Abstract
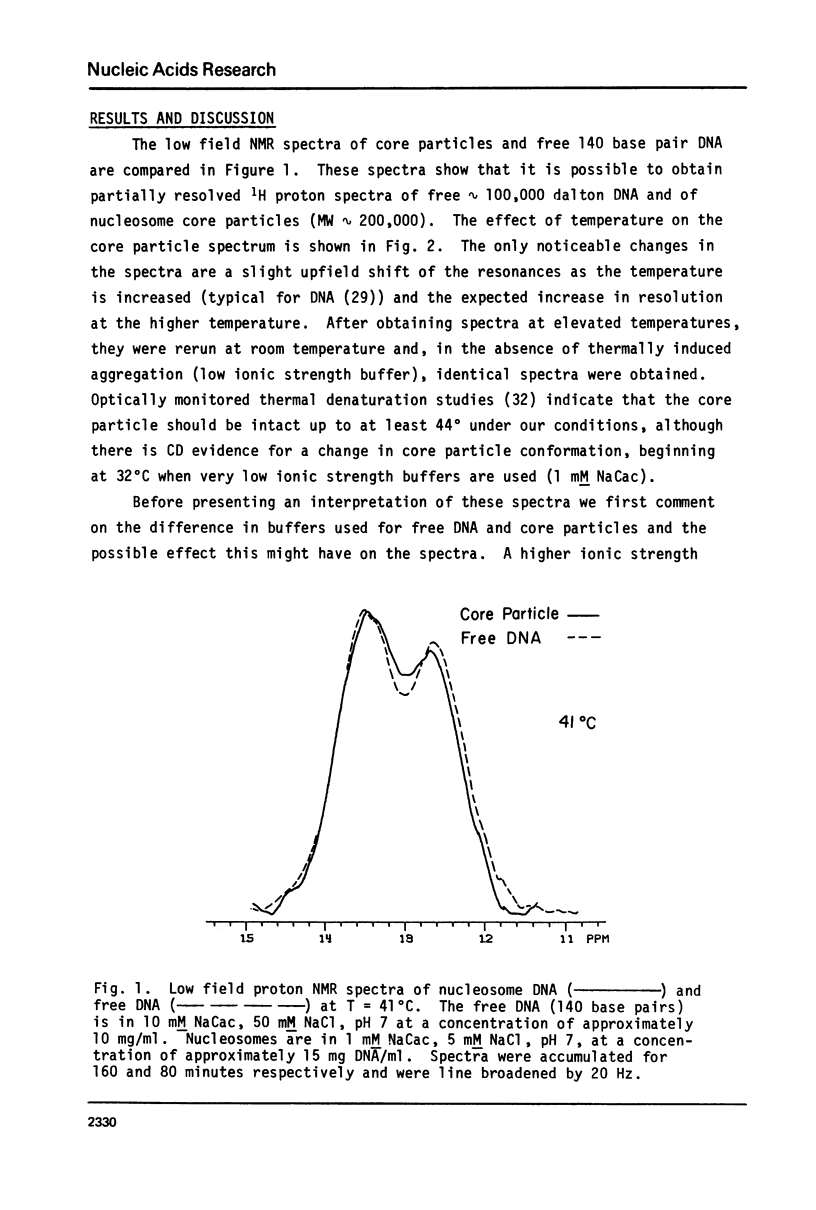
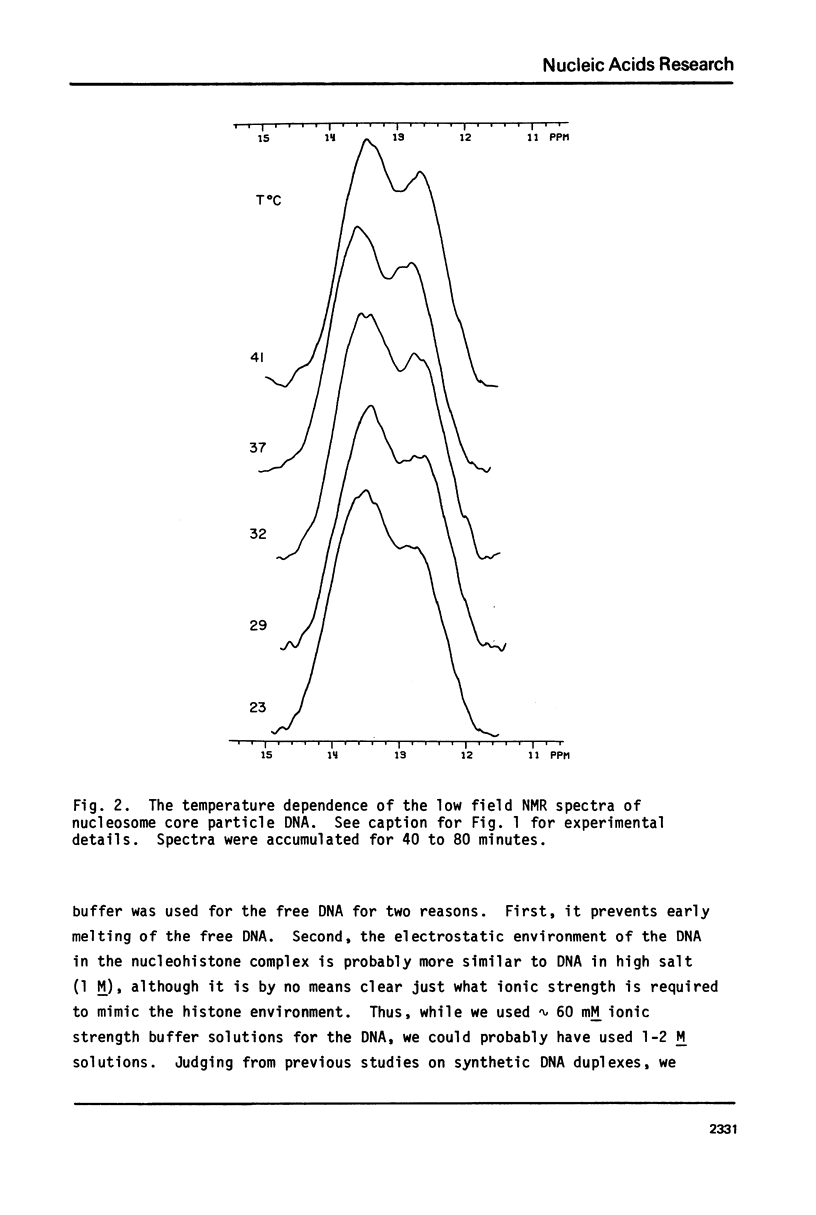
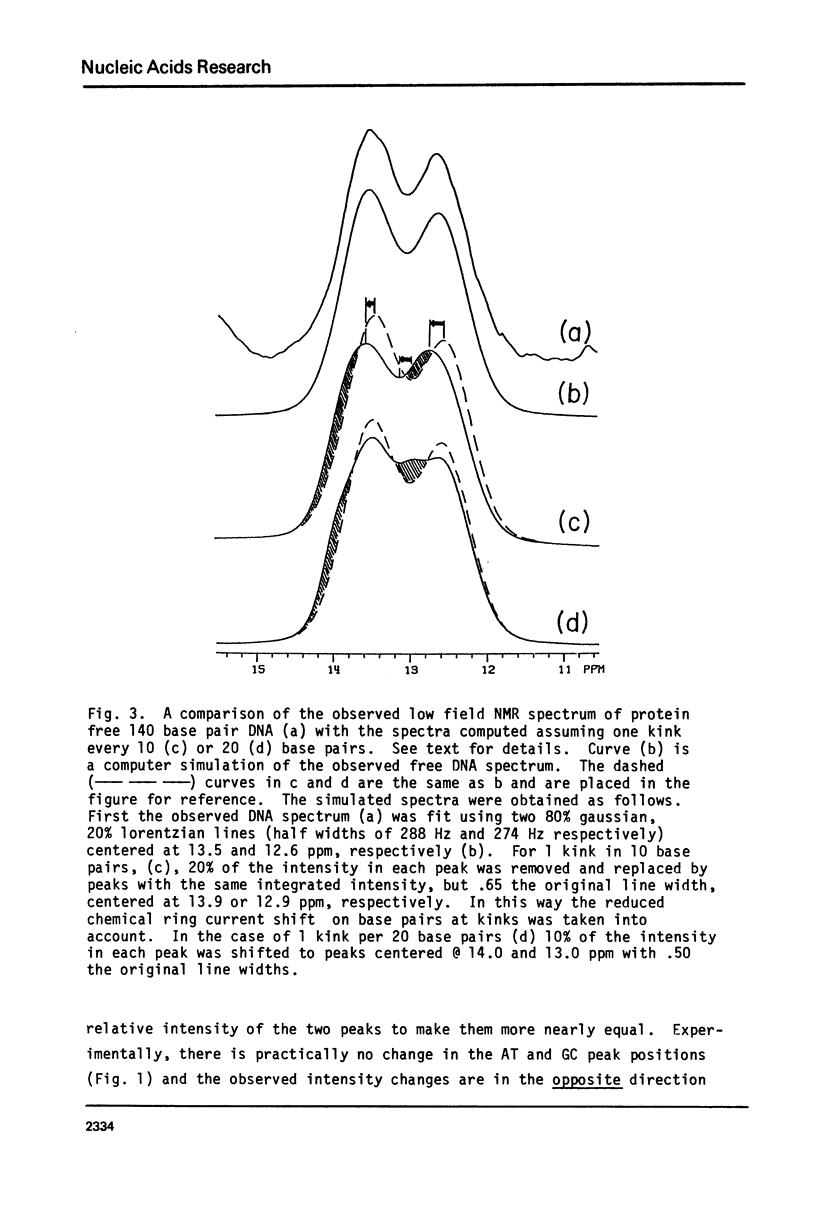
In this study 1H NMR has been used to investigate the conformational state of DNA in nucleosome core particles. The nucleosome core particles exhibit partially resolved low field (10-15 ppm) spectra due to imino protons in Watson-Crick base pairs (one resonance per GC or AT base pair). To a first approximation, the spectrum is virtually identical with that of protein-free 140 base pair DNA, and from this observation we draw two important conclusions: (i) Since the low field spectra of DNA are known to be sensitive to conformation, the conformation of DNA in the core particles is essentially the same as that of free DNA (presumably B-form), (ii) since kinks occurring at a frequency at 1 in 10 or 1 in 20 base pairs would result in a core particle spectrum different from that of free DNA we find no NMR evidence supporting either the Crick-Klug or the Sobell models for kinking DNA around the core histones. Linewidth considerations indicate that the rotational correlation time for the core particles is approximately 1.5 X 10(-7) sec, whereas the end-over-end tumbling time of the free 140 base pair DNA is 3 X 10(-7) sec.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arter D. B., Schmidt P. G. Ring current shielding effects in nucleic acid double helices. Nucleic Acids Res. 1976 Jun;3(6):1437–1447. doi: 10.1093/nar/3.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Kan L. S., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. I. Proton magnetic resonance studies on the nonexchangeable protons of ribosyl ApApGpCpUpU. Biochemistry. 1975 Nov 4;14(22):4847–4863. doi: 10.1021/bi00693a012. [DOI] [PubMed] [Google Scholar]
