Abstract
Bacteriophages are increasingly being utilized and considered for various practical applications, ranging from decontaminating foods and inanimate surfaces to human therapy; therefore, it is important to determine their concentrations quickly and reliably. Traditional plaque assay (PA) is the current “gold standard” for quantitating phage titers. However, it requires at least 18 h before results are obtained, and they may be significantly influenced by various factors. Therefore, two alternative assays based on the quantitative real-time polymerase chain reaction (QPCR) and NanoSight Limited (NS) technologies were recently proposed for enumerating phage particles. The present study compared the three approaches' abilities to quantitate Listeria monocytogenes-, Escherichia coli O157:H7- and Yersinia pestis-specific lytic phages quickly and reproducibly. The average coefficient of variation (CVS) of the PA method including all three phages was 0.15. The reproducibility of the PA method decreased dramatically when multiple investigators performed the assays, and mean differences of as much as 0.33 log were observed. The QPC R method required costly equipment and the synthesis of phage-specific oligonucleotide primers, but it determined phage concentrations faster (within about 4 h) and more precisely than did PA (CVS = 0.13). NS technology required costly equipment, was less precise (CVS = 0.28) than the PA and QPCR methods, and only worked when the phages were suspended in clear medium. However, it provided results within 5 min. After the overall correlation is established with the PA method, either of the two assays may be useful for quickly and reproducibly determining phage concentrations.
Key words: bacteriophage, phage, plaque assays, phage titer
Introduction
Viruses that kill bacteria by intracellular lysis were first identified during the early part of the 20th century by Frederick Twort and Felix d'Herelle who called them bacteriophages (i.e., Greek for “bacteria-eaters”) or phages.1 Since their discovery, phages were used for various practical applications, including in human and veterinary medicine (reviewed in refs. 2–4). They also served as model microorganisms for some of the most significant discoveries in the field of molecular biology, including deciphering the genetic code and the discovery of the transduction phenomenon.5 Although therapeutic applications of bacteriophages were largely forgotten in the west after the widespread acceptance of antibiotics during the 1940s and 1950s, the emergence of antibiotic-resistant bacteria and the increasing popularity of environmentally-friendly, “green” technologies has rekindled interest in practical uses for phages. Indeed, several bacteriophage-based products recently were approved for food safety applications and are being commercialized in the United States and western Europe.6 Also, at least two phage preparations were examined during recent clinical trials in the United States and United Kingdom,7,8 and more trials are being planned (http://clinicaltrials.gov/ct2/results?term=bacteriophage). One of the key requirements for developing “phage technology” for practical applications is the ability to quantitate viable bacteriophages accurately and reproducibly, so that rigorous studies of their potencies, minimal effective doses, side effects, etc., can be performed.
Traditionally, three methods have been used to quantitate bacteriophages: (1) plaque counts on agar plates seeded with the bacteria in which the phages can propagate, (2) a dilution method, where bacterial lysis is used as an indicator of phage presence and (3) measuring the length of time required to lyse a standardized bacterial suspension.9 However, only the first method (originally developed by d'Herelle in 1917) has been generally useful for determining actual phage titers, and it has been used—with minor modifications—since then in numerous laboratories throughout the world. The basis of that traditional phage assay (PA) involves the interaction of a single lytic phage particle and a permissive bacterium, which results in the host bacterium's lysis and the release of newly formed phage progeny. When mixtures of the phages and their specific bacterial host cells in molten soft agar are poured onto the surface of a base layer of nutrient-containing agar supporting bacterial growth, the host cells resume their growth. In areas where phages are not present, the bacteria grow to the stationary phase and form a confluent, opaque layer or “lawn” in the soft agar overlay. However, in areas where phages are present, the phage progeny released from each infected bacterium will lyse neighboring bacteria and produce a growing zone of liberated phages, which eventually becomes visible to the naked eye as a clear circular area or “plaque” in the otherwise confluent lawn. The plaques are counted, and the phage concentration/titer is commonly expressed as the number of plaque-forming units (PFU)/mL of the assayed preparation. Although the PA is currently considered the “gold standard” for determining phage concentrations, it is not without drawbacks, including (1) poor reproducibility; e.g., a change in the salt concentration of the nutrient agar can change the viable titer of T2 phage preparations by more than ca. 1,000-fold, and assaying the same phage against different bacterial host strains will often yield a different titer against each host and (2) a relatively long time (usually 18 to 24 h) is required to complete the assay.9,10
Phage particles also can be enumerated by transmission electron microscopy (TEM) after negative staining or by epifluorescence microscopy after staining with DNA fluorochromes. However, those techniques also have limitations; e.g., epifluorescence microscopy suffers from significant background problems and the equipment for TEM is too expensive to make that method commonly available.10 Recently, two new rapid assays were proposed for determining phage concentrations: (1) a quantitative real-time PCR (QPCR)-based approach,11 and (2) a nanoparticle tracking analysis (NTA)-based approach using NanoSight Limited (NS) technology. The QPCR method is based on measuring the total fluorescence generated during each PCR cycle as the phage-containing specimen is amplified; i.e., performing time-dependent analysis of phage DNA molecule accumulation. The underlying idea behind the assay is that establishing the correlation between DNA counts (as determined by the QPCR) and the number of viable phage particles (as determined by PA) will enable one to use the former to determine phage concentrations in aqueous solutions. The NTA-based approach utilizes laser-illuminated optical microscopy for direct, real-time visualization of nanoparticles in a clear liquid. The nanoparticles are detected as light-scattering centers moving under Brownian motion, and they are counted in a few seconds or a few minutes. NTA using NS technology has been used to analyze various nanoparticles in suspension and, more recently, at least one bacteriophage preparation has been examined by that procedure (www.nanosight.com/applications/biological-nanoparticles/bacteriophage). The goal of the current study was to compare the speed and precision of the classical PA, the QPCR-based approach and NTA using NS-based technology for determining phage concentrations.
