Abstract
Soft-ripened cheeses belong to the type of food most often contaminated with Listeria monocytogenes, and they have been implicated in several outbreaks of listeriosis. Bacteriophages represent an attractive way to combat foodborne pathogens without affecting other properties of the food. We used the broad host range, virulent Listeria phage A511 for control of L. monocytogenes during the production and ripening phases of both types of soft-ripened cheeses, white mold (Camembert-type) cheese, as well as washed-rind cheese with a red-smear surface (Limburger-type). The surfaces of young, unripened cheese were inoculated with 101–103 cfu/cm2 L. monocytogenes strains Scott A (serovar 4b) or CNL 103/2005 (serovar 1/2a). Phage was applied at defined time points thereafter, in single or repeated treatments, at 3 × 108 or 1 × 109 pfu/cm2. With Scott A (103 cfu/cm2) and a single dose of A511 (3 × 108 pfu/cm2) on camembert-type cheese, viable counts dropped 2.5 logs at the end of the 21 day ripening period. Repeated phage application did not further inhibit the bacteria, whereas a single higher dose (1 × 109 pfu/cm2) was found to be more effective. On red-smear cheese ripened for 22 days, Listeria counts were down by more than 3 logs. Repeated application of A511 further delayed re-growth of Listeria, but did not affect bacterial counts after 22 days. With lower initial Listeria contamination (101–102 cfu/cm2), viable counts dropped below the limit of detection, corresponding to more than 6 logs reduction compared to the control. Our data clearly demonstrate the potential of bacteriophage for biocontrol of L. monocytogenes in soft cheese.
Key words: Listeria monocytogenes, bacteriophage, food safety, soft-ripened cheese
Introduction
Cheese is considered to be one of the foods most frequently contaminated with Listeria monocytogenes. About 30% of the major food related outbreaks of L. monocytogenes can be traced back to contaminated cheeses.1 The incidence of L. monocytogenes on cheese was found to be in a range from 1% and 22%, with an average contamination frequency of 7.4%.2–10 Soft-ripened cheeses seem to be more frequently contaminated than semi-hard or hard cheeses,9,11 although there are some contrary reports.6 Soft cheeses provide appropriate growth conditions for Listeria,12,13 not only because of the psychrotrophic and halotolerant nature of the organism,14 but also because they are commonly consumed as they are, i.e., without any bactericidal treatment (cooking, heating) to eliminate contaminations introduced throughout the production and ripening period. The significant pH increase during the surface ripening of these dairy products also strongly supports growth of Listeria species. Therefore, appropriate control of L. monocytogenes on these products is very important, and new strategies to achieve this goal are highly desirable.
The application of bacteriophages for specific killing of undesirable contaminants such as Listeria represent a promising approach to enhance food safety. Phages offer an extreme specificity for their hosts, are widely distributed in the environment, and form part of the natural microbiological flora found in foods.15–18 This renders them ideal candidates for the control of pathogens without interfering with the resident microflora or starter culture organisms required for production of fermented foods. Listeria phage A511 is a large, virulent (i.e., obligately lytic) SPO1-like phage from the Myoviridae taxonomic group.19 Phage A511 has a broad host range within the genus Listeria and is strictly specific for this genus.20 The aim of this work was to evaluate usefulness of A511 for the control of L. monocytogenes during the entire 3-week ripening phases of both white-mold and washed-rind type soft cheeses. Our data demonstrate the potential of phage as ideal candidates to significantly decrease development of the pathogen in the cheese surfaces.
Results
Two different soft-ripened cheese models were established in the laboratory in order to evaluate the potential of A511 for controlling L. monocytogenes on the cheese surfaces. The surface pH changes were monitored and found characteristic for the ripening processes of white mold cheeses and washed rind, red-smear cheeses, respectively.1,21–23 White mold cheeses featured an initial pH of 5.6 ± 0.2, which increased after 3 days and reached a maximum of pH 7.6 ± 0.2 after 20 days. On the red-smear cheeses, the initial pH (5.4 ± 0.1) increased to pH 7.8 ± 0.2 at the end of the ripening period of 22 days. The population of Listeria Scott A and CNL 103/2005 on the non-phage treated control cheeses closely reflected the pH changes: growth started when the pH rose above 6.0. After 20 days, counts of strain Scott A reached approximately 105 cfu/cm2 on white mold cheese, (Fig. 1), whereas red-smear cheeses supported growth up to 107 cfu Listeria per cm2 (Figs. 2 and 3). Interestingly, strain CNL 103/2005 yielded significantly higher counts (up to 108 cfu/cm2) on red-smear cheese surfaces (Fig. 2B).