- Cotter R. I., Lilley D. M. The conformation of DNA and protein within chromatin subunits. FEBS Lett. 1977 Oct 1;82(1):63–68. doi: 10.1016/0014-5793(77)80886-7. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Hilbers C. W., Shulman R. G. Nuclear magnetic resonance study of hydrogen-bonded ring protons in Watson-Crick base pairs. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2899–2901. doi: 10.1073/pnas.70.10.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Burd J. F., Larson J. E., Wells R. D. High resolution proton nuclear magnetic resonance investigation of the structural and dynamic properties of d(C15A15)-d(T15G15). Biochemistry. 1977 Feb 8;16(3):541–551. doi: 10.1021/bi00622a031. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Goodwin D. C., Brahms J. Form of DNA and the nature of interactions with proteins in chromatin. Nucleic Acids Res. 1978 Mar;5(3):835–850. doi: 10.1093/nar/5.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein D. G., Findlay J. B., Momii R. K., Luxon B. A., Kar D. Temperature dependence of the 31P chemical shifts of nucleic acids. A prode of phosphate ester torsional conformations. Biochemistry. 1976 Aug 24;15(17):3796–3803. doi: 10.1021/bi00662a023. [DOI] [PubMed] [Google Scholar]
- Hanlon S., Glonek T., Chan A. Comparison of the phosphorus magnetic resonance and circular dichroism properties of calf thymus DNA and chromatin. Biochemistry. 1976 Aug 24;15(17):3869–3875. doi: 10.1021/bi00662a034. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Appleby D. W., Bradley C. H. 31P magnetic resonance of DNA in nucleosome core particles of chromatin. Nature. 1978 Mar 9;272(5649):134–138. doi: 10.1038/272134a0. [DOI] [PubMed] [Google Scholar]
