63α transgenic mice.
(A). Transgenic construct using the 300-bp (−4296 to −4009 bp) Col10a1 distal promoter and the 330-bp (−220 to +110 bp) Col10a1 basal promoter to drive the LacZ gene was as illustrated (top). Paraffin embedded sections of distal scapula from X-gal-stained Tg-4x300 transgenic mice at E15.5 (embryonic) and P1 (postnatal) stages were counterstained with nuclear fast red. Blue staining indicating β-galactosidase activity is exclusively throughout the hypertrophic zone (bottom) as previously described (Zheng et al., 2009). XBP: Col10a1 basal promoter. (B). Same Col10a1 distal promoter and a shorter Col10a1 basal promoter (−220 to +45 bp) elements were used to generate P63-expressing transgenic construct. A HA- and a Flag-tag at either 5′- or 3′-end were cloned in-frame with human TAP63α cDNA. Shorter basal promoter (with the 3′-sequence ending at exon I) was used to allow transgene expression with the endogenous splicing acceptor site. ShXBP: shorter Col10a1 basal promoter. (C). PCR genotyping using human P63 and HA- or Flag fragment specific primers indicated that we have successfully generated four Col10a1-TA63α transgenic founders (left, lanes 33, 52, 54, and 72). Transgene expression in offspring of Col10α1-TAP63α transgenic mouse line 72 was confirmed by RT-PCR analysis of total RNAs prepared from limb tissues using the HA- and human P63-specific primers (right, lanes 2, 3, 5, and 6).
2.3 Skeletal phenotypic analysis
Mouse skeletons from transgenic (TG) or wild-type (WT) littermates at embryonic day 15.5 (E15.5), 17.5 (E17.5), and postnatal day 1 (P1) stages were prepared and stained with Alcian blue and Alizarin red staining according to published protocol with modification (Ovchinnikov D, 2009). Briefly, both embryonic and P1 mice were eviscerated and fixed in 95% ethanol for at least 24 hours. These mouse skeletons were then transferred to staining solution consisting of 0.03% Alcian blue 8GX (Sigma, St. Louis, MO) in 80% ethanol and 20% acetic acid. After 24 hours, the skeleton samples were rinsed with 95% ethanol and 95% ethanol/2% KOH (v/v=1:1) and counterstained overnight with 0.03% Alizarin Red S (Sigma, St. Louis, MO) in 1% KOH and water. Clearing of the samples was conducted by placing them in 1% KOH/20% glycerol for at least two days before transferring into glycerol/95% ethanol (v/v=1:1). Samples were then treated with 80% glycerol and finally stored at 100% glycerol. To analyze the skeletal phenotypes, mouse offspring from both Col10a1-TAP63α transgenic mouse lines were genotyped and confirmed by transgene expression analysis. The occurrence and signal intensity of the alizarin red staining indicating ossification status were analyzed for limbs, digits and mouse tail bones (ossified caudal vertebrate numbers) and compared between TG and WT controls. At least three lines from Col10a1-TAP63α TG mice or WT littermates were stained and analyzed.
2.4. Transgene and marker gene expression
Total RNAs from limb tissues of mice at the E17.5 and p1 stages from three Col10a1-TAP63α transgenic mouse lines were prepared. After reverse transcription, the RT products were used for expression profiling of transgene (Flag-tag or P63) and following apoptotic (Bax, Bcl-2 ), bone/cartilage-specific (Col10a1, Runx2, and Sox9) and mineralization related marker genes (Alp, Ank, Enpp1, and Phospho1) using qRT-PCR as mentioned above. The primer sequence and amplicon size were as listed (Table 1).
2.5. Hematoxylin/Eosin and von Kossa staining
Mouse hind limbs from Col10a1-TAP63α mouse offspring were used. Specifically, mouse limbs at E17.5 or P1 stages were collected and fixed in 10% formalin and stored in 70% ethanol. These mouse limbs were then subjected to dehydration, paraffin embedding and sectioning without decalcification. H & E staining was performed using standard protocols. von Kossa staining was performed on the paraffin embedded sections based on previous protocol with modifications (Bonewald et al., 2003). Briefly, xylene and ethanol treated slides were incubated with 5% silver nitrate and exposed to an ultraviolet light for 30–60 minutes. After rinse with distilled water, slides were treated with 5% sodium thiosulfate for 2–3 minutes and counterstained with 0.1% nuclear fast red. At least 30 longitudinal (sagittal) sections (5-μm-thick) of the limb growth plate from both TG and WT littermates were collected and analyzed with the microscope (Nikon Eclipse 80i, Nikon Instruments Inc., Melville, NY USA) and the Qcapture Suite software (version, 2.95.0, Quantitative Imaging Corp., USA).
2.6. Immunohistochemical analysis
Relevant mouse limb sections were subjected to immunohistochemical analysis using following primary antibodies: anti-p63 (H-129, sc-8344, Santa Cruz) which recognizes the p63α isoforms (Romano et al., 2006), anti-Sox9 (H-90, sc-20095, Santa Cruz), and anti-Flag (#2368, Cell Signaling). The procedures were based on manufacture suggested protocol with modifications. Briefly, after de-paraffin and rehydration, the selected TG or WT limb sections underwent a series of pretreatments before incubation with the primary antibodies (4°C, overnight). The pretreatments include hot citrate buffer incubation (0.01 M, pH 6.0, 95°C, 10 min) to retrieve antigen, hydrogen peroxide treatment (3% H2O2 in 100% ethanol, 5 min) to quench the endogenous peroxidase, and blocking with 30% normal horse serum (30 min). The concentrations used for the primary antibodies were at 1:300 (anti-P63), 1:3000 (anti-Sox9) and 1:500 (anti-Flag) dilutions respectively. Non-immune mouse IgG was used as a negative control. After washing with the 1xTBST (Tris Buffered Saline with 0.1% Tween-20), tissue sections were further incubated with biotinylated secondary antibody (anti-rabbit IgG, Santa Cruz). Detection was using the reagents, and protocol as instructed in the ABC kit (Elite PK-6200 Universal, VECTOR laboratories). Slides were counterstained with hematoxylin before microscopic analysis as described above.
