Abstract
Budding yeasts adhere to biotic or abiotic surfaces and aggregate to form biofilms, using wall-anchored glycoprotein adhesins. The process is paradoxical: adhesins often show weak binding to specific ligands, yet mediate remarkably strong adherence. Single-molecule atomic force microscopy, genomics, biochemistry, and cell biology have recently explained the puzzle, with Candida albicans Als adhesins as the paradigm. The strength of adhesion results partly from force-activated amyloid-like clustering of hundreds of adhesin molecules to form arrays of ordered multimeric binding sites. The various protein domains of eukaryotic adhesins cooperate to facilitate this fascinating new mechanism of activation.
Human and microbial amyloids
Protein amyloids (see Glossary) are justly infamous as markers of neurodegenerative diseases such as Alzheimer's, Parkinson's, and vCJD (mad cow disease), and so have been considered an abnormal state of proteins. However, it is clear that many sequences in the proteome can form amyloids [1], and that the ability to form ordered amyloid arrays can have positive functional consequences as well. In mammalian cells, functional amyloids include high-density storage granules of peptide hormones in exocytotic vesicles [2], protein templates for melanin deposition [3], and perhaps as factors in synaptic remodeling and learning [4]. In microbes, the list of functional amyloids is growing fast. Gram-negative adhesive curli and chaplins are discussed in an accompanying article by Blanco et al. [5]. Amyloids are also widely found in biofilms, although we are not yet sure what their role is [6]. In yeasts, heritable amyloid prions consisting of transcription regulators regulate gene expression and have been characterized both as disease states and as sources of evolutionary variability [7-10]. Amyloid interactions also mediate assembly of fungal hydrophobin cell coats, and are responsible for heterokaryon incompatibility and meoiotic drive in Podospora anserina [11, 12].
The recent discovery of functional amyloids in yeast cell adhesion proteins (adhesins) adds a new way of thinking about the roles of amyloids [13-16]. Cell surface clusters of amyloid-interacting adhesins can explain a long-standing paradox: there is very strong cell-cell and cell-substrate adhesion of fungi, but on the molecular level the assayed adhesins have weak ligand binding together with broad ligand specificity. Amyloid formation also helps to explain the unexpectedly complex domain structure of yeast adhesins.
Yeast cell adhesion proteins
Yeast adhesins mediate flocculation, invasive growth, biofilm, mat, and flor formation, and host-pathogen binding; and they share a basic architecture [17-20]. These adhesins bind carbohydrate ligands through N-terminal lectin domains or peptide ligands through tandem immunoglobulin (Ig)-like domains. C-terminal to these domains are variable numbers of β-sheet tandem repeat domains, long highly glycosylated extensions of up to 1000 amino acids, and at the C terminus, modified glycosylphosphatidylinisotol (GPI) anchors form covalent bonds to cell wall glucan fibers [17, 21]. Potential amyloid-forming sequences are found in various regions in the different adhesins; we are just beginning to identify those that are functional in vivo [14-16].
Candida albicans Als adhesins as a paradigm
The Candida albicans Als proteins are typical yeast adhesins. There are multiple isoforms encoded at 8 diploid loci in most strains [20]. Als adhesins share the common design features, shown in cartoon form in Figure 1a. Each Als protein has two tandem Ig-like adhesion domains [20, 22, 23]. This is followed by an evolutionarily conserved Thr-rich β-sheet domain with a 7-residue functional amyloid sequence, which mediates the molecular interactions described in this article. The Thr-rich domain is followed by highly glycosylated β-sheet tandem repeats (TR), and a long Ser-Thr-rich highly glycosylated stalk regions preceding the modified GPI anchor [17, 20, 24]. Als proteins bind to a wide variety of ligands with low affinity, but can mediate tight cell adhesion to both biotic and abiotic substrates [20, 25-28].