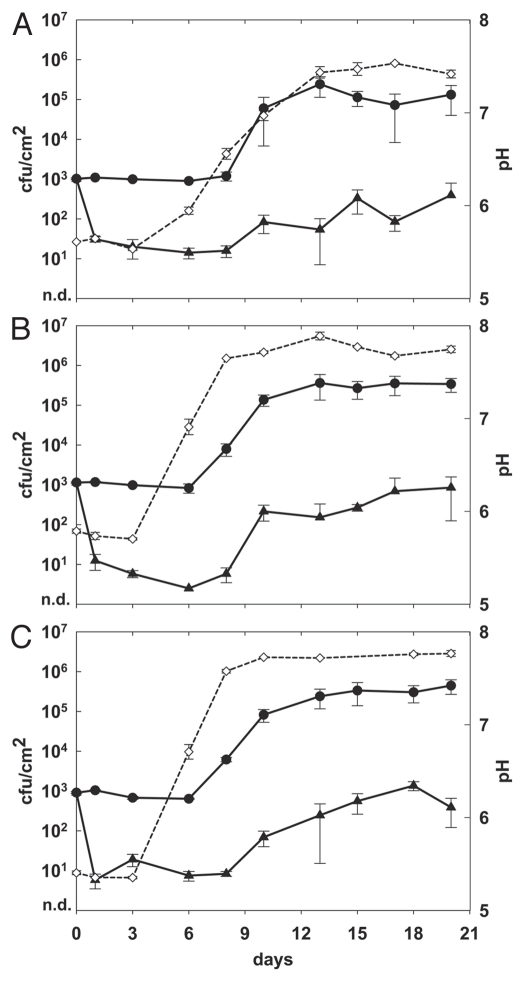
Figure 1.
Effect of phage A511 on growth of L. monocytogenes strain Scott A on white mold soft cheese, using different phage application protocols. Cheeses were spiked with 1 × 103 cfu/cm2 on day 0, and phage A511 was applied to the test samples: 3 × 108 pfu/cm2 after 1 h (A), 3 × 108 pfu/cm2 after 1 h and 20 h (B), 1 × 109 pfu/cm2 after 1 h (C). Cheeses were ripened for 10 days at 12–13°C and 95% relative humidity of the air, and then stored for another 10 days at 6°C (closed circles, control; closed triangles, with phage A511; open diamonds and dotted line, surface pH).
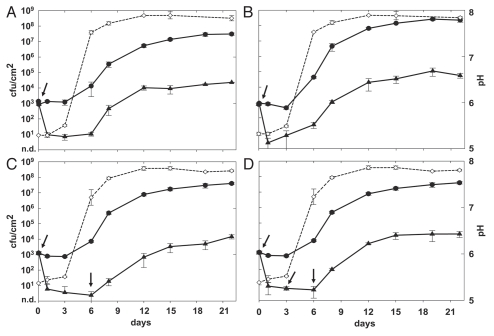
Figure 2.
Effect of phage A511 on growth of L. monocytogenes strain CNL 103/2005 (B) and Scott A (A, C and D) on red-smear soft cheese, using different phage application protocols. Cheeses were spiked with 1 × 103 cfu/cm2 on day 0, and phage A511 (3 × 108 pfu/cm2) was applied to the test samples: after 1 h (A and B), after 1 h and 6 d (C), and after 1 h, 3d and 6 d (D) (indicated by arrows). Cheeses were ripened for 11 days at 12–13°C and 95% relative humidity of the air, and then stored for another 11 days at 6°C (closed circles, control; closed triangles, with phage A511; open diamonds and dotted line, surface pH).
Figure 3.
Effect of phage A511 on growth of L. monocytogenes strain Scott A on red-smear soft cheese at different initial contamination levels. Cheeses were spiked on day 0 with 1 × 103 cfu/cm2 (A), 1 × 102 cfu/cm2 (B), 1 × 10 cfu/cm2 (C) and phage A511 (3 × 108 pfu/cm2) was applied after 1 h to the test samples. Cheeses were ripened for 11 days at 12–13°C and 95% relative humidity of the air, and then stored for another 11 days at 6°C (closed circles, control; closed triangles, with phage A511; open diamonds and dotted line, surface pH).
To confirm that phage A511 added to the smear brine would not affect the added starter and ripening microflora consisting of Gram-positive bacteria, yeasts and molds, total aerobic plate counts were determined over one entire red-smear cheese ripening trial. We found no differences between phage-treated and control cheeses (data not shown).