2.7. Statistical Analysis
One-way ANOVA and student t-tests were used to assess the significance across experimental groups (TG vs WT). Values (ossified caudal vertebrate numbers) are presented as means ± SEM (standard error of mean). Real-time RT-PCR data was analyzed using 2−ΔΔ Ct values of each sample (Livak KJ, Schmittgen TD, 2001). p< 0.05 was considered to be statistically significant.
3. Results
3.1. mRNA transcripts of p53 family members in MCT cells
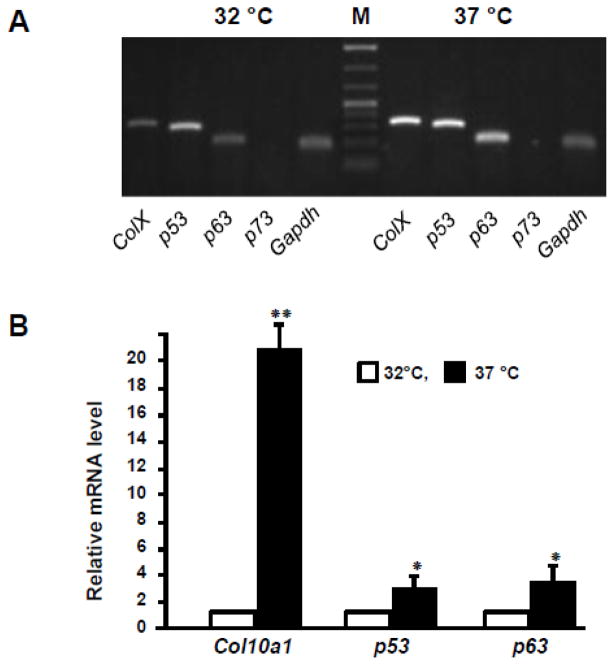
To investigate the putative role of p53 family members during chondrogenesis, we examined the mRNA level of p53, p63, and p73 in MCT cells, a cell model of chondrocyte differentiation and maturation that has been described previously (Lefebvre et al., 1995). As expected, the mRNA transcript of type X collagen gene (Col10a1 or ColX) is much more abundant in hypertrophic MCT cells (37°C) by semi-quantitative RT-PCR analysis (30 cycles). We also observed more abundant levels of p53 and p63 mRNA transcripts in hypertrophic MCT cells (37°C) compared to cells grown in permissive temperature (32°C). No p73 mRNA transcript was detected in either proliferative or hypertrophic MCT cells with designated primers (Table 1 and Fig. 1A). Meanwhile, qRT-PCR confirmed the significantly upregulated mRNA level (more than 20-fold) of Col10a1, while both p53 and p63 were 2–3 fold upregulated in hypertrophic MCT cells compared to that in proliferative MCT cells as normalized to Gapdh (Fig. 1B). These results together suggest a potential function of p53 family members, especially p53 and p63, during chondrocyte differentiation and maturation in vitro.
Fig 1. mRNA levels of Col10a1 and p53 family members in MCT cells.
(A). Total RNAs were prepared from both proliferative (32 °C) and hypertrophic (37 °C) MCT cells using the TRIzol reagent. Reverse transcribed cDNA were subjected to RT-PCR analysis and the results showed that Col10a1, p53 and p63 transcripts were much more abundant in hypertrophic MCT cells than in proliferative MCT cells. No p73 amplicon was detected using the designated pair of PCR primers. (B). Real-time RT-PCR analysis showed that Col10a1 mRNA transcript is more than 20-fold upregulated in hypertrophic MCT cells than in proliferative MCT cells as normalized to Gapdh. Meanwhile, both p53 and p63 showed 2–3 fold upregulation in hypertrophic MCT cells compared to proliferative MCT cells. *: p<0.05; **: p<0.01. M: Marker.
3.2. Establishment of Col10a1-TAP63α transgenic mouse lines
We have previously reported Col10a1 promoter analysis by transgenic studies that used four copies of the 300 bp Col10a1 distal promoter (−4296 to −4009) and its basal promoter (−220 to +110) to drive the LacZ gene (Fig. 2A, top, Tg-4x300-LacZ) (Zheng et al., 2003). These Col10a1 promoter elements were able to direct exclusive reporter expression (X-gal blue staining) throughout the hypertrophic zone of the growth plate (distal scapula) in transgenic mice at both E15.5 and P1 stages (Fig. 2A, bottom). To study the in vivo role of P63 during skeletal development, we have performed P63 gain-of- function studies by generating a transgenic construct in which HA- and flag-tagged TAP63α cDNA was driven by the same hypertrophic chondrocyte-specific Col10a1 distal promoter and a shorter basal promoter as illustrated (Fig. 2B, Tg-4x300-P63). The shorter basal promoter which ends at exon I was used so as to avoid the requirement of additional splicing acceptor site. PCR genotyping using HA fragment and human P63 specific primers (Table 1) showed that we have successfully generated four Col10a1-TA63α transgenic founders (Fig. 2C, left, lanes 33, 52, 54, and 72). All these transgenic founders except founder 54 undergo germline transmission after PCR genotyping of the offspring of each of these four founders (data not shown). We also examined the transgene (HA- and Flag-tagged human TAP63α) expression in these lines at both E17.5 and P1 stages after genotyping (data not shown). Here we show the representative results of transgene expression in offspring of Col10α1-TAP63α transgenic mouse line 72. As illustrated, transgene expression was confirmed in transgenic pups by RT-PCR analysis of total RNAs prepared from limb tissues using the HA- and human P63-specific primers (Fig. 2C, right, lanes 2, 3, 5, and 6).