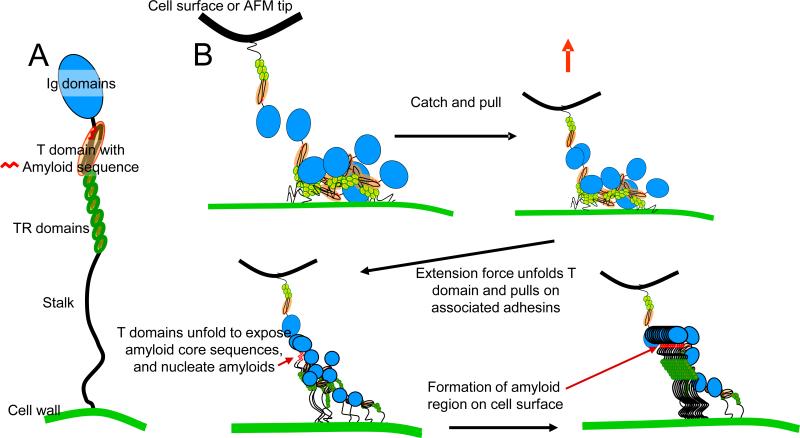
Figure 1.
Als proteins: multidomain adhesion proteins that form amyloids. (a) Model illustrating the domains of Als protein. (b) Model showing how homotypic binding of Als adhesins and mechanical stimulus lead to formation of cell surface amyloid nanodomains activating cell adhesion.
A remarkable trait of Als adhesins is that their different protein domains synergize to bind an extremely broad range of ligands. The N-terminal Ig-like regions bind to broad consensus “τφ+” peptides (τ, a residue common in turns; φ, a bulky hydrophobic residue; +, Lys or Arg), which constitute 2-5% of tripeptide sequences in the proteome [25]. Because this motif is usually buried in native proteins, they become much better ligands after denaturation, consistent with demonstrations that peptide flexibility is important, and that C. albicans preferentially binds to damaged regions of tissues and denatured proteins [22, 25, 29]. The list of peptide and protein ligands of Als proteins is impressively long, currently including dozens of different sequences in proteins and synthetic peptides [23, 25, 26, 29-32]. Als proteins bind to other Als proteins, as well as to non-related proteins on the surfaces of other yeasts and bacteria to form aggregates and co-aggregates, and may well be a factor in the high level of co-infection of C. albicans with other yeasts and bacteria [30, 33-37]. At least one Als protein can also bind to fucose-containing glycans, a finding that further expands the range of potential ligands in host tissues [27]. The peptide and glycan interactions have Kd values near 10-4-10-3 M, with rapid dissociation rates [27]. Binding to macromolecules is somewhat stronger, with Kd values ranging from 10-5-10-6 M, values considered to be ‘weak’ [27].
The TR domains of Als adhesins mediate hydrophobic effect interactions that are even weaker, and may have little or no specificity [24, 38]. Each TR domain is a 36 residue β-sheet fold with large surface-exposed regions of aliphatic and aromatic amino acid side chains [24, 38-40]. These regions mediate adhesion to hydrophobic surfaces to initiate biofilms, and also interact with each other to promote Als protein homotypic binding (Figure 1) [13, 16, 19, 24, 37, 38]. Therefore, the Als adhesins bind many ligands, but with binding energies of a few kcal/mol. Weak ligand binding affinities and hydrophobic-effect interactions of tandem repeat domains are shared by other adhesin families, including other C. albicans adhesins, the Saccharomyces Flo family mannose-specific lectins and the Candida glabrata Epa galectins [17, 41, 42].
Cellular activation
Als adhesins mediate binding of yeast to ligand-coated beads. About 5 min after the cells bind to beads, other yeasts begin to bind to the yeast already bound to beads as an increased ‘stickiness’ propagates around the cell surface of the bound cells [39, 43, 44]. Cell-cell binding is strong enough to deform the cells and is resistant to high shear forces in vortex mixing. This activation is not affected by inhibitors of gene expression or signal transduction, or even by heat-killing the cells. Instead, the process is perturbed by agents that perturb protein secondary structural changes or amyloid formation [13, 43, 44].
Amyloid formation
Recent work demonstrates that Als activation is a consequence of the formation of cell surface arrays of hundreds of closely spaced molecules. The close packing results in the physical equivalent of multivalency. Because the binding sites are close to each other, a ligand which dissociates from one adhesin molecule is much more likely to rebind to another [45]. This local concentration effect mirrors the interaction of pentavalent IgM and its ligands, a phenomenon known as antibody avidity [46]. The formation of amyloid adhesion domains leads to a large gain in free energy of cell binding, with a concomitant decrease in macroscopic Kd value, but retains the extremely broad range of effective ligands. Depending on the degree of ordering and the local concentration of ligands and adhesins, the effective Kd value and strength of adhesion for the cell surface as a whole increase exponentially with the number of interacting pairs of molecules [46, 47]. For a system where there are several hundred adhesin-ligand pairs, the chances of dissociation become vanishingly small. Indeed, physicians sampling oral and vaginal Candida colonies may need to scrape hard enough to dislodge attached epithelial cells. Thus, the formation of amyloid regions in Als-expressing yeast cells generates strong interactions from adhesins with weak ligand binding.