Addition of 3 × 108 pfu/cm2 A511 on white mold soft cheese contaminated with Listeria Scott A resulted in a decrease of viable counts by more than 2 orders of magnitude (logs) during the first 6–8 days of production (Fig. 1A). However, re-growth of Listeria occurred when the cheese surface pH rose above 6.5. Nevertheless, compared to the controls, Listeria growth on the phage-treated samples was significantly slower. Repeated application of phage at 3 × 108 pfu/cm2 (Fig. 1B), or a single higher dose application of 1 × 109 pfu/cm2 (Fig. 1C) further reduced the Listeria population during the initial 8–10 day period, by approximately 90% compared to the single lower dose treatment (Fig. 1A). Considering final counts obtained at the end of the 3 week ripening periods, the single high dose application (Fig. 1C) yielded the largest difference (3.1 log, p < 0.05) between phage-treated cheeses and the controls, compared to using less phage in a single (Fig. 1A) or repeat application scheme (Fig. 1B) (2.5 to 2.6 logs difference; p < 0.05).
On the rind-washed, red-smear soft cheeses, the effects obtained by single or repeated phage challenge were similar. Viable Listeria counts decreased by approximately 2 orders of magnitude within the first 6 days after A511 application at 3 × 108 pfu/cm2 (Fig. 2A and B). Thereafter, re-growth occurred and approximately 104 cfu/cm2 bacteria were present at the end of the ripening phase on day 22. Compared to the controls, reduction was 3.1 log for Scott A (Fig. 2A), and 3.6 log for CNL 103/2005 (Fig. 2B). To further improve the killing efficacy of phage, additional trials were performed: application of 3 × 108 pfu/cm2 of A511 twice during the rind-washing process (smearing) on days 0 and 6 resulted in a lower bacterial counts between days 6–12 (Fig. 2C), compared to the single dose treatment. When A511 was added at three time points (day 0, 3, 6), significantly lower Listeria numbers were observed between days 8–19 (Fig. 2D). However, final counts obtained after 22 days by the 3 phage application protocols were not significantly different (p > 0.05).
Additional experiments were performed to determine the role of the initial contamination level on the efficacy of phage treatment of the cheese surfaces (Fig. 3). When cheeses received 1 × 103 cfu/cm2 Scott A (Fig. 3A), application of phage resulted in an approximately 3 log difference after 22 days. When lower numbers were used 1 × 102 cfu/cm2 (Fig. 3B) and 1 × 101 cfu/cm2 (Fig. 3C), differences in cell counts of more than 6 logs were achieved, and no viable cells could be recovered by direct plating after day 6 (detection limit 1 cfu/cm2). Thus, initial killing efficacy of the phage and the final difference in viable cell count was significant better when the target cell concentration was 1 × 102 cfu/cm2 or below.
By the end of the trial periods, the number of infective A511 particles on the surface of the white mold soft cheeses dropped by approximately 1.5 log pfu/cm2, whereas phage infectivity was less affected (less than 1 log reduction) on the surface of red-smear soft cheeses.
Listeria clones recovered from phage-treated cheeses were also tested for possible development or selection of insensitivity/resistance to A511. While all 10 Scott A isolates remained fully sensitive to A511 infection, three out of ten (30%) of the CNL 103/2005 clones recovered from day 22 cheese samples that received A511 showed a phage-insensitive phenotype.
Discussion
Because of the relevance of soft-ripened cheeses in sporadic or epidemic cases of listeriosis, phage application for control of L. monocytogenes during processing and ripening of these products represents an attractive alternative to increase food safety. To ensure that the cheese models set up in the laboratory properly reflect the characteristic steps during the different ripening processes, several parameters were monitored. In all trials, the changes in surface pH were found typical for the ripening of white mold cheeses and red-smear cheeses, respectively.1,21–23 Addition of phage had no influence on total aerobic plate counts and cheeses that received phage were indistinguishable from the controls regarding odor, texture and general appearance, and they were overall comparable to commercially available products.