Results and Discussions
Plaque assay (PA).
PA studies were performed within a seven-day time period. The CVS ranged from 0.11 to 0.24, 0.06 to 0.26 and 0.06 to 0.22 for ECML-117, List-36 and YpP-G, respectively. The mean CVS were tightly grouped: 0.15 for ECML-117, 0.14 for List-36 and 0.13 for YpP-G. The overall range of the CVS was 0.06 to 0.26, with a mean value of 0.15 (Table 1). The CVS may be used to estimate the highest and lowest values in a 90% CI for any given sample (using the Z value for 90% confidence), by the following formula (derived from ref. 14): VH = V × (1 + CVS × Z90) and VL = V × (1 - CVS × Z90), where V = sample value, VH = highest value for 90% confidence, VL = lowest value for 90% confidence and Z90 = Z value for 90% confidence (Z = 1.6445).15 For example, a phage preparation with a PA-determined titer of ca. 1 × 108 PFU/mL and a CVS of ca. 0.15, would have a low interval of ca. 7.5 × 107 PFU/mL and a high interval of ca. 1.3 × 108 PFU/mL, with 90% confidence. To put this into further perspective, phage specimens whose titers (determined by the same investigator in the same laboratory) are within that range may be considered to be identical to a titer of ca. 1 × 108 PFU/mL.
Table 1.
CVS for the PA, NS-based and QPC R-based methods
| Method | Phage | Expected titer (PFU/mL) | Phage-specific CVS | Overall CVS | ||||
| 1 × 108 | 1 × 109 | 1 × 1010 | Range | Average | Range | Average | ||
| ECML-117 | 0.11–0.23 | 0.12–0.14 | 0.12–0.24 | 0.11–0.24 | 0.15 | |||
| PA | List-36 | 0.14–0.26 | 0.13–0.23 | 0.06–0.08 | 0.06–0.26 | 0.14 | 0.06–0.26 | 0.15 |
| YpP-G | 0.22 | 0.12 | 0.06 | 0.06–0.22 | 0.13 | |||
| ECML-117 | 0.22 | 0.28 | 0.30 | 0.22–0.30 | 0.27 | |||
| NS | List-36 | 0.27 | 0.86a | 0.27 | 0.27 | 0.27 | 0.22–0.35 | 0.28 |
| YpP-G | 0.30 | 0.28 | 0.35 | 0.28–0.35 | 0.31 | |||
| ECML-117 | 0.09 | 0.09 | 0.08 | 0.08–0.09 | 0.09 | |||
| QPCR | List-36 | 0.20 | 0.11 | 0.19 | 0.11–0.20 | 0.17 | 0.08–0.20 | 0.13 |
| YpP-G | 0.15 | 0.09 | 0.15 | 0.09–0.15 | 0.13 | |||
The outlier excluded from further analysis. n = 390. For the PA, EC ML-117 and List-36 phages: 30 specimens per expected titer per phage = 180. For the YpP-G phage: 10 specimens per expected titer = 30. For all other assays and phages: 10 specimens per phage per expected titer = 180. Total data points for all specimens: 390.
The CVS was reduced as the phage concentration increased: they averaged 0.18, 0.14 and 0.12 for phage preparations containing 108, 109 and 1010 PFU/mL, respectively. The data presented in Table 1 provide insight into the expected variations and the possible range of titration errors when evaluating PA-determined phage titers obtained by different investigators in the same laboratory.