We propose that a 5 to 7-residue core sequence forms the amyloids through repeating interactions between identical sequences in Als molecules [1, 7, 48]. The Als core sequences are unusual in their amino acid composition: about 70% Ile, Val, and non-glycosylated Thr, all β-branched aliphatic residues. (These sequences score highly positively in the β-aggregate predictor TANGO [49] and a ‘steric zipper’ geometric screen [1], but not in the sequence-based predictor, WALTZ [50].) This composition is conserved in diverse adhesins from different yeasts and different gene families [14-16]. The hydrophobicity of Ile and Val probably promotes rapid amyloid formation, and the lack of ionizable groups is consistent with the observed pH and ionic insensitivity of adhesion activation [39]. Thus, the amyloid-like properties are complemented by high hydrophobicity, and are different from other amyloid sequences that are rich in Asn, Gln, and other polar residues [1, 50].
Als-mediated adhesion is activated by force-induced amyloids
The key discovery of amyloid nanodomains owes much to recent progress in atomic force microscopy (AFM) techniques, which can detect, localize, and force-probe single adhesin molecules on the surface of live cells [51, 52]. Such single-molecule AFM experiments directly demonstrate Als protein clusters on the cell surface following application of extension force to single molecules (Figure 2)[53], through a process modeled in Figure 1b.
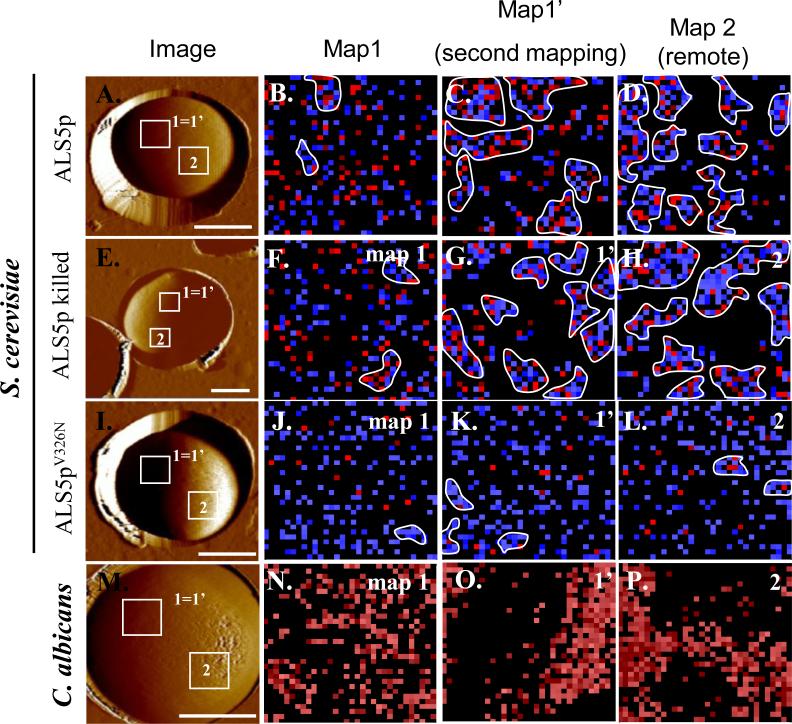
Figure 2.
Induction of surface nanodomains follows application of extending force to individual Als molecules on the surface of yeasts. On the left are AFM images of yeast trapped in a microporous membrane. The second column shows maps of position of Als proteins within the 1 μm square marked “1=1’.” Blue pixels show interactions of <150 pN and red pixels show interactions resistant to forces >150 pN. Clusters of 10 or more contiguous colored pixels are outlined in white [13]. The third column shows clustering of the Als molecules in a second mapping of the same region of the cell walls, and the last column shows similar clustering in a remote region (Map 2). Clustering is a result of surface amyloid formation, as shown by lack of clustering in cells expressing the non-amyloid V326N form of Als5p. Reprinted with permission from Garcia et al. [13].