Not surprisingly, the onset of growth of Listeria Scott A and CNL 103/2005 on the cheese surfaces closely reflected the changes in surface pH, which is the major limiting factor for growth of the organism in these environments.1,12,21,22,24 Interestingly, we noted that Scott A grew significantly slower than CNL 103/2005. The latter had been isolated in 2005 from a listeriosis outbreak in Switzerland, involving white mold soft cheese.25 Thus, it appears to be better adapted to this specific environment than strain Scott A, which is a clinical isolate,26 and had been held under laboratory conditions for many years. Similar observations regarding L. monocytogenes strain-specific growth differences have been reported for other cheese models and related studies.12,13,21,22,24,27
Our data indicate that application of phage A511 is able to eradicate L. monocytogenes cells from the cheese surfaces to levels below 1 cfu/g, when the contamination rate at the beginning of the ripening phase was at 100 cfu/cm2 or below. At a higher level (103 cfu/cm2), a still very significant reduction of approx. 3 logs was observed on red-smear and white mold soft cheeses. This correlates well with earlier reports where a single higher dose of Listeria phage P100 (6 × 107 pfu/cm2) was sufficient to eliminate an initial contamination of 2 × 101 cfu/cm2 L. monocytogenes from the surface of red-smear soft cheese,24 whereas repeated application of lower doses (2 × 106 pfu/cm2) resulted in a reduction of 2–3 logs. Altogether, these findings again underline the important role of phage concentration for efficacy of using phage in foods.28 This is especially true for foods featuring a large, uneven and moisture-absorbing surface (such as cheese), which severely limits free diffusion of the virus particles in a thin water film.28 Our results also correlate well with other studies using phage to target food-borne bacteria.29–31
Efficacy of phage treatment appears to be highest at target cell concentrations at or below 100 cfu/cm2. However, it should be realized that this finding does not diminish its effectivity or affect its applicability in commercial cheese production, since the initial contamination levels actually found on soft-ripened cheese are usually very low.6,9 Nevertheless, it is also important to note that phage application during soft cheese production and ripening must be precisely timed to correlate with the possible time point(s) of Listeria entry into the product. If phage would only be applied late in the ripening phase, i.e., after bacterial growth has been initiated (after day 3 to 6), the bacterial population may have reached higher cell densities which might reduce efficacy of phage treatment. Critical consideration should also be given to optimal distribution of the phage on the cheese surface, in order to provide highest probability for contact between virus and target bacteria. Clearly, this is dependent on the surface properties and matrix of the cheeses. Mature soft cheese features a highly complex flora in a thick multilayered biofilm-like structure consisting of bacteria, mold mycelium, cheese proteins, lipids and approximately 2–4% NaCl. The target bacteria embedded within this matrix are at least partially shielded from diffusing liquid, and therefore are also protected from phage. Such limited accessibility explains the finding that repeated application of phages did not improve overall killing efficacy.
Stability of phage is another parameter which may be important for success of phage application. Infectivity of phage A511 particles was less affected on red-smear cheese than on the white mold cheese surface, which might have contributed to the somewhat lesser efficacy of A511 on the latter product. It is likely that proteolytic activity of starter and ripening culture microorganisms could affect integrity and infectivity of the phage particles. Similar observations have been made for enteroviruses,32 and this effect is also assumed to be responsible for deactivation of bacteriocin activity on cheese surfaces.23 Although this effect may not be generalized for all foods and application schemes, phage stability and infectivity is an important consideration regarding its use in industrial processes.
Several studies exist on biocontrol of L. monocytogenes in Camembert-like23,33,34 and red-smear soft cheeses.21,22,24,35–38 In most cases, however, not phages but “anti-Listeria cultures” or bacteriocins were used, and the different experimental setups and various types of cheeses employed make it difficult to compare these data. Important parameters which affect the results include the Listeria strain(s) and contamination levels used, type of phage or bacteriocin applied, time point of application, type of cheese, ripening conditions and packaging conditions. Nevertheless, it can be concluded that control of Listeria contamination by phage A511 generally yielded satisfactory results that seem superior to most other approaches.