- Kearns D. R. High-resolution nuclear magnetic resonance studies of double helical polynucleotides. Annu Rev Biophys Bioeng. 1977;6:477–523. doi: 10.1146/annurev.bb.06.060177.002401. [DOI] [PubMed] [Google Scholar]
- Klevan L., Dattagupta N., Hogan M., Crothers D. M. Physical studies of nucleosome assemble. Biochemistry. 1978 Oct 17;17(21):4533–4540. doi: 10.1021/bi00614a027. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C., Roux B. Nucleosomes arrangement in chromatin. Nucleic Acids Res. 1978 Nov;5(11):4431–4449. doi: 10.1093/nar/5.11.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins D. E., Bryan P. N., Harrington R. E., Hill W. E., Olins A. L. Conformational states of chromatin nu bodies induced by urea. Nucleic Acids Res. 1977 Jun;4(6):1911–1931. doi: 10.1093/nar/4.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Hilbers C. W. Proton nuclear magnetic resonance investigations of fraying in double-stranded d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2651–2656. doi: 10.1021/bi00683a014. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Mutagen-nucleic acid complexes at the polynucleotide duplex level in solution: intercalation of proflavine into poly(dA-dT) and the melting transition of the complex. Biopolymers. 1977 Dec;16(12):2739–2754. doi: 10.1002/bip.1977.360161212. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Proton and phosphorus NMR studies of d-CpG(pCpG)n duplexes in solution. Helix-coil transition and complex formation with actinomycin-D. Biopolymers. 