An AFM probe is derivatized with an antibody to an epitope tag in Als5p, or with homologous Als5p protein itself, then tapped over the surface to find regions where Als molecules bind to the tip. As the AFM probe is withdrawn, force-extension curves for single Als molecules show successive unfolding of domains, and the extension lengths are consistent with initial unfolding of the Thr-rich domain, then the tandem repeats, and finally the Ig-like domains [40].
In an initial mapping of a 1μm2 area of the cell surface, there was a random pattern of Als5p molecules (Figure 2, second column) [13, 53]. However, upon re-mapping the same area, the Als5p molecules were clustered on the surface of the wall (Figure 2, third column). The adhesins are densely packed on the cell surface (Figure 1b), and we estimate about 5 molecules per pixel [45][17]. However, we propose that most of the molecules are cryptic, because they are bound to other adhesin molecules on the cell surface.
How do amyloids form on the cell surface?
The clustering is remarkable since the adhesins are anchored to the wall matrix. Amyloid interactions are possible because the long stalk regions allow each Als molecule to swivel around its attachment point to explore an area at least 240 nm in diameter [54][[17, 40, 53]. Therefore, each tethered molecule can interact with hundreds of others.
A model in Figure 1b shows how homotypic interactions between Als molecules on the surface of a cell could lead to amyloid nanodomain formation after stimulation by extension forces in the AFM or through cell-cell interactions. In the initial unactivated state, Als proteins would bind to each other through rapidly dissociable hydrophobic effect interactions of the TR domains, and probably ligand binding of Ig domains [22, 25, 28, 55]. Extension of single adhesin molecules by AFM should lead to partial unfolding of the Thr-rich domains, exposing the amyloid sequences, which sequentially immediately follow the Ig-like domains [22, 39]. This exposure makes a thermodynamic trap: a nearby molecule that has a transient exposure of the amyloid domain is likely to interact through its amyloid sequences, beginning to nucleate formation of the cell surface nanodomain [1, 48].
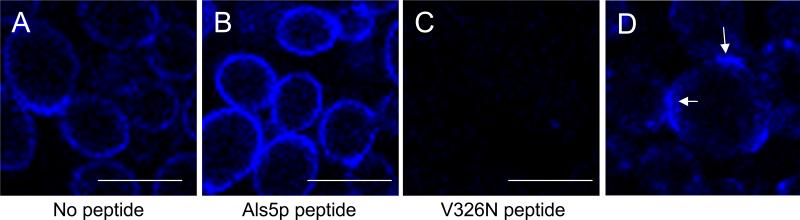
Importantly, both AFM and confocal microscopy demonstrate that the observed Als nanodomains result from amyloid interactions since the nanodomains do not form in Saccharomyces cerevisiae cells expressing Als5p with the non-amyloid-forming V326N single-site mutation (Figure 2 and Figure 3)[13, 40, 53]. Anti-amyloid dyes and a specific anti-amyloid peptide inhibit cellular aggregation as well as formation of nanoadhesion domains [13, 14]. Cells expressing non-amyloid Als5pV326N are rescued for adhesion activation and for nanodomain formation by inclusion of a specific amyloid-forming peptide. Thus single-molecule, genetic and chemical experiments show that the amyloid-forming region is important and functional in formation of nanoadhesion domains and subsequent formation of strong adhesive bonds between cells.
Figure 3.
Confocal imaging of amyloid patches on aggregated C. albicans Day 286. These are activated in a cell adhesion assay and stained with thioflavin T, 100 nM. (a) and (d) aggregated cells. (b) Fluorescence was enhanced after incubation with an amyloid-promoting peptide from Als5p. (c) Anti-amyloid V326N peptide inhibits aggregation and extinguishes fluorescence [13]. (d) Regions of enhanced fluorescence are marked where cells are tightly apposed. Bars represent 8 μm. Reprinted with permission from Garcia et al. [13].
Two lines of evidence suggest that the trigger for amyloid formation results from partial unfolding of an Als Thr-rich domain to expose the Als5p amyloid core sequence I325VIVATT331 and its homologs in other Als proteins (Table 1) [15, 16]. First, the Thr-rich domain appears to be the first to unfold in single molecule AFM [40, 53]. Second, soluble full-length Als5p forms amyloids in vitro under native-like conditions, suggesting that the core sequence can be exposed in these same conditions [14, 15]. These ideas support the model of Figure 1b, in which the Thr-rich domain is initially folded, but can partially unfold to expose the amyloid-forming sequence and initiate of amyloid nanodomains formation on the cell surface.