One of the most critical issues regarding the phage-against-pathogens concept is the possible emergence of phage resistance. In our hands, none of the strain Scott A clones re-isolated from cheeses treated with phage A511 revealed resistance against the phage. However, with strain CNL 103/2005, some phage insensitive clones could be recovered at the end of the ripening period after 22 days. Although the precise reasons for this phenotype are not clear, it is likely that the bacteria modified the specific surface carbohydrates responsible for phage attachment (Huber T and Loessner MJ, unpublished data). In other studies, no phage resistant Listeria emerged on phage-treated ready-to-eat foods over 6 to 13 days,31 or red-smear cheese over 3 weeks.24 Resistance to a specific phage added to food was also not observed with Salmonella Enteritidis on fresh-cut fruit during a 7 day period,39 or Campylobacter jejuni on chicken skin after 10 days.40 However, some phage-insensitive Brochothrix thermosphacta emerged on pork adipose tissue 8 days after phage was added.41 Altogether, the available data indicate that development of phage resistance on foods seems to be variable and hard to predict, but the probability and frequency seems to be relatively low. If there are no negative effects on fitness or growth rate, resistant bacteria might gain an advantage over non-resistant bacteria during the period that phage is present, and may eventually dominate the bacterial population on phage treated foods. However, if maintenance costs for phage resistance are high, such cells may persist only as a minor fraction of the entire population, in line with what has been observed for CNL 103/2005 in this study. Similar explanations were offered as a result of in vitro models.42 Moreover, a phage resistance-phenotype is also likely to revert without the selective pressure, i.e., in the absence of phage.43
In conclusion, biocontrol by phage represents a promising “green” strategy against contamination of soft cheese by Listeria monocytogenes. Phage application must be integrated into a specifically tailored hygiene concept, and its success will depend on several factors: (i) the initial phage concentration must be sufficiently high (in the range of 108 pfu/cm2), and will have to be optimized for each product and its particular production scheme; (ii) phage needs to be applied at an early stage, to prevent development of L. monocytogenes above a certain threshold (approximately 100 cfu/cm2) which might reduce efficacy of phage treatment; (iii) to avoid the emergence of phage-resistant bacteria, phage-treated products should not be allowed to re-enter the production cycle. This may not be valid for white-mold cheeses. However, with respect to red-smear soft cheese production, this requirement will specifically exclude the traditional practice of “old-young smearing”, where the rind microflora from mature cheeses is used to wash the young, “green” cheeses. This procedure has long been recognized as being responsible for the recycling of Listeria contamination on these products.22 Instead, it appears necessary to install appropriate production schemes using separate, strictly controlled, pathogen-free batches of cheeses whose surface flora is exclusively used to inoculate the young cheeses and therefore minimizes initial pathogen contamination.
Materials and Methods
Microorganisms.
Listeria monocytogenes Scott A (serovar 4b) and CNL 103/2005 (serovar 1/2a), and L. ivanovii WSLC 3009 (serovar 5) were grown in half-concentrated (0.5x) brain heart-infusion medium (BHI 1/2) (Biolife) at 30°C, for 16 h. Strain CNL 103/2005 was isolated from a swiss white-mold soft cheese (“Tomme”) which caused a listeriosis outbreak.25 For phage indicator strain L. ivanovii WSLC 3009 cmr, chloramphenicol (7.5 µg/ml) (Sigma) was added to the media. Phage A511 lysates were prepared and purified as previously described in reference 31. In brief, the phage was amplified on log-phase cells of WSLC 3009 in liquid culture, with shaking at 30°C. Following complete lysis of the host cells, bacterial debris was removed by centrifugation, and phage concentrated and washed using tangential-flow ultrafiltration. Final concentration of the phage suspension was 3 × 1011 pfu/ml, and it was stored at 4°C until use.
Ripening of soft cheeses.
Unripened white mold soft cheeses and red-smear soft cheeses (22 cm diameter, 1.7 kg weight, 55% fat in dry matter), were obtained immediately post-brining from a large cheese producer in Switzerland, and transported to the laboratory. Samples of the unripened cheeses were tested for occurrence of Listeria spp. according to IDF-Standard 143A:1995.44 The cheeses were then subjected to appropriate ripening conditions as described previously in reference 21 and 34. For this, the whole cheeses were divided into 12 equal pieces, which were further cut horizontally in half, to yield samples of approximately 80 g each with surface area (on each flat side) of approximately 24 cm2. Cheeses were then placed on stainless steel racks in large glass desiccators, and relative humidity inside the chambers was adjusted to 95% by addition of 7.95% (w/v) sodium chloride solution into the bottom of the desiccators.
Listeria contamination and phage application.
Overnight cultures of L. monocytogenes strains were diluted 1:5 in fresh medium, incubated for 2–3 h at 30°C until an OD600 of approximately 0.4 was reached, and diluted in PBS (100 mM NaCl, 20 mM Na2HPO4, pH 7.4) to the desired concentrations. Using sterile nitrile gloves, cheeses were surface-inoculated with L. monocytogenes to yield final concentrations of 1 × 101, 1 × 102 or 1 × 103 cfu/cm2, on a total treated surface of approximately 24 cm2. This procedure was developed empirically and checked in every case by preparation of a sample and surface plating as described below. Treated cheeses were placed in the desiccators and stored for 1 h at 12–13°C. Then, phage A511 was applied in the same fashion to the white mold cheese samples subjected to phage treatement, in 3 different application schemes: (i) after 1 h as a single dose of 3 × 108 pfu/cm2; (ii) after 1 h and 20 h at two single doses of 3 × 108 pfu/cm2 each; (iii) after 1 h as a single dose of 3 × 109 pfu/cm2. With the red-smear cheeses, phage A511 was added to the smear brine used to wash the cheese rind, containing NaCl (3.0% w/v), and a commercially used culture mix containing Brevibacterium casei, Brevibacterium linens, Debaryomyces hansenii, Candida utilis, and Geotrichum candidum at approximately 3 × 107 cfu/ml (OFR9, Danisco, Denmark). Red-smear cheeses were then washed (suspension manually rubbed in using sterile nitrile gloves) a total of three times (day 0, 3 and 6). Phage A511 was applied at 3 × 108 pfu/cm2, either (i) once (1 h after contamination with Listeria), (ii) twice (1 h after the contamination and on day 6) or (iii) three times (1 h after contamination, on day 3, and on day 6). Cheeses were ripened in the desiccators for 11 days at 12°C. Then, they were aseptically removed, packed in aluminum foil and stored at 6°C for another 11 days until the end of the ripening period.