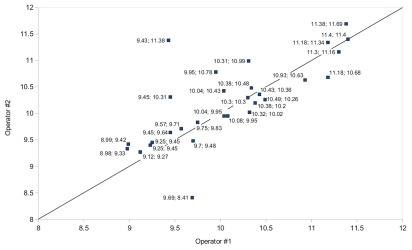
The reproducibility/precision of the PA-based method among various laboratories was poor. For example, only one dataset (for ECML-117 at an expected concentration of 1010 PFU/mL) was not significantly different (p > 0.05) when the phages were assayed in two different participating laboratories (Intralytix in Maryland and EPI UF in Florida). However, all other matched means were significantly different from one another (p < 0.05). Thus, in order to determine the range of possible variance in phage titers determined by PA, 10 Listeria phages (including List-36) were analyzed by the PA assay. The purpose of the analyses was to determine the variance between paired mean PA values obtained on separate days by different investigators during a time period when the phage preparations should be completely stable; i.e., when changes in their titers should not occur. We chose to compare the mean values because they represent the working values upon which decisions are made, rather than the individual triplicate values obtained by each titration. Twenty-nine paired titrations were performed, within seven days of one another, by at least two different investigators quantitating the 10 different Listeria phages. Each titration value was subjected to a log10 transformation and the calculated difference values between the titrations yielded 29 different values, one for each pair of titrations. Paired two tailed t test was used to calculate the means, standard errors of the means and 95% CI (Fig. 1). We found that the (1) mean difference between titrations was −0.1238 log, (2) standard error of the mean difference was 0.1008 log, (3) upper 95% CI of the mean difference between titrations was 0.3303 log and (4) lower 95% CI of the mean difference between titrations was 0.0827 log. Our data suggest that titrations whose log10 transformation values are within 0.33 log of one another may be considered equivalent since the differences are within the 95% CI of the mean differences between the titrations. Furthermore, if the absolute values of the differences between the titrations were used to calculate the mean differences, the range was even larger at ca. 0.5 log. This analysis took both inter-operator and day-to-day variations into account. To put the above numbers into practical perspective: if investigators in one laboratory determine the mean titer is 1 × 108 PFU/mL, investigators in other laboratories may expect the same phage preparation's titer will range from 4.7 × 107 to 2.1 × 108 PFU/mL (based on mean difference of 0.33 log), assuming that all participants use the same host strain and standardized PA for their analyses.
Figure 1.
Range of possible variance in phage titers determined by the PA. The diagonal line represents perfect agreement. The numeric values before and after the semi-colons were obtained by two different investigators.
The possible effect of the agar overlay's volume on phage titer enumeration was examined during additional titration studies performed with each phage preparation (ca. 1010 PFU/mL), using agar overlay volumes of 2, 3 and 4 mL. The values were not significantly different from one another and from the results obtained with the initial 1 mL agar overlay (data not shown). Thus, within those parameters, the volume of the agar overlay does not significantly influence the results.
QPCR-based analyses.
The CVS for the QPCR-based analyses ranged from 0.08 to 0.09, 0.11 to 0.20 and 0.09 to 0.15 for ECML-117, List-36 and YpP-G, respectively (Table 1). The mean CVS were 0.09, 0.17 and 0.13 for ECML-117, List-36 and YpP-G, respectively (the differences were not statistically significant [p > 0.05]). The overall range of the CVS for the phage titers determined by the QPCR-based method was 0.08 to 0.20, with a mean value of 0.13 (Table 1). The CVS and the above-described formula may be used to calculate the range of titration values obtained by the QPCR-based method. For example, a phage preparation with a QPCR method-determined titer of ca. 1 × 108 PFU/mL and a CVS of ca. 0.13 (Table 1), would have a low interval of ca. 7.9 × 107 PFU/mL and a high interval of ca. 1.2 × 108 PFU/mL, with 90% confidence.
The QPCR-based method, which had the lowest CVS, was the most precise of the three assays we examined. However, since that assay involves amplifying phage DNA, we were concerned that it may overestimate the concentration of viable phage particles by detecting free phage DNA and/or DNA inside damaged phage capsids rather than the DNA in viable phage particles. Although, we did observe that the phage concentrations estimated by the QPCR method were higher than those obtained by PA, we found that was not the case with all phages. Thus, the correlation between the results obtained by QPCR (which measures the DNA of both viable and nonviable phages) and PA (which only measure viable phage particles) must be established for each phage before using the QPCR-based assay to quantitate the number of its viable particles in a specific preparation. Moreover, the correlation may need to be updated every time the production, purification and general handling practices that might affect phage viability are changed.
Some shortcomings of the QPCR-based assay include its: (1) requirement for expensive equipment (although real-time PCR machines are becoming increasingly common in many laboratories), (2) requirement for optimizing PCR amplification conditions and ongoing calibration with known standards analyzed concurrently with the test samples and (3) need for phage-specific oligonucleotide primers. The latter necessitates obtaining at least partial phage sequences and poorly designed primers may form self-dimers and decrease PCR efficiency.11 The advantages of the QPCR-based assay include its (1) good precision/reproducibility which correlates well with that of the PA, (2) its requirement for only a few reagents, (3) high throughput (many phage preparations may be analyzed concurrently when the assay is performed in 96-well microtiter plates), (4) fairly fast turn-around time of 3 to 4 h and (5) potential for greater precision than PA performed by investigators in different laboratories (Table 2).
Table 2.