Table 1.
Ile-, Val-, and Thr-rich potential amyloid sequences from yeast and mammalian CAMsa
| Accession number | Sequence | Position in ORFb | TANGO β-aggregate %c | %IVTd | |
|---|---|---|---|---|---|
| Yeast proteins | |||||
| C. albicans Alsl Als3, Als5 | EAK99144, EAK93472, EAK99139 | IVIVATT | 325 | 93 | 83 |
| S. cerevisiae Flo1 | AAC09499 | TVIVI | 308, 1547 | 43 | 100 |
| S. cerevisiae Flo11 | NP_012284 | VVSTTV | 1036, 1367 | 75 | 83 |
|
Mammalian proteins | |||||
| Integrin α4 | NP 000876 | VFVYI | 340 | 94 | 60 |
| YIVLFY | 502 | 69 | 33 | ||
| N-CAM | AAB31836 | IVTIVGL | 660 | 98 | 71 |
| e-Cadherin | CAA78353.1 | VGVFII | 202 | 98 | 67 |
| VISVVTT | 325 | 83 | 87 | ||
| VVITTL | 391 | 62 | 83 | ||
| Galectin 4 | AAP35679.1 | LVFIVLA | 103 | 99 | 43 |
| Galectin 12 | EAW74155.1 | YVTTIF | 49 | 99 | 67 |
| SigLec | AAF34702.1 | SVFVTAL | 146 | 87 | 43 |
Another fascinating discovery is the ability of the nanodomains to grow and propagate across the entire cell. Using AFM and fluorescence microscopy, nanodomains were formed within ~30 min after force activation and migrated at a speed of ~20 nm·min−1, indicating that domain formation and propagation are slow, time-dependent processes [13, 53]. The amyloid nanodomains recruit nearby Als molecules to activate more nanodomains around the entire cell surface, and might even spread from cell to cell (Figures 1b, Figure 2 last column, and Figure 3)[13, 43, 53], so the yeast cells aggregate homotypically. There is circumstantial evidence that amyloid formation strengthens cell-to-cell bonding (Figure 3d) [13, 14, 16], because anti-amyloid small molecules or peptides causes disaggregation, consistent with amyloid bonds between cells.
Synergistic roles of the different Als domains in amyloid adhesion
Formation of amyloid adhesion nanodomains is an emergent property, based on the activities of the various domains in Als (and also in other yeast adhesins) [13-16]. The initial adhesion to a surface is through Ig-domain-mediated binding to peptide ligands or hydrophobic effect surface binding of Als Tandem repeats [23]. In the C-terminal region of each adhesin, the long, glycosylated ‘stalk’ elevates the N-terminal and mid-regions away from the cell surface and allows the flexibility needed for adhesin molecules to interact and form amyloids in vivo on the cell surface [21, 28]. These essential domain properties are somewhat variable, so that different Als adhesins are specialized to mediate diverse biological functions including adhesion, aggregation, biofilm formation, Fe acquisition, and cell wall architecture [18-20].
Amyloids: a general and versatile mechanism for activating cell adhesion?
Other yeast adhesins also show activation by amyloid formation [14]. In surveys, most adhesins had predicted amyloid forming regions, with amino acid composition rich in β-branched aliphatic amino acids, similar to the Als sequences [14]. Several of these sequences were tested as peptides or as secreted proteins, including the S. cerevisiae flocculins Flo1p and Muc1p/Flo11p. All tested sequences formed amyloids. Furthermore, similar to Als aggregation, Flo1p-mediated flocculation and Muc1p-mediated flocculation were inhibited by anti-amyloid dyes [14]. Thus, in limited tests, amyloid formation is a common strategy for formation of robust adhesions in yeast cell adhesion proteins.
A metazoan connection?