3.3. Transgene expression in Col10a1-TAP63α transgenic mice
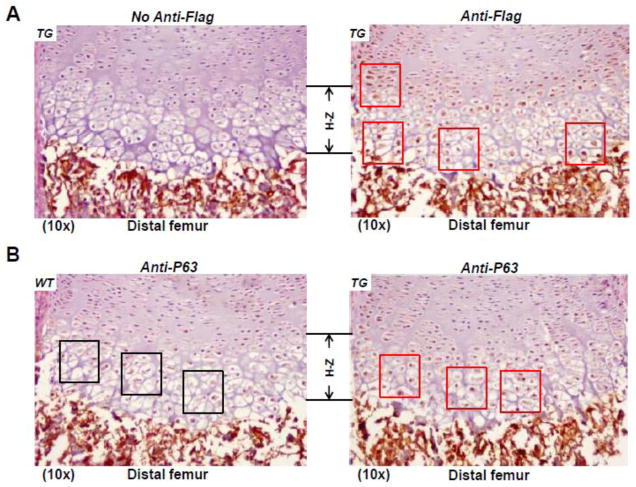
As described above, transgene expression was confirmed by RT-PCR analysis using mouse limb tissues of three Col10a1-T AP63α transgenic mouse lines that were established from founders 33, 52 and 72. To examine the targeting efficiency of transgene gene driven by the cell-specific Col10a1 control elements, we have performed immunohistochemical staining using anti-Flag antibody on sagittal sections of distal femur of TG mice at the P1 stage. While no staining was detected in the growth plate when control serum was used (Fig. 3A, left), specific brown staining showing transgene expression was primarily restricted to pre-hypertrophic and hypertrophic chondrocytes (Fig. 3A, right, red squares)whenanti-Flag antibody was used. Meanwhile, immunohistochemistry using anti-P63 antibody was also performed on sagittal sections of distal femur of both TG and WT littermates at the P1 stage. While endogenous p63 was shown in the proliferative and (pre-) hypertrophic zone of WT mice (Fig. 3B, left), more intense brown staining indicating increased P63 expression was detected in the prehypertrophic and hypertrophic zone of TG mice (Fig. 3B, right, red squares). These results together suggest that the cell-specific Col10a1 control elements (Zheng et al., 2009) direct target transgene (Flag-tagged TAP63α) expression primarily in the (pre-)hypertrophic region.
Fig 3. Transgene expression in Col10α1-TAP63α transgenic mice.
(A). Sagittal sections of distal femur from transgenic mouse limbs at the P1 stage were subjected to immuno-histochemical staining using anti-Flag antibody. Transgene expression was primarily observed in the pre-hypertrophic and hypertrophic region (red squares, right panel). Left panel shows no antibody control. TG: transgenic. (B). Sagittal sections of distal femur from both TG and WT mouse limbs at the P1 stage were subjected to immuno-histochemistry analysis using anti-P63 antibody. P63 was overexpressed in the pre-hypertrophic and hypertrophic region of TG mice (red squares, right panel) compared to WT littermates (black squares, left panel).
3.4. Accelerated ossification in Col10α1-TAP63α transgenic mice
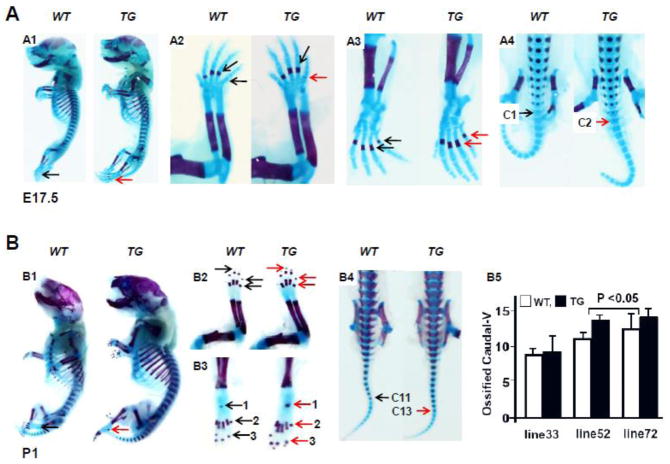
Skeletal phenotypes of Col10α1-TAP63α TG mice were analyzed using Alcian blue and Alizarin red staining and compared to the WT controls. Mouse embryos at E15.5, E17.5 and mice at P1 stage from each of the Col10α1-TAP63α transgenic mouse lines 33, 52, and 72 were analyzed. No obvious skeletal difference was observed between TG and WT littermates at E15.5 based on results of whole skeletal staining from all three Col10α1-TAP63α transgenic mouse lines (data not shown). The skeletal consequences from line 72 and the comparison between TG and WT littermates at both E17.5 and P1 stages were as illustrated (Fig. 4). At E17.5, the TG mouse embryos are slightly larger in size than the WT controls (Fig. 4, A1), slightly accelerated ossification was also observed as indicated by more intense alizarin red staining in the fore limb phalange and digits (Fig. 4, A2), hind limb metatarsal and digits (Fig. 4, A3), and the ossified caudal vertebrates in tail bones (Fig. 4, A4). For mice at the P1 stage, TG mice are much larger in size as shown by whole skeleton preparation (Fig. 4, B1). Enhanced ossification signs (alizarin red staining) were also observed in fore limb (phalanges and digits, Fig. 4, B2), hind limb (calceneus, metatarsal and digits, Fig. 4, B3), and tail bones (caudal vertebrates, Fig. 4, B4). We observed similar enhanced ossification in TG embryos (E17.5) and P1 mice as well in line 52 compared to WT littermates (data not shown), while no skeletal phenotypic difference was shown between TG and WT controls in line 33 at both E17.5 and P1 stages (data not shown). We also calculated the ossified caudal vertebrates of P1 mice for these Col10α1-TAP63α transgenic mouse lines. While no difference was observed between TG and WT littermates of line 33 (p>0.05), TG mice in both line 52 and line 72 have more ossified caudal vertebrates than that of WT littermates (p<0.05, Fig. 4, B5), suggesting the putative function of TAP63α during endochondral bone formation.