Key differences and pros and cons of the PA, QPC R-based and NS-based methods
| Characteristics | Method | ||
| PA | QPCR-based | NS-based | |
| Handling requirements | Needs many dilution steps requiring numerous pipetting steps. Requires manual (visual) plaque recognition and counting. | Specimen pretreatment is required. Requires only minimal pipetting steps. | Requires multiple software and hardware adjustments. Specimen chamber must be cleaned after each reading. |
| Time requirements | 18 to 24 h | 3 to 4 h | ≤5 min |
| Cost considerations | Specialized equipment not required and there is only a moderate reagent cost per result. | There is an upfront equipment cost of $40,000 to $50,000, but a low reagent cost per result. | There is an upfront equipment cost of $40,000 to $60,000, but no additional reagent costs. |
| Coefficient of variation (CVS) | 0.15 | 0.13 | 0.28 |
| Optimal concentration range | 30 to 300 PFU/mL | 106 to 108 PFU/mL | 107 to 109 PFU/mL |
| Additional pros | Involves direct enumeration of viable phage particles. | Reproducibility among different laboratories is expected to be good. | Reproducibility among different laboratories is expected to be good. |
| Additional cons | Results are affected by numerous factors and are poorly reproducible among different laboratories. | Abnormal storage conditions likely to affect phage viability may deleteriously impact correlation with PA Requires the synthesis of phage-specific oligonucleotide primers. | Abnormal storage conditions likely to affect phage viability may deleteriously impact correlation with PA Results only are accurate when the phages are in clear solutions rather than in cloudy ones; e.g., in the host bacterium's broth culture. |
NS-based analyses.
The CVS of the NS-based assays ranged from 0.22 to 0.30, 0.27 to 0.86 and 0.28 to 0.35 for ECML-117, List-36 and YpP-G, respectively (Table 1). The mean CVS values were 0.27 for ECML-117 and List-36, and 0.31 for YpP-G. One specimen of List-36 had an abnormally high CVS of 0.86 at the expected concentration of 109 PFU/mL. After that one sample was rejected using Dixon's Q test with 99% confidence (Q = 0.80), the overall range of the CVS values were calculated to be 0.22 to 0.35, with a mean CVS of 0.28 for the NS method (Table 1). Thus, the CVS values and the above-described formula may be used to calculate the range of titration values obtained by the NS method. For example, a NS-obtained value of ca. 1 × 108 PFU/mL and an overall CVS of 0.28 (Table 1) would have a low interval of ca. 5.4 × 107 PFU/mL and a high interval of ca. 1.5 × 108 PFU/mL, with 90% confidence. In other words, NS-obtained values in the range of 5.4 × 107 to 1.5 × 108 PFU/mL may be considered equivalent to 1 × 108 PFU/mL.
The NS method was found to be affected by background particle distributions, and many commercially available media for propagating bacteria (including BHI broth and LB broth) elicited high levels of “background noise,” which made it very difficult (if not impossible) to interpret the results. Diluting phage specimens to a concentration of ca. 1 × 107 PFU/mL, using 0.9% saline or deionized water, did not entirely alleviate the problem. Diluting further to reduce the background noise was not feasible because the optimal reading of the NS system is in the range of 107 to 109 PFU/mL.
The NS- and QPCR-based assays measure both viable and nonviable phage particles simultaneously. Thus, NS and PA data must be obtained for each phage and its production and storage protocols before using the NS-based method to determine viable concentrations of that phage. Some drawbacks of the NS method include: (1) it requires fairly expensive equipment (although prices are likely to come down in the future), (2) it only reliably detects phages with a size larger than 40 to 50 nm (which is a minor concern since most phages used for food safety, environmental decontamination and clinical applications are larger than that), (3) the phages need to be suspended in a clear medium (which is a significant limiting factor because it negates one of the most attractive potential applications of the technology: using it to optimize phage production) and (4) in order for the results to be reliable, the phage concentrations must be within a range of 107 to 109 PFU/mL. However, NS provides results within an impressive ≤5 min timeframe, which is significantly faster than does PA and the QPCR method (18 to 24 h and 3 to 4 h, respectively), and its performance does not require any additional reagents. Once optimized, it is likely that the NS-based method will be reproducible among various laboratories, with accuracy comparable to PA performed by various investigators (but significantly faster).
Comparison of phage titers determined by the PA and by the QPCR- and NS-based assays.
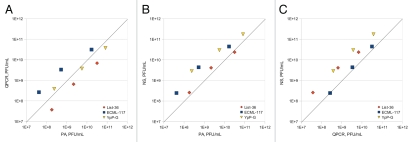
At the present time, the PA is considered to be the gold standard for determining phage titers and it is the only method among the three approaches we examined that directly enumerates viable phage particles (QPCR and NS methods quantitate both viable and nonviable phages). Therefore, as previously mentioned, correlations must be established between the data obtained by the PA and the other two assays before using only the QPCR and/or NS methods to determine viable phage concentrations. Furthermore, even after the correlations are established, it may be prudent to continue to use PA to verify the outcomes of the QPCR- and NS-based analyses, at least for a period of time until the “correction factor” and reproducibility of testing is established with absolute certainty for the bacteriophage under investigation. The data presented below and in Figure 2 provide preliminary insight into the correlation among the phage titers determined by the three assays.
Figure 2.