The mosaic modular structure of metazoan cell adhesion molecules (CAMs) and the presence of potential amyloid-forming TANGO-positive sequences are tantalizing clues that mammalian cell adhesion molecules might have the ability to form amyloids as well. Additionally, yeast surface amyloid nanodomains are densely packed cell adhesion molecules reminiscent of metazoan desmosomes and spot junctions, as well as clusters of integrins or cadherins [56]. Many mammalian CAMs have analogous or homologous domains which occur in a similar order from the N-terminus towards the C-terminus: Ig or lectin domains, Thr-rich mucin domains, small β-sheet-rich TRs (e.g. EGF domains) and poorly structured extracellular juxtamembrane ‘stalk-like’ regions [23]. Indeed, many of these CAMs have predicted amyloid forming sequences with composition similar to the Ile, Val, Thr-rich sequences of the fungal adhesins (Table 1). Thus, these cell adhesion proteins might be activated in a similar manner (Box 1), but this hypothesis is so far untested.
Conclusions
The complex domain structure of C. albicans Als adhesins is typical of fungal adhesins. Complementary single-molecule, genetic and chemical experiments have revealed that there is a synergistic relationship between the activities of the ligand binding domains, amyloid forming regions, and the less specific hydrophobic effect TR domains. The TR domains are responsible for initial fast, weak binding with hydrophobic surface, including homotypic binding to other TR domains. The peptide-binding Ig domains form bonds with diverse peptide ligands, with intermediate strengths of adhesion. Finally, the cells might be ‘cemented’ together and ’glued’ to the substrate through strong homotypic amyloid bonds, which make nanodomain arrays of hundreds to thousands of activated interacting cell adhesion proteins. The formation of amyloid domains allows formation of strong cell-cell and cell-substrate bonds to an astounding variety of adhesive ligands to mediate biofilm formation as well as fungal-host and fungal-fungal interactions.
Box 1. Outstanding questions.
What is the structure of the amyloid-like domains on the cell surface?
What is the influence of the amino acid composition of the amyloid core sequences? What is the effect of length and flexibility of the stalk regions?
Can we design anti-amyloid treatments to disrupt adhesion in host-pathogen interactions and ameliorate disease?
What are the roles of adhesin amyoids in biofilm, flor, and mat formation?
Can we integrate the tools of biology and nanotechnology, as discussed here, to discover amyloid-like adhesion domains in other pathogens?
Acknowledgements
Work at Brooklyn College was supported by NIH grant SC1 GM 083756. Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y.F.D. and D.A. are Senior Research Associate and Research Fellow of the FRS-FNRS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Adhesin: a microbial cell adhesion protein or glycoprotein. The yeast adhesins are 600-1600 amino acids in length, are highly N- and O-glycosylated, and are covalently crosslinked to cell wall polysaccharides
Amyloid: an insoluble fibrous aggregate of proteins characterized by assembly of identical or near-identical short sequences in many molecules to form ordered regions of β-sheets orthogonal to the fiber axis. Amyloid-forming sequences are common, but usually buried within protein domains and inaccessible for amyloid interactions. Amyloids are associated with cytotoxicity in neurodegenerative diseases of humans.
GPI anchor: a glycosyl phosphatydylinositol phospholipid covalently attached to the C- terminal carboxylate group of an extracellular protein. In most eukaryotes, the lipid fatty acid chains are intercalated into the plasma membranes. In fungi, the GPIs of some proteins are cleaved in the glycan moiety, leaving the lipid portion in the membrane. The reducing end of the protein-associated carbohydrate moiety is covalently added to a hydroxyl group in the cell wall glucan.
Protein domain: in the sense used here, a compactly and independently folded region within a protein sequence. Such folded domains are typically about 100-300 residues. Long proteins such as yeast adhesins are composed of many sequential domains. This is a more restricted definition than common usage of ‘domain’ as any sequence fragment with an identifiable function.