The cheese surface pH was monitored over the entire ripening periods, at 3–5 different spots on each cheese, using a surface pH electrode (Mettler Toledo).
Quantitative determination of listeria and bacteriophage.
Listeria viable plate counts (cfu/ml) and A511 phage counts (pfu/ml) were determined after each treatment of the cheeses, and at regular time intervals thereafter. For this purpose, 20 g samples were removed from the cheese surfaces with the aid of a sterile knife, placed in sterile polypropylene bags and homogenized in 180 ml citrate buffer using a stomacher laboratory blender (Seward Medical). For quantitative determination of Listeria counts below 103 cfu/cm2, pour plates (Oxford agar) were used with 10 ml aliquots of the homogenates, and surface plating was performed on 145 mm Oxford agar plates with 1 ml aliquots of the homogenates. For quantitative viable counts above 103 cfu/cm2, 0.1 ml aliquots of decimal dilutions of the homogenates were plated on 90 mm plates of Oxford agar. All plating was performed in duplicates, and plates were incubated for 48 h at 35°C until typical Listeria colonies could be enumerated. Infective phage recovered from the foods were enumerated as described earlier in reference 31, employing a chloramphenicol-resistant L. ivanovii (WSLC 3009 cmr) as phage indicator to enable direct plating and prevent contamination of the plates by other bacteria. To determine the possible emergence and frequency of phage-resistant bacteria in the challenged food samples, Listeria clones recovered from phage-treated red smear cheeses via selective agar plates were tested for A511 susceptibility. For this purpose, ten colonies of each of the Listeria strains were randomly picked from the day 22 plates, and streaked on non-selective BHI agar. Broth cultures prepared from single colonies (0.2 ml aliquots) were then mixed with 4 ml of molten soft agar to prepare double layer soft agar plates. After solidifying, ten µl drops of different A511 dilutions (109, 106 and 104 pfu/ml) were placed on the soft agar surface, and the plates were incubated at 30°C for 16–20 h until plaques could be seen.
Statistics.
For each measurement and time point, three cheese samples were analyzed. The bacterial and phage counts were determined by duplicate plating. Results are presented as means, and standard deviation of the means is indicated by error bars. Student's t-test (unpaired, two-tailed, heteroscedastic) was used to determine the significance of cell count differences between controls and phage treated samples, based on an alpha-level of 5% (p < 0.05).
Acknowledgements
We thank Monique Herensperger and the late Uschi Schuler for excellent technical assistance, and are grateful to the large swiss dairy company for supplying the freshly produced unripened cheeses.
References
- 1.Ryser ET. Incidence and behavoir of Listeria monocytogenes in cheese and other fermented dairy products. In: Ryser ET, Marth EH, editors. Listeria, listeriosis and food safety. New York: Marcel Dekker Inc.; 1999. pp. 411–503. [Google Scholar]
- 2.Reports on trends and sources of zoonotic agents in the European Union and in Norway: European Commission Health & Consumer Protection Directorate-General 2001. Anonymous. [Google Scholar]
- 3.Beckers HJ, Soentoro PSS, Delgou-van Asch EHM. The occurrence of Listeria monocytogenes in soft cheeses and raw milk and its resistance to heat. Int J Food Microbiol. 1987;4:249–256. doi: 10.1016/0168-1605(87)90041-9. [DOI] [Google Scholar]
- 4.Eppert I, Lechner E, Mayr R, Scherer S. Listeria and coliforms in soft cheeses made from raw and pasteurized milk. Arch Lebensmittelhyg. 1995;46:85–88. [Google Scholar]
- 5.Farber JM, Johnston MA, Purvis U, Loit A. Surveillance of soft and semi-soft cheeses for the presence of Listeria spp. Int J Food Microbiol. 1987;5:157–163. doi: 10.1016/0168-1605(87)90033-X. [DOI] [Google Scholar]
- 6.Loncarevic S, Danielssontham ML, Tham W. Occurrence of Listeria monocytogenes in soft and semisoft cheeses in retail outlets in Sweden. Int J Food Microbiol. 1995;26:245–250. doi: 10.1016/0168-1605(95)00105-S. [DOI] [PubMed] [Google Scholar]