Correlation between the results obtained by the PA and by the NS- and QPCR-based assays for determining phage titers. The symbols represent matched-pair, ten-point data set means plotted on a log scale. The diagonal lines represent perfect agreement. (A) Results obtained with the NS method and the PA. (B) Results obtained with the QPC R method and the PA. (C) Results obtained with the NS- and QPC R-based assays.
In all instances, R2 values were calculated with n = 3. In general, the QPCR-based assay under-reported the number of viable phage particles determined by the PA for List-36, over-reported the number for ECML-117 and yielded mixed results for YpP-G (Fig. 2A). Among the three phages examined, the strongest correlation observed between the QPCR- and PA-determined means of the titers was R2 = 1 for YpP-G with an average difference of 50%, followed by R2 = 0.99 for List-36 with an average difference of 76% and R2 = 0.98 for ECML-117 with an average difference of 38%. Thus, the values determined by the QPCR method were ca. 0.5-fold different (higher or lower), 0.76-fold lower and 3.9-fold higher for YpP-G, List-36 and ECML-117, respectively, than those determined by the PA. Although the percent- and/or fold-differences (i.e., the “correction factor”) seem high, the R2 values determined for all three phage preparations were excellent (ranging from a very high value of 0.98 for ECML-117, to a perfect match of 1 for YpP-G), which suggests that the results obtained by the QPCR and PA methods correlate very well with one another. In other words the viable phage titers obtained by the QPCR-based assay are essentially identical to those obtained by the PA, for the same sample, when an appropriate adjustment is made with the above-determined “correction factor.” We expected (as mentioned above) that the QPCR-based assay would over-report the data obtained by the PA, which turned out to be the case for many, but not for all, of the specimens examined. Also, half of the readings obtained for YpP-G were over-reported and half-were under-reported. At the present time, it is unclear why some readings obtained by the QPCR method were under-reported, but possible contamination of the phage specimens or PCR reaction mixture, which interfered with PCR-mediated amplification, may be one explanation.
The NS-based assay over-reported the number of viable phage particles determined by the PA, except when List-36 was enumerated at a concentration of 1010 PFU/mL, which was slightly under-reported (Fig. 2B). Among the three phages examined, the strongest correlation between the NS- and PA-determined means of the titers was R2 = 1 for YpP-G with an average difference of 544%, followed by R2 = 0.98 for List-36 with an average difference of 52% and R2 = 0.98 for ECML-117 with an average difference of 461% (Fig. 2B). In other words, the phage titers determined by the NS method were ca. 0.5-fold higher, 4.6-fold higher and 5.4-fold higher for List-36, ECML-117 and YpP-G, respectively, than they were when the phage preparations were quantitated by the PA. A possible explanation for these differences is that NS enumerated viable phage particles, non-viable phage particles and background noise where as PA only enumerated viable phage particles. The correlation between the phage titers determined by the NS method and PA was very good, with an R2 ranging from a very high value of 0.98 for List-36 and ECML-117, to a perfect R2 of 1 for YpP-G. Thus, as with the QPCR-determined method, we expect that the titers obtained by the NS-based assay adjusted by the above-determined average “correction factor” will very accurately predict PA-determined titers.
The NS method over-reported the number of viable phage particles determined by the QPCR-based assay, except when ECML-117 was enumerated at a concentration of ca. 108 PFU/mL. Among the three phages examined, the strongest correlation between the results obtained with the NS and QPCR methods was R2 = 1 for ECML-117 with an average difference of 27%, followed by R2 = 0.99 for List-36 (with an average difference of 452%) and YpP-G (with an average difference of 568%) (Fig. 2C).
Materials and Methods
Bacteriophages.
Three different bacteriophages were used during our studies: (1) Listeria monocytogenes-specific bacteriophage List-36 (ATCC #PTA-5376), (2) Escherichia coli O157:H7-specific bacteriophage ECML-117 (ATCC #PTA-7950) and (3) Yersinia pestis-specific bacteriophage YpP-G. List-36 and ECML-117 are part of the commercial phage preparations ListShield™ and EcoShield™, respectively (www.intralytix.com). YpP-G is a component of a diagnostic phage preparation for detecting Y. pestis, which was produced in the former Soviet Union and was kindly supplied by Dr. Nikoloz Tsertsvadze (Georgian National Center for Disease Control; Tbilisi, Georgia). Based on the classification scheme of Ackermann and Berthiaume,12 List-36, ECML-117 and YpP-G belong to the Siphoviridae (List-36), Myoviridae (ECML-117) and Podoviridae (YpP-G) families of double-stranded DNA-containing bacteriophages, respectively. Studies of ECML-117 and List-36 were done at Intralytix and the Emerging Pathogens Institute of the University of Florida in Gainesville (EPI-UF), under BSL-2 biosafety conditions required for their bacterial host species; i.e., E. coli O157:H7 and L. monocytogenes, respectively. Studies of YpP-G were done at the EPI-UF, under BSL-3 biosafety conditions required for its host bacterium (Y. pestis). ECML-117 and List-36 phages were filter purified and buffer exchanged via diafiltration into sterile PBS and 100 mM saline, respectively, while YpP-G phages were filter purified in the original growth media.