Literature Cited
- 1.Goldschmidt L, et al. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maji SK, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Si K, et al. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Blanco LP, et al. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012 doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen P, et al. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 7.Shewmaker F, et al. Structural insights into functional and pathological amyloid. J. Biol. Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel A, et al. Amyloid of the Candida albicans Ure2p prion domain is infectious and has an in-register parallel beta-sheet structure. Biochemistry. 2011;50:5971–5978. doi: 10.1021/bi200142x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberti S, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 11.Baxa U, et al. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv. Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- 12.Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin. Cell Dev. Biol. 2011;22:460–468. doi: 10.1016/j.semcdb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Garcia MC, et al. A role for amyloid in cell aggregation and biofilm formation. PloS One. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsook CB, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otoo HN, et al. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell. 2008;7:776–782. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobbs AH, et al. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae Reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell. 2010;9:1622–1634. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dranginis AM, et al. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell. 2011;10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoyer LL, et al. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family--a sticky pursuit. Med. Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frieman MB, Cormack BP. The omega-site sequence of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol Microbiol. 2003;50:883–96. doi: 10.1046/j.1365-2958.2003.03722.x. [DOI] [PubMed] [Google Scholar]
- 22.Salgado PS, et al. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15775–15779. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan QT, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank AT, et al. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot. Cell. 2010;9:405–414. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotz SA, et al. Degenerate peptide recognition by Candida albicans adhesins Als5p and Als1p. Infect Immun. 2004;72:2029–34. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaur NK, et al. Candida albicans and Saccharomyces cerevisiae expressing ALA1/ALS5 adhere to accessible threonine, serine, or alanine patches. Cell Commun Adhes. 2002;9:45–57. doi: 10.1080/15419060212187. [DOI] [PubMed] [Google Scholar]
- 27.Donohue DS, et al. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol. Microbiol. 2011;80:1667–1679. doi: 10.1111/j.1365-2958.2011.07676.x. [DOI] [PubMed] [Google Scholar]
- 28.Loza L, et al. Functional analysis of the Candida albicans ALS1 gene product. Yeast. 2004;21:473–82. doi: 10.1002/yea.1111. [DOI] [PubMed] [Google Scholar]
- 29.Gaur NK, Klotz SA. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology. 2004;150:277–84. doi: 10.1099/mic.0.26738-0. [DOI] [PubMed] [Google Scholar]
- 30.Klotz SA, et al. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med. Mycol. 2007;45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 31.Klotz SA, et al. Inhibition of adherence and killing of Candida albicans with a 23-Mer peptide (Fn/23) with dual antifungal properties. Antimicrob Agents Chemother. 2004;48:4337–41. doi: 10.1128/AAC.48.11.4337-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida RS, et al. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klotz SA, et al. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 2007;59:401–406. doi: 10.1016/j.diagmicrobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Thorn JL, et al. Postmortem candidaemia: marker of disseminated disease. J. Clin. Pathol. 2010;63:337–340. doi: 10.1136/jcp.2009.070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klotz SA, Lipke PN. The Perfect Adhesive. Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology. 2010:838–844. [Google Scholar]
- 37.Silverman RJ, et al. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauceo JM, et al. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot. Cell. 2006;5:1664–1673. doi: 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaur NK, Klotz SA. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun. 1997;65:5289–94. doi: 10.1128/iai.65.12.5289-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alsteens D, et al. Unfolding individual Als5p adhesion proteins on live cells. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verstrepen KJ, et al. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–90. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frieman MB, et al. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 2002;46:479–492. doi: 10.1046/j.1365-2958.2002.03166.x. [DOI] [PubMed] [Google Scholar]
- 43.Rauceo JM, et al. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun. 2004;72:4948–55. doi: 10.1128/IAI.72.9.4948-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaur NK, et al. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect Immun. 1999;67:6040–7. doi: 10.1128/iai.67.11.6040-6047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen ZM, et al. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J Biol Chem. 2001;276:15768–75. doi: 10.1074/jbc.M010421200. [DOI] [PubMed] [Google Scholar]
- 46.Fitzgerald J, Lugovskoy A. Rational engineering of antibody therapeutics targeting multiple oncogene pathways. MAbs. 2011;3:299–309. doi: 10.4161/mabs.3.3.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantor CR, Schimmel PR. The behavior of biological macromolecules. W. H. Freeman; 1980. pp. 878–893. [Google Scholar]
- 48.Jahn TR, et al. The common architecture of cross-beta amyloid. J. Mol. Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Escamilla AM, et al. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 50.Maurer-Stroh S, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 51.Dufrene YF. Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Microbiol. 2008;6:674–680. doi: 10.1038/nrmicro1948. [DOI] [PubMed] [Google Scholar]
- 52.Muller DJ, et al. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 53.Alsteens D, et al. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jentoft N. Why are proteins O-glycosylated? Trends Biochem. Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 55.Sheppard DC, et al. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–9. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- 56.Turner ML. Cell adhesion molecules: a unifying approach to topographic biology. Biol. Rev. Camb. Philos. Soc. 1992;67:359–377. doi: 10.1111/j.1469-185x.1992.tb00729.x. [DOI] [PubMed] [Google Scholar]