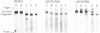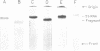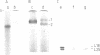Abstract
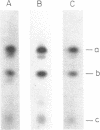
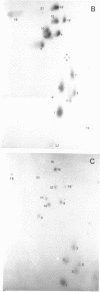
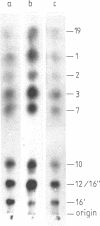