Equipment calibration, culture media and reagents.
Prior to use, all equipment and pipettes were inspected and calibrated. Reconstituted culture media and most of the reagents were prepared at the sites taking part in the studies. Luria-Bertani (LB) Broth and Luria-Bertani Agar were purchased as powders from Neogen Corporation (LB broth, 7279B, LB agar, 7213B). The soft agar overlay with which the phages and their bacterial host species were mixed contained sterile isotonic saline from Fisher BioReagents (0.9% saline, 50-700-066), Tris-HCl buffer, pH 7.5 (50 ml/L) from Teknova (T1075) and agar-agar (7 g/L) from EMD Chemicals (1.01614). Brain-heart infusion (BHI) broth and agar were purchased from Becton Dickinson (BHI broth, 237200, BHI agar, 241810).
Preparation of phage specimens for assaying.
PA was used to determine the initial titers of all three bacteriophage preparations, after which they were diluted (with sterile isotonic saline) to yield preparations with phage concentrations of 1010, 109 and 108 PFU/mL. Mixing was done with a Mini-Vortexer VM-3000 (VWR Scientific, 945300) after each dilution step. Aliquots of the diluted List-36 and ECML-117 phage preparations were stored at Intralytix for subsequent analysis and they were also express-mailed (on wet ice) to the EPI-UF. Studies of YpP-G were done only at the EPI-UF.
Plaque-based assays (PA).
PA was performed essentially as described by Adams.9 Briefly, LB agar in Petri dishes were streaked (for purity and well-isolated colonies) with thawed specimens of the bacterial host strains (stored in 70% LB broth/30% glycerol in a −80°C freezer) and the inoculated media were incubated overnight at 30°C, 37°C and 28°C for the L. monocytogenes, E. coli and Y. pestis hosts, respectively. Aliquots (10 mL) of sterile LB broth were inoculated with one colony of the appropriate host strains and incubated (2 to 6 h, with shaking) until the optical density reached an OD600 of 0.2 to 0.3. The phage-host cell interaction mixtures consisted of 10 aliquots (1 mL each) of each diluted phage preparation (103, 102 and 101 PFU/mL) and 100 µL of the appropriate host culture in sterile, 10-mL graduated Falcon culture tubes. The phage-specific bacterial host-soft agar overlays were prepared by incubating (10 min, room temperature) the phage-host mixtures and gently mixing them with aliquots (3 mL) of molten (45° to 50°C) soft agar (0.7% w/v). After the individual mixtures were quickly but very gently poured onto appropriate solid culture media in Petri dishes (10 cm diameter), the dishes were rotated gently upright on a flat surface to distribute the mixtures evenly, after which they were incubated (room temperature, 15 min) until the top layer solidified. Subsequently, the Petri dishes were inverted and incubated (18 to 20 h) at the appropriate temperature. Petri dishes exhibiting between 30 to 300 well isolated plaques were used to calculate the phage titers and for subsequent statistical analyses.
QPCR-based assays.
The assays were done at the EPI-UF. Primer sets specific to each phage were required for the assays, and they were designed after determining full-genome sequences of each phage, using the Primer3 software.13 After identifying the required primer sequences, the primers were synthesized by Invitrogen Corporation on a fee-for-service basis. The primer pairs for each bacteriophage were as follows: Ec3F (5′-GCG ATA GTT GCA TCC GTC TT-3′) and Ec3R (5′-GCA GGA AGT TGT GAC CGA CT-3′) for ECML-117; Lp3F (5′-GCA GAA GCT TCG GTA CCT TTA-3′) and Lp3R (5′-CAA GCC GTG GTA CGG TTT AT-3′) for List-36; and Yp1F (5′-CGC GGT ACT CTA GGA TGA GC-3′) and Yp1R (5′-CTA TTG GGG AAG GGG GTT TA-3′) for YpP-G.
The assays were performed, with minor modifications, as described by Edelman and Barletta.11 Briefly, aliquots (500 µL) of each undiluted phage stock were incubated (37°C, 90 min, on a heat block) with proteinase K (1 mg/50 µL), after which the mixtures were immediately subjected to another heat treatment (70°C, 30 min). After the heat treatments, two sets of ten replicates were prepared for each of the samples that contained ca. 107 PFU/mL prior to the heat treatments. The QPCR assay reaction mixtures (25 µL) consisted of 12.5 µL of iQ SYBR Green Supermix (Bio-Rad 170-8880), forward primer (10 mM in 0.5 µL), reverse primer (10 mM in 0.5 µL), template phage DNA preparation (2 µL) and sterile MilliQ water (9.5 µL). Amplification was performed with a iQ5 detection system Real-Time PCR Detection System (Bio-Rad, 170-9780). QPCR cycling was done at 95°C (5 min), followed by 35 3-step cycles at 95°C (30 s), 50°C (20 s) and 72°C (45 s), and a final cycle at 72°C (5 min). Real-time data capture was performed with Bio-Rad's iQ5 Standard Edition Optical System software, version 2.0.148.060623.