Fig 4. Accelerated ossification in Col10α1-TAP63α transgenic mice.
(A). Alcian blue and Alizarin red staining of mouse embryos at E17.5 stage. A1: Whole skeletal staining showed slightly oversize of TG over WT littermate controls. A2: Fore limb, faint alizarin red staining can be seen in the last phalange of TG mouse embryo (red arrow), but not in the WT mouse (black arrow). A3: Hind limb, the metatarsal bones show more intense ossification signs in TG vs WT embryos. A4: Tails, ossification signs were clearly shown in the second caudal vertebrate of TG mouse embryo (red arrow) compared to the first caudal vertebrate of WT mouse embryo (black arrow). (B). Skeletal staining of Col10α1-TAP63α TG and WT mice at P1 stage. B1: Whole skeletal staining, Col10α1-TAP63α TG mice are generally larger than its WT littermates. B2: Fore limbs, more intense alizarin red staining was observed in the terminal digits and phalange bones of the TG mice (red arrows) compared to WT littermates (black arrows). B3: Hind limbs, more obvious ossification signs were observed in the calceneus (1), the metatarsal bones (2) and terminal digits (3) of the TG mice (red arrows) compared to WT littermates (black arrows). B4: Tail bones, alizarin red staining showed ossifications signs up to 11th caudal vertebrate in WT mouse tail (black arrow), compared to the clear alizarin red staining in the 13th caudal vertebrae of TG mouse tail (red arrow). B5: Ossified caudal vertebrates were calculated and subjected to statistical analysis for three Col10α1-TAP63α transgenic mouse lines. TG mice of line 52 [10.8±0.50 (WT) and 13.2±0.44( TG), p<0.001], and line 72 [11.7±1.50 (WT) and 13.4±0.54 (TG), p<0.05] showed more ossified caudal vertebrates than that of WT littermates, while no difference was observed between TG and WT littermates of line 33 [9.1±0.38 (WT) and 9.6±1.81 (TG)] (p>0.05).
3.5. Increased matrix mineralization in Col10α1-TAP63α transgenic mice
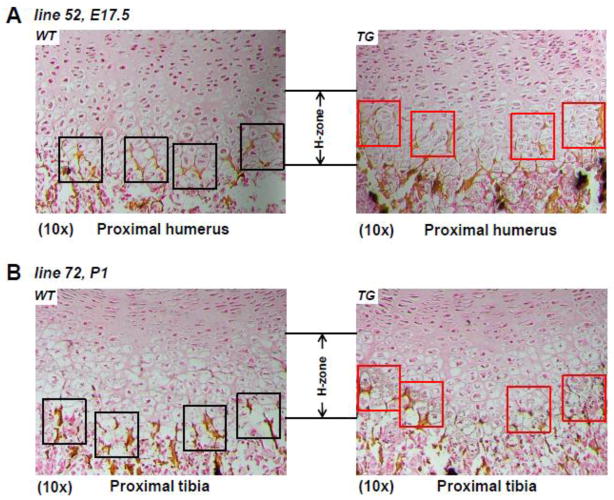
To determine the status of matrix mineralization in TG mouse lines 52 and 72 that show accelerated ossification, we have performed von Kossa staining on limb sections of these mice at both E17.5 and P1 stages. Figure 5A shows von Kossa staining of sagittal sections of proximal humerus of both TG and WT littermates at E17.5 stage from transgenic mouse line 52. The results showed that matrix mineralization starts in the terminal hypertrophic chondrocytes of WT littermates (Fig. 5A, left, black squares), while increased von Kossa staining was observed in the upper hypertrophic zone of TG mice (Fig. 5A, right, red squares). Von Kossa staining was also performed in sagittal sections of proximal tibia of both TG and WT littermates at P1 stage from transgenic mouse line 72. The results also suggest an earlier mineralization process in the middle hypertrophic zone of TG mice (Fig. 5B, right, red squares) compared to the WT littermate controls (Fig. 5B, left, black squares).
Fig 5. Increased mineralization in C ol10α1-TAP63α transgenic mice.
(A). Sagittal sections of proximal humerus from both TG and WT mouse embryos of line 52 at the E15.5 stage were subjected to von Kossa staining. More von Kossa staining (black and silver dots as well as dark brown staining) suggesting increased mineralization was shown in the hypertrophic zone of TG mice (right red squares) compared to WT littermate controls (left, black squares). (B). Von Kossa staining was also performed in sagittal sections of proximal tibia of both TG and WT littermates at P1 stage from transgenic mouse line 72. The results also suggest an earlier mineralization process in the middle hypertrophic zone of TG mice (right, red squares) compared to the WT littermate controls (left, black squares). H-zone: hypertrophic zone.