1976 Mar;15(3):533–558. doi: 10.1002/bip.1976.360150310. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Tonelli A. E. Assignment of the proton Nmr chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxy oligonucleotides in aqueous solution. Biopolymers. 1974;13(10):1943–1964. doi: 10.1002/bip.1974.360131003. [DOI] [PubMed] [Google Scholar]
- Ramm E. I., Vorob'ev V. I., Birshtein T. M., Bolotina I. A., Volkenshtein M. V. Circular dichroism of DNA and histones in the free state and in deoxyribonucleoprotein. Eur J Biochem. 1972 Feb 15;25(2):245–253. doi: 10.1111/j.1432-1033.1972.tb01690.x. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Suau P., Kneale G. G., Braddock G. W., Baldwin J. P., Bradbury E. M. A low resolution model for the chromatin core particle by neutron scattering. Nucleic Acids Res. 1977 Nov;4(11):3769–3786. doi: 10.1093/nar/4.11.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Rushizky G. W., Simpson R. T. DNase-sensitive sites in nucleosomes. Their relative suspectibilities depend on nuclease used. J Biol Chem. 1977 May 10;252(9):3003–3006. [PubMed] [Google Scholar]
- Whitlock J. P., Jr Staphylococcal nuclease and pancreatic DNase cleave the DNA within the chromatin core particle at different sites. J Biol Chem. 1977 Nov 10;252(21):7635–7639. [PubMed] [Google Scholar]
- Wong K. L., Kearns D. R. Investigation of the base-pairing structure of the anticodon hairpin from E. coli initiator tRNA by high-resolution nmr. Biopolymers. 1974;13(2):371–380. doi: 10.1002/bip.1974.360130212. [DOI] [PubMed] [Google Scholar]