Crude phage DNA (used to create standard curves) and the heat-treated phage preparations were analyzed simultaneously by agarose gel electrophoresis, and the latter were amplified via standard PCR. Aliquots (5 µL) of the crude phage DNA preparations and each of the heat-treated phage specimens (containing ca. 1010 PFU/mL prior to heating) were analyzed by electrophoresis in a 1% agarose gel to confirm band sizes. PCR reaction mixtures (50 µL) consisted of 10x-concentrated XPCR buffer (5 µL), MgCl2 (50 mM in 2 µL), dNTPs (10 mM in 2 µL), forward primer (10 mM in 1 µL), reverse primer (10 mM in 1 µL), crude phage DNA preparation (2 µL) and a Taq-Pro Red DNA polymerase preparation (0.5 unit in 0.5 µL; TaqPro Red Core Kit; Denville Scientific, CB4060). PCR cycling was done at 94°C (5 min), followed by 35 3-step cycles at 94°C (45 s), 48°C (30 s) and 72°C (45 s), and a final cycle at 72°C (5 min). The PCR products were purified with a Geneclean Spin Kit (MP Biomedical, 1101-200) and DNA was eluted with sterile MilliQ water (30 µL). The DNA standards were serially diluted down to 101 copies/2 µL. The most relevant and encompassing standard range was used during the QPCR-based assays of the test specimens.
NTA/NS technology-based assays.
The assays were performed exclusively at the EPI-UF, according to the recommended protocols of the required machine's manufacturer (NanoSight, LM20). Briefly, aliquots (300 to 400 µL) of 10 replicates (1-mL) of diluted phage preparations (108 PFU/mL) in sterile 10-mL culture tubes were injected aseptically into the NanoSight LM20 machine's specimen chamber until the liquid reached the nozzle's tip. The specimens were tracked and measured at room temperature for 60 s with a manual shutter and the brightness, gain and blur ranged between 3 to −6, 0.5 to 2.0 and 3 × 3 to 5 × 5, respectively. The data were captured and analyzed with NTA Build 127 software (version 2.0).
Data collection and statistical analyses.
The PA-based data were collected by visual examination of the plates and manual plaque counting. Data collected during the QPCR- and NS-based assays were processed with Bio-Rad's iQ5 2.0 Standard Edition Optical System software (version 2.0.148.060623) and NTA Analytical software (version 2.0), respectively. Duplicate QPCR results were averaged and all data were saved to Open Office Calc spreadsheets. Calculations of confidence intervals (CI), coefficient(s) of variation (CVS); i.e., the population's standard deviation divided by its mean,14 and power relation coefficients of determination (R2) between the three assaying methods were completed in Open Office Calc. software. Paired two-tailed t test and one-way analysis of variance (ANOVA) calculations were performed with the GraphPad InStat (version 3.05) program (GraphPad Software, www.graphpad.com) and Open Office Calc and the supplemental OooStat statistics package (version 0.5) (written by David Hitchcock and released under the GNU general public license), respectively.
Summary
Bacteriophages are increasingly being used and/or considered for use in many practical applications. For example, at least two bacteriophage-based products are currently being marketed for food safety applications, to reduce or eliminate contamination of various food products with the foodborne bacterial pathogen L. monocytogenes.6 In addition, bacteriophage preparations are being considered for use as natural, “green” decontaminating agents effective in reducing or eliminating contamination of various inanimate surfaces naturally or intentionally (for bioterrorist attacks) contaminated with pathogenic bacteria, including class A agents (e.g., Y. pestis); i.e., those capable of causing outbreaks and epidemics with significant human morbidity and mortality.16 Also, lytic bacteriophages are being increasingly considered in the West as therapeutic modalities, for preventing and treating human diseases caused by bacteria resistant to currently available antibiotics.17 Finally, phages continue to be used as important diagnostic modalities and/or research tools in many research laboratories around the world.18 All of those applications require phage preparations whose titers have been rigorously characterized by an appropriate assay. The present study was designed to compare the speed and precision of the classical PA assay with those of two recently developed methods: the QPCR- and NTA/NS-based assays.
The pros and cons of each assay are summarized in Table 2. Each approach has advantages and disadvantages. In general, however, both of the new methods appear to be viable alternatives to the PA for quantifying high-concentration (>106 PFU/mL) phage preparations. Both are significantly faster to perform than the classical PA as the QPCR-based assay and the NS protocol yield results within 3 to 4 h and 5 min, respectively. The faster turnaround time for the NS-based assay is counterbalanced by the reduced precision when compared to the QPCR-based assay. However, we believe the NS method's precision may be increased by, for example, performing it with monodisperse phage specimens that are not contaminated with phage aggregates. The manufacturer of the equipment used to perform the assay suggested (www.nanosight.com/faq/validity-of-method) that a reproducibility within 2 to 3% may be achieved under ideal conditions.