3.6. Expression profiling of relevant marker genes in Col10α1-TAP63α transgenic mice
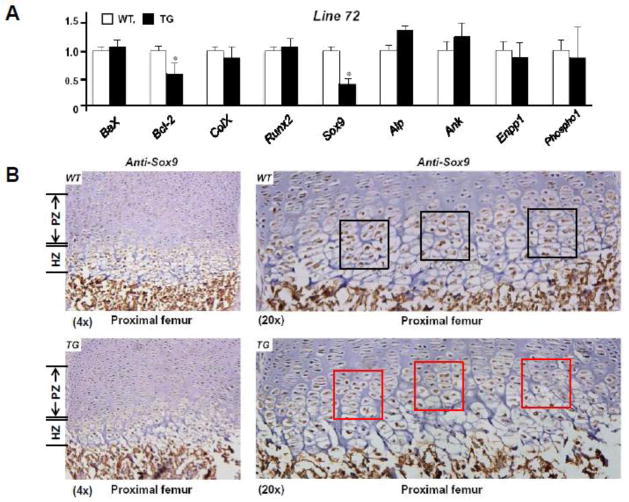
To investigate the putative mechanisms of the accelerated mineralization/ossification observed in Col10α1-TAP63α TG mice, we examined the relative expression of several apoptotic and chondrogenic related marker genes (such as Bax, Bc-2, Col10a1, Runx2 and Sox9) in all three Col10α1-TAP63α transgenic mouse lines. The representative marker gene expression in line 72 at the P1 stage was as illustrated. While no obvious fold-change of the mRNA transcripts was shown between TG and WT controls for Bax, Col10a1 and Runx2 genes, we detected significant reduction of Bcl-2 and Sox9 mRNA transcripts in TG mice compared to the WT littermates by qRT-PCR analysis (p < 0.05, Fig. 6A). Reduction of Bcl-2 and Sox9 mRNA transcripts was also observed in TG mice of line 52, but not in line 33 (data not shown). Some bone mineralization related marker genes (such as Alp, Ank, Enpp1 and Phospho1) were also examined by qRT-PCR analysis. While no notable difference was observed between TG and WT mice for genes Enpp1 and Phospho1, Alp and Ank mRNA transcripts did show 20–30% increase in lines 52 and 72 (Fig. 6A). We have also performed immunohistochemical staining using anti-Sox9 antibody and mouse limb sections from line 72 at the P1 stage. Decreased Sox9 expression was observed in the proliferative and hypertrophic zone of TG mice (Fig. 6B, bottom panels) compared to WT controls (Fig. 6B, top panels). Sox9 downregulation was also detected in the growth plate region of TG mice from line 52 by immunohistochemical analysis (data not shown).
Fig 6. Relevant marker gene expression in Col10α1-TAP63α transgenic mice.
(A). Total RNAs from mouse hind limbs of both line 52 and line 72 were prepared and reverse transcribed for expression analysis using qRT-PCR approach. Illustrated is the relative mRNA level from representative line 72. Bax, Col10a1 and Runx2 in TG mice is similar to that of WT controls. Meanwhile, significant reduction of Bcl-2 (WT vs. TG: 1.00 ± 0.12 vs. 0.62 ± 0.20, n = 7, p < 0.05) and Sox9 (WT vs. TG: 1.00 ± 0.09 vs. 0.42 ± 0.11, n = 7, p < 0.05) transcripts were detected in TG mice compared to the WT littermates. Data ColX: Col10a1. (B). Sagittal sections of proximal femur from both TG and WT mouse limbs at the P1 stage were subjected to immunohistochemistry analysis using anti-Sox9 antibody. Decreased Sox9 expression was observed in the proliferative and hypertrophic zone in TG mice (bottom, left) compared to WT controls (top, left, 4x magnifications). Higher (20x) magnifications of the same panels of images clearly showed less brown staining and signal intensity indicating decreased Sox9 expression in cells of prehypertrophic and hypertrophic zone (bottom, right, red squares) of the TG mice compared to the littermate controls (top, right, black squares). PZ: proliferative zone; HZ: hypertrophic zone.
4. Discussion
As a p53 family member, P63 was originally thought to be another tumor suppressor. However, unlike P53, few mutations in P63 gene have been identified and associated with human cancers (van Bokhoven et al., 2001; Finlan LE, Hupp TR, 2007). Instead, P63 has been widely recognized as a critical transcription factor for proper epithelial development and maintenance (Perez CA, Pietenpol. JA., 2007). It was not until more than a decade ago, when p63 was shown to be important for limb development, as mice lack p63 are either absent or have severely truncated limbs. This severe limb defect is most likely caused by abnormal ectodermal-mesenchymal signaling, suggesting a role of P63 during apical ectodermal ridge (AER) induction (which develops as early as embryonic day 9.5) and maturation that are essential for early limb development (Fernandez-Tern M, Ros MA., 2008). Interestingly, the limb defect may also be attributed to abnormal maturation and apoptosis of cartilage as indicated previously (Trink et al., 2007). This makes P63 a possible candidate for chondrocyte apoptosis, a known process essential for endochondral bone formation. However, how P63 plays a role upon endochondral bone formation during early skeletal development, has not been elucidated.