- 7.Pak SI, Spahr U, Jemmi T, Salman MD. Risk factors for L. monocytogenes contamination of dairy products in Switzerland 1990–1999. Prev Vet Med. 2002;53:55–65. doi: 10.1016/S0167-5877(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 8.Pini PN, Gilbert RJ. The occurrence in the UK of Listeria species in raw chickens and soft cheeses. Int J Food Microbiol. 1988;6:317–326. doi: 10.1016/0168-1605(88)90025-6. [DOI] [PubMed] [Google Scholar]
- 9.Rudolf M, Scherer S. High incidence of Listeria monocytogenes in European red smear cheese. Int J Food Microbiol. 2001;63:91–98. doi: 10.1016/s0168-1605(00)00413-x. [DOI] [PubMed] [Google Scholar]
- 10.Terplan G, Schoen R, Springmeyer W, Degle I, Becker H. Occurrence, behavior and significance of Listeria in milk and dairy products. Arch Lebensmittelhyg. 1986;37:131–137. [Google Scholar]
- 11.Ryser ET. Foodborne Listeriosis. In: Ryser ET, Marth EH, editors. Listeria, listeriosis and food safety. New York: Marcell Dekker Inc.; 1999. pp. 299–358. [Google Scholar]
- 12.Back JP, Langford SA, Kroll RG. Growth of Listeria monocytogenes in Camembert and other soft cheeses at refrigeration temperatures. J Dairy Res. 1993;60:421–429. doi: 10.1017/s0022029900027758. [DOI] [PubMed] [Google Scholar]
- 13.Ryser ET, Marth EH. Behavior of Listeria monocytogenes during manufacture and ripening of brick cheese. J Dairy Sci. 1989;72:838–853. doi: 10.3168/jds.S0022-0302(89)79176-1. [DOI] [PubMed] [Google Scholar]
- 14.McClure PJ, Kelly TM, Roberts TA. The effects of temperature, pH, sodium chloride and sodium nitrite on the growth of Listeria monocytogenes. Int J Food Microbiol. 1991;14:77–91. doi: 10.1016/0168-1605(91)90039-R. [DOI] [PubMed] [Google Scholar]
- 15.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H. Phage-host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy JE, Bitton G. Bacteriophages in foods. In: Goyal SM, Gerba CP, Bitton G, editors. Phage Ecology. New York: John Wiley and Sons; 1987. pp. 289–316. [Google Scholar]
- 17.Whitman PA, Marshall RT. Isolation of psychrophilic bacteriophage-host systems from refrigerated food products. Appl Microbiol. 1971;22:220–223. doi: 10.1128/am.22.2.220-223.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitman PA, Marshall RT. Characterization of two psychrophilic Pseudomonas bacteriophages isolated from ground beef. Appl Microbiol. 1970;22:463–468. doi: 10.1128/am.22.3.463-468.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zink R, Loessner MJ. Classification of virulent and temperate bacteriophages of Listeria spp. on the basis of morphology and protein analysis. Appl Environ Microbiol. 1992;58:296–302. doi: 10.1128/aem.58.1.296-302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loessner MJ, Busse M. Bacteriophage typing of Listeria species. Appl Environ Microbiol. 1990;56:1912–1918. doi: 10.1128/aem.56.6.1912-1918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppert I, Valdes-Stauber N, Goetz H, Busse M, Scherer S. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin-producing Brevibacterium lines as evaluated in situ on soft cheese. Appl Environ Microbiol. 1997;63:4812–4817. doi: 10.1128/aem.63.12.4812-4817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loessner M, Guenther S, Steffan S, Scherer S. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl Environ Microbiol. 2003;69:1854–1857. doi: 10.1128/AEM.69.3.1854-1857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan J, Harmark K, Davidson BE, Hillier AJ, Gordon JB, Wilcock A, et al. Inhibition of Listeria monocytogenes by piscicolin 126 in milk and Camembert cheese manufactured with a thermophilic starter. J Appl Microbiol. 1997;82:273–280. doi: 10.1046/j.1365-2672.1997.00349.x. [DOI] [PubMed] [Google Scholar]
- 24.Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study and application. Regul Toxicol Pharmacol. 2005;43:301–312. doi: 10.1016/j.yrtph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Bille J, Blanc DS, Schmid H, Boubaker K, Baumgartner A, Siegrist HH, et al. Outbreak of human listeriosis associated with tomme cheese in northwest Switzerland 2005. Euro Surveill. 2006;11:91–93. [PubMed] [Google Scholar]