The current study was limited because, with the exception of the PA, we quantitated only three bacteriophages with the assays. However, the results obtained when we analyzed the three phages and compared their titers determined by (1) the PA and the QPCR-based method, (2) the PA and the NS-based method and (3) the QPCR- and NS-based assays correlated well with each other (Fig. 2), which suggests that a similar trend may be applicable to other bacteriophages. However, the CVS and correction factors did vary for the three different phages, which indicates that whenever a new phage is characterized, initial testing must be performed to establish the phage's specific relationship between the classical PA and either of the two new assays. After the overall correlation (including the CVS, correction factor and R2) is established, either of the new assays may be useful for quickly and reproducibly determining phage concentrations. The NS method may be considered to be superior to the QPCR-based assay because of the former's very high speed and its lack of a requirement for phage-specific primers and other expensive reagents. In addition, it enables simultaneous assessment of a phage preparation's purity and degree of aggregation, which will be valuable when the preparation is being quality-tested for, for example, clinical applications. Given the poor reproducibility and potentially very large error range of 0.33 log of the traditional PA assay, the availability of the two new complementary methods may be very useful for future basic and applied research with bacteriophages.
Acknowledgements
We are grateful to Tamar Abuladze and Leroy Voelker for their help with the plaque assays. We thank Dr. Nikoloz Tsertsvadze for providing us with the YpP-G phage preparation and Drs. David M. Anderson and Arnold Kreger for their editorial assistance. B.A., C.C. and A.S. hold an equity stake in Intralytix, Inc., a Maryland corporation developing therapeutic phage preparations. The US Environmental Protection Agency, through its Office of Research and Development, partially funded (contract #EP-C-08-031, to A.S.) and collaborated in the research described in this publication. Additional funding was provided by Intralytix, Inc., and by SBIR award W911QY-07-C-0125 from the United States Army (to A.S.).
References
- 1.Duckworth DH. Who discovered bacteriophage? Bacteriol Rev. 1976;40:793–802. doi: 10.1128/br.40.4.793-802.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers WC. Bacteriophage therapy. Annu Rev Microbiol. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 3.Sulakvelidze A, Barrow P. Phage therapy in animals and agribusiness. In: Kutter E, Sulakvelidze A, editors. Bacteriophages: Biology and Applications. Boca Raton, FL: CRC Press; 2005. pp. 335–380. [Google Scholar]
- 4.Sulakvelidze A, Kutter E. Bacteriophage therapy in humans. In: Kutter E, Sulakvelidze A, editors. Bacteriophages: Biology and Application. Boca Raton, FL: CRC Press; 2005. pp. 381–436. [Google Scholar]
- 5.Cairns J, Stent GS, Watson JD. Phage and the origins of molecular biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1966. [Google Scholar]
- 6.Sulakvelidze A, Pasternack G. Industrial and regulatory issues in bacteriophage applications in food production and processing. In: Sabour PM, Griffiths MW, editors. Bacteriophages in the control of food- and waterborne pathogens. Washington, DC: ASM Press; 2010. pp. 297–326. [Google Scholar]
- 7.Wright A, Hawkins CH, Anggard EE, Harper DR. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibioticresistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol. 2009;34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 8.Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A. Bacteriophage therapy of venous leg ulcers in humans: results of a Phase I safety trial. J Wound Care. 2009;18:237–238. doi: 10.12968/jowc.2009.18.6.42801. [DOI] [PubMed] [Google Scholar]
- 9.Adams MH. Bacteriophages. London: Interscience Publishers, Ltd.; 1959. Enumeration of bacteriophage particles; pp. 27–34. [Google Scholar]
- 10.Carlson K. Working with bacteriophages: common techniques and methodological approaches. In: Kutter E, Sulakvelidze A, editors. Bacteriophages: Biology and Application. Boca Raton, FL: CRC Press; 2004. pp. 437–494. [Google Scholar]
- 11.Edelman DC, Barletta J. Real-time PCR provides improved detection and titer determination of bacteriophage. Biotechniques. 2003;35:368–375. doi: 10.2144/03352rr02. [DOI] [PubMed] [Google Scholar]
- 12.Ackermann HW, Berthiaume L. Atlas of virus diagrams. Boca Raton, FL: CRC Press; 1995. [Google Scholar]
- 13.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 14.Quinn GP, Keough MJ. Experimental design and data analysis for biologists. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- 15.Mahajan BK. Methods in Biostatistics. New Delhi, India: Jaypee Brothers Publishers; 2002. [Google Scholar]
- 16.Abuladze T, Li M, Menetrez MY, Dean T, Senecal A, Sulakvelidze A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli and ground beef by Escherichia coli O157:H7. Appl Environ Microbiol. 2008;74:6230–6238. doi: 10.1128/AEM.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel K. Old dogma, new tricks—21st Century phage therapy. Nat Biotechnol. 2004;22:31–36. doi: 10.1038/nbt0104-31. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths MW. Phage-based methods for detection of bacterial pathogens. In: Sabour PM, Griffiths MW, editors. Bacteriophages in the control of food- and waterborne pathogens. Washington, DC: ASM Press; 2010. pp. 31–59. [Google Scholar]