We have examined the mRNA levels of p53 family members in an established cell model of mouse chondrocytes, the MCT cells (Lefebvre et al., 1995). It was previously demonstrated that MCT cells proliferate well in permissive temperature (32°C) of SV40 large T antigen. These cells undergo hypertrophy or maturation which is characterized by significant up-regulation of Col10a1 expression upon growth arrest (grown in 37°C). Indeed, Col10a1 mRNA transcript is much more abundant (more than 20-fold) in hypertrophic MCT cells than in proliferative MCT cells (Fig. 1A, 1B). Surprisingly, both p53 and p63 mRNA transcripts were (2–3 fold) upregulated in hypertrophic MCT cells compared to that in proliferative MCT cells (Fig. 1A, 1B). P63 has been well demonstrated to be a regulatory protein that regulates target gene expression and cellular processes (Perez CA, Pietenpol. JA., 2007). The upregulated p63 expression may contribute to the enhanced Col10a1 expression and the terminal differentiation of hypertrophic MCT cells, and therefore, play a role in the endochondral pathway.
To investigate the potential in vivo function of P63 upon late embryonic skeletal development, we have performed P63 gain of function studies using the transgenic approach. We selectively target TAP63α in hypertrophic chondrocytes using the cell-specific Col10a1 control elements that we previously described (Zheng et al., 2009). The α-isoform is the longest isoform of P63 subtypes. TAP63 has previously been shown to transactivate or repress target gene expression to cause cellular responses. In our Col10a1-TAP63α transgenic studies, while no notable difference of skeletal phenotypic consequence was observed between TG and WT controls in line 33, the TG mice from lines 52 and 72 did show accelerated ossification in long bone, digit and tail bones at both E17.5 and P1 stages compared to their WT littermates (Fig. 5). Earlier mineralization was also observed in the growth plate of Col10a1-TAP63α TG mice compared to their littermate controls (Fig. 5). These results suggest a putative function of TAP63α upon endochondral bone formation.
We have performed expression analysis of some chondrogenic and apoptotic related marker genes (Bax, Bcl-2, Col10a1, Runx2 and Sox9) in our transgenic mice. No notable difference was detected between TG and WT littermates of line 33 of Col10α1-TAP63α mice (data not shown). Interestingly, qRT-PCR analysis detected significant reduction of Bcl-2 and Sox9 transcripts in TG mice of lines 52 and 72 at both E17.5 and P1 stages (Fig. 6A and data not shown). Meanwhile, Alp and Ank mRNA transcripts increased by 20–30% in lines 52 and 72 (Fig. 6A), though not statistically significant. However, immunohistochemical analysis confirmed the decreased Sox9 expression in the proliferative and hypertrophic zone of lines 52 and 72 of Col10α1-TAP63α TG mice (Fig. 6B). These results together may suggest a potential mechanism of chondrocyte maturation/apoptosis or matrix mineralization that contributes to the accelerated ossification in two of the Col10α1-TAP63α mouse lines (Fig. 4).
Chondrocyte hypertrophy and apoptosis are essential steps of endochondral bone formation. Apoptosis in hypertrophic chondrocytes may be linked to mineralization of the extracellular matrix (Mello et al., 2002; Shapiro et al., 2005). Multiple apoptotic marker genes such as Bax and Bcl-2 have been shown to play a role in chondrocyte cell death by maintaining the ratio of Bcl-2 to Bax to determine cell fate (Mello et al., 2002; Cheung et al., 2003; Oshima et al., 2008). Bax overexpression causes accelerated cell death while Bcl-2 null mice have accelerated endochondral ossification (Boot-Handford et al., 1998; Mello et al., 2002). In our Col10a1-TAP63α mouse lines 52 and 72, while Bax did not show a difference, significant reduction of Bcl-2 mRNA transcripts was detected in TG mice compared to WT controls. This will cause the ratio change of Bcl-2 over Bax and possibly leading to the phenotypic consequence.
In the mean time, we also detected decreased level of Sox9 mRNA transcripts and protein in TG mice of these two lines by qRT-PCR and immunohistochemical analysis. Sox9 inactivation has recently been associated with chondrocyte apoptosis, suggesting its anti-apoptotic function (Ikegami et al., 2011). Downregulation of Bcl-2 and Sox9 expression may promote apoptosis of hypertrophic chondrocytes that are essential for subsequent matrix mineralization and osteoprogenitor cell deposition during endochondral ossification. Indeed, we detected increased level of Alp (30%), and Ank (20%) transcripts in TG mouse limb tissues of lines 52 and 72. Alp (alkaline phosphatase), a known early osteogenic marker, has previously been implicated to play a crucial role in promoting mineralization of the extracellular matrix (Yadav et al., 2011), whereas Ank (progressive ankylosis protein) has been demonstrated to be a positive regulator of bone formation and remodeling (Kim et al., 2010). Although the increase is not statistically significant, possibly due to the fact that the total RNAs were prepared from whole hind limbs, not from hypertrophic chondrocytes enriched limb tissue, we did observe earlier matrix mineralization as demonstrated by von Kossa staining in the hypertrophic zone of Col10a1-TAP63α TG mice compared to their littermate controls. In addition, SOX9 has been shown to inhibit the transactivation of RUNX2 (Cheng et al., 2010), a master transcription factor both for osteoblast differentiation and chondrocyte maturation (Otto et al., 1997; Kim et al., 1999; Inada et al., 1999; de Crombrugghe et al., 2001; Takeda et al., 2001; Hinoi et al., 2006; Komori et al., 2010). SOX9 is dominant over RUNX2 function during endochondral ossification (Zhou et al., 2005). Therefore, the accelerated ossification in our Col10a1-TAP63α mice could be partially attributed to Sox9 downregulation which will release the inhibition of Runx2 function to promote chondrocyte maturation and bone formation.