- 26.Fleming DW, Cochi SL, MacDonald KL, Brondum J, Hayes PS, Plikaytis BD, et al. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N Engl J Med. 1985;312:404–407. doi: 10.1056/NEJM198502143120704. [DOI] [PubMed] [Google Scholar]
- 27.Ryser ET, Marth EH. Fate of Listeria monocytogenes during manufacture and ripening of camembert cheese. J Food Prot. 1987;50:372–378. doi: 10.4315/0362-028X-50.5.372. [DOI] [PubMed] [Google Scholar]
- 28.Hagens S, Loessner M. Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr Pharm Biotechnol. 2010;11:58–68. doi: 10.2174/138920110790725429. [DOI] [PubMed] [Google Scholar]
- 29.Greer GG. Effects of phage concentration, bacterial density and temperature on phage control of beef spoilage. J Food Sci. 1988;53:1126–1127. [Google Scholar]
- 30.Leverentz B, Conway WS, Janisiewicz W, Camp MJ. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. J Food Prot. 2004;67:1682–1686. doi: 10.4315/0362-028x-67.8.1682. [DOI] [PubMed] [Google Scholar]
- 31.Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophages for biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol. 2009;75:93–100. doi: 10.1128/AEM.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cliver DO, Herrmann JE. Proteolytic and microbial inactivation of enteroviruses. Water Research. 1972;6:797–805. [Google Scholar]
- 33.Maisnier-Patin S, Deschamps N, Tatini SR, Richard J. Inhibition of Listeria monocytogenes in camembert cheese made with a nisin-producing starter. Lait. 1992;72:249–263. [Google Scholar]
- 34.Sulzer G, Busse M. Growth inhibition of Listeria spp. on Camembert cheese by bacteria producing inhibitory substances. Int J Food Microbiol. 1991;14:287–296. doi: 10.1016/0168-1605(91)90120-e. [DOI] [PubMed] [Google Scholar]
- 35.O'Sullivan L, O'Connor EB, Ross RP, Hill C. Evaluation of live-culture-producing lacticin 3147 as a treatment for the control of Listeria monocytogenes on the surface of smear-ripened cheese. J Appl Microbiol. 2006;100:135–143. doi: 10.1111/j.1365-2672.2005.02747.x. [DOI] [PubMed] [Google Scholar]
- 36.Cao-Hoang L, Chaine A, Grégoire L, Waché Y. Potential of nisin-incorporated sodium caseinate films to control Listeria in artificially contaminated cheese. Food Microbiol. 2010;27:940–944. doi: 10.1016/j.fm.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Izquierdo E, Machioni E, Aoude-Werner D, Hasselmann C, Ennahar S. Smearing of soft cheese with Enterococcus faecium WHE 81, a multi-bacteriocin producer, against Listeria monocytogenes. Food Microbiol. 2009;26:16–20. doi: 10.1016/j.fm.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Monnet C, Bleicher A, Neuhaus K, Sarthou AS, Leclercq-Perlat MN, Irlinger F. Assessment of the antilisterial activity of microfloras from the surface of smear-ripened cheeses. Food Microbiol. 2010;27:302–310. doi: 10.1016/j.fm.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Leverentz B, Conway WS, Alavidze Z, Janisiewicz WJ, Fuchs Y, Camp MJ, et al. Examination of bacteriophage as a biocontrol method for Salmonella on fresh-cut fruit: A model study. J Food Prot. 2001;64:1116–1121. doi: 10.4315/0362-028x-64.8.1116. [DOI] [PubMed] [Google Scholar]
- 40.Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF. Application of host specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl Environ Microbiol. 2003;69:6302–6306. doi: 10.1128/AEM.69.10.6302-6306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer GG, Dilts BD. Control of Brochothrix thermosphacta spoilage of pork adipose tissue using bacteriophages. J Food Prot. 2002;65:861–863. doi: 10.4315/0362-028x-65.5.861. [DOI] [PubMed] [Google Scholar]
- 42.Levin BR, Bull JJ. Phage therapy revisited: The population biology of a bacterial infection and its treatment with bacteriophage and antibiotics. American Naturalist. 1996;147:881–898. [Google Scholar]
- 43.O'Flynn G, Ross RP, Fitzgerald GF, Coffey A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol. 2004;70:3417–3424. doi: 10.1128/AEM.70.6.3417-3424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milk and Milk products—detection of Listeria monocytogenes. International Dairy Federation; 1995. Anonymous. IDF standard 143A:1995. [Google Scholar]