In conclusion, we have, for the first time, investigated the potential function of TAP63α, the longest P63 isoform, during early skeletal development in vivo by selectively target TAP63α expression in hypertrophic chondrocytes. Our results support that TAP63a play a positive role upon endochondral ossification, possibly through interaction with genes (such as Alp, Ank, Bcl-2, Sox9, etc.) relevant to matrix mineralization, chondrocyte maturation or apoptosis. Further elucidating the role of p63 isoforms and the underlying mechanisms upon different skeletal developmental stages will help to identify potential therapeutic targets for P63 related skeletal diseases.
Highlights.
P63 is upregulated in hypertrophic MCT cells compared to proliferative MCT cells.
We have generated transgenic mice overexpressing TAP63α in hypertrophic chondrocytes.
Enhanced ossification was observed in these transgenic mice at E17.5 and P1 stages.
The result suggests a positive role of TAP63α in late embryonic skeletal development.
Acknowledgments
We are grateful to Dr. Kotaro Sena and Mr. David Karwo for technical help on histological analysis of the transgenic mice at the Rush soft tissue and decalcified hard tissue histology sub-core. We thank Dr. Benoit de Crombrugghe for the MCT cells. The Col10a1-TAP63α transgenic mice were generated within the Transgenic Production Service core facility (directed by Dr. Roberta Franks) at the University of Illinois at Chicago (UIC). This work was supported by the 2008 Pilot Project of Rush University Medical Center (Q.Z.), and the Bear Necessities Pediatric Cancer Foundation (Q.Z.).
Abbreviations
- RT-PCR
Reverse transcription polymerase chain reaction
- q-(RT-)PCR
quantitative or real-time (RT-) polymerase chain reaction
- Gapdh
glyceraldehydes-3-phosphate dehydrogenase
- Col10α1
type X collagen, alpha 1
- mTrp53/63/73
murine transformation related protein 53/63/73
- Bax
BCL-2-associated X protein
- Bcl-2
B-cell leukemia (lymphoma) 2
- Runx2
runt-related transcription factor 2
- Sox9
SRY-box containing gene 9
- Alp
Alkaline phosphatase
- Ank
progressive ankylosis gene
- Enpp1
ectonucleotide pyrophosphatase/phosphodiesterase 1
- Phospho1
phosphatase, orphan 1
- H & E staining
Hematoxylin/Eosin
- MCT cells
Mouse chondrocytes immortalized with SV40 large T antigen
- XBP
Col10a1 basal promoter
- WT
Wild-type
- TG
Transgenic
- E17.5
Embryonic day 17.5
- P1
Postnatal day one
- TBST
Tris Buffered Saline
- ANOVA
Analysis of variance between groups
Footnotes
Disclosures: All the authors state that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonewald LF, Harris SE, Rosser J, Dallas MR, Dallas SL, Camacho NP, Boyan B, Boskey A. von Kossa staining alone is not sufficient to confirm that mineralization in vitro represents bone formation. Calcif Tissue Int. 2003 May;72(5):537–47. doi: 10.1007/s00223-002-1057-y. Epub 2003 May 6. [DOI] [PubMed] [Google Scholar]
- Boot-Handford RP, Michaelidis TM, Hillarby MC, Zambelli A, Denton J, Hoyland JA, Freemont AJ, Grant ME, Wallis GA. The bcl-2 knockout mouse exhibits marked changes in osteoblast phenotype and collagen deposition in bone as well as a mild growth plate phenotype. Int J Exp Pathol. 1998 Oct;79(5):329–35. doi: 10.1046/j.1365-2613.1998.790411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Genever PG. SOX9 determines RUNX2 transactivity by directing intracellular degradation. J Bone Miner Res. 2010;25:2404–2013. doi: 10.1002/jbmr.174. [DOI] [PubMed] [Google Scholar]
- Cheung JO, Grant ME, Jones CJ, Hoyland JA, Freemont AJ, Hillarby MC. Apoptosis of terminal hypertrophic chondrocytes in an in vitro model of endochondral ossification. Pathol. 2003 Nov;201(3):496–503. doi: 10.1002/path.1462. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001 Dec;13(6):721–7. doi: 10.1016/s0955-0674(00)00276-3. Review. [DOI] [PubMed] [Google Scholar]
- Dietz S, Rother K, Bamberger C, Schmale H, Mössner J, Engeland K. Differential regulation of transcription and induction of programmed cell death by human p53-family members p63 and p73. FEBS Lett. 2002 Aug 14;525(1–3):93–9. doi: 10.1016/s0014-5793(02)03093-4. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M, Ros MA. The Apical Ectodermal Ridge: morphological aspects and signaling pathways. Int J Dev Biol. 2008;52(7):857–71. doi: 10.1387/ijdb.072416mf. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Hupp TR. p63: the phantom of the tumor suppressor. Cell Cycle. 2007 May 2;6(9):1062–71. doi: 10.4161/cc.6.9.4162. Epub 2007 May 19. [DOI] [PubMed] [Google Scholar]
- Hattori T, Muller C, Gebhard S, Bauer E, Pausch F, Schlund B, Bosl MR, Hess A, Surmann-Schmitt C, von der Mark H, de Crombrugghe B, von der Mark K. SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification. Development. 2010;137:901–911. doi: 10.1242/dev.045203. [DOI] [PubMed] [Google Scholar]
- Hinoi E, Bialek P, Chen YT, Rached MT, Groner Y, Behringer RR, Ornitz DM, Karsenty G. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 2006;20:2937–2942. doi: 10.1101/gad.1482906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami D, Akiyama H, Suzuki A, Nakamura T, Nakano T, Yoshikawa H, Tsumaki N. Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development. 2011;138:1507–1519. doi: 10.1242/dev.057802. [DOI] [PubMed] [Google Scholar]
- Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999 Feb;80(2):159–70. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Minashima T, McCarthy EF, Winkles JA, Kirsch T. Progressive ankylosis protein (ANK) in osteoblasts and osteoclasts controls bone formation and bone remodeling. J Bone Miner Res. 2010 Aug;25(8):1771–83. doi: 10.1002/jbmr.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Murao H, Yamamoto K, Deng JM, Behringer RR, Nakamura T, Akiyama H. Generation of transgenic mice for conditional overexpression of Sox9. J Bone Miner Metab. 2011 Jan;29(1):123–9. doi: 10.1007/s00774-010-0206-z. Epub 2010 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010 Jan;339(1):189–95. doi: 10.1007/s00441-009-0832-8. Epub 2009 Aug 1. Review. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Garofalo S, de Crombrugghe B. Type X collagen gene expression in mouse chondrocytes immortalized by a temperature-sensitive simian virus 40 large tumor antigen. J Cell Biol. 1995;128:239–245. doi: 10.1083/jcb.128.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mello MA, Tufan AC, Daumer KM, Bruna Pucci, Lafond T, Hall DJ, Tuan S. Regulation of chondrogenesis and cartilage maturation in vitro: role of TGF-β1, thyroid hormone, and Wnt signaling. In: Shapiro IM, et al., editors. The growth plate. IOS Press; 2002. pp. 37–52. [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999 Apr 22;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Mills AA. P63: oncogene or tumor suppressor? Curr Opin Genet Dev. 2006;16:38–44. doi: 10.1016/j.gde.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002 Jun 15;16(12):1446–65. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Akiyama T, Hikita A, Iwasawa M, Nagase Y, Nakamura M, Wakeyama H, Kawamura N, Ikeda T, Chung UI, Hennighausen L, Kawaguchi H, Nakamura K, Tanaka S. Pivotal role of Bcl-2 family proteins in the regulation of chondrocyte apoptosis. J Biol Chem. 2008 Sep 26;283(39):26499–508. doi: 10.1074/jbc.M800933200. Epub 2008 Jul 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D. Alcian blue/alizarin red staining of cartilage and bone in mouse. Cold Spring Harb Protoc. 2009;(3):pdb.prot5170. doi: 10.1101/pdb.prot5170. [DOI] [PubMed] [Google Scholar]
- Perez CA, Pietenpol JA. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle. 2007 Feb 1;6(3):246–54. doi: 10.4161/cc.6.3.3801. Epub 2007 Feb 3. Review. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, Puisieux A, Hainaut P, Caron de Fromentel C. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008 Feb;29(2):273–81. doi: 10.1093/carcin/bgm258. Epub 2007 Nov 28. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Birkaya B, Sinha S. Defining the regulatory elements in the proximal promoter of DeltaNp63 in keratinocytes: Potential roles for Sp1/Sp3, NF-Y, and p63. J Invest Dermatol. 2006 Jul;126(7):1469–79. doi: 10.1038/sj.jid.5700297. Epub 2006 Apr 27. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Adams CS, Freeman T, Srinivas V. Fate of the hypertrophic chondrocyte: microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res C Embryo Today. 2005 Dec;75(4):330–9. doi: 10.1002/bdrc.20057. [DOI] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trink B, Osada M, Ratovitski E, Sidransky D. p63 transcriptional regulation of epithelial integrity and cancer. Cell Cycle. 2007 Feb 1;6(3):240–5. doi: 10.4161/cc.6.3.3803. Epub 2007 Feb 3. Review. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, Merkx GF, Tenconi R, Fryns JP, Verloes A, Newbury-Ecob RA, Raas-Rotschild A, Majewski F, Beemer FA, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates JR, Neri G, Brunner HG. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001 Sep;69(3):481–92. doi: 10.1086/323123. Epub 2001 Jul 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, Rechthand P, Califano J, Moon CS, Ratovitski E, Jen J, Sidransky D, Trink B. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003 May 15;63(10):2351–7. [PubMed] [Google Scholar]
- Wu G, Osada M, Guo Z, Fomenkov A, Begum S, Zhao M, Upadhyay S, Xing M, Wu F, Moon C, Westra WH, Koch WM, Mantovani R, Califano JA, Ratovitski E, Sidransky D, Trink B. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res. 2005 Feb 1;65(3):758–66. [PubMed] [Google Scholar]
- Yadav MC, Simão AM, Narisawa S, Huesa C, McKee MD, Farquharson C, Millán JL. Loss of skeletal mineralization by the simultaneous ablation of PHOSPHO1 and alkaline phosphatase function: a unified model of the mechanisms of initiation of skeletal calcification. J Bone Miner Res. 2011 Feb;26(2):286–97. doi: 10.1002/jbmr.195. Epub 2010 Aug 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999 Apr 22;398(6729):714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003 Sep 1;162(5):833–42. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Sebald E, Zhou G, Chen Y, Wilcox W, Lee B, Krakow D. Dysregulation of chondrogenesis in human cleidocranial dysplasia. Am J Hum Genet. 2005;77:305–312. doi: 10.1086/432261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Keller B, Zhou G, Napierala D, Chen Y, Zabel B, Parker A, Lee B. Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J Bone Miner Res. 2009;24:1022–1032. doi: 10.1359/JBMR.081249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci USA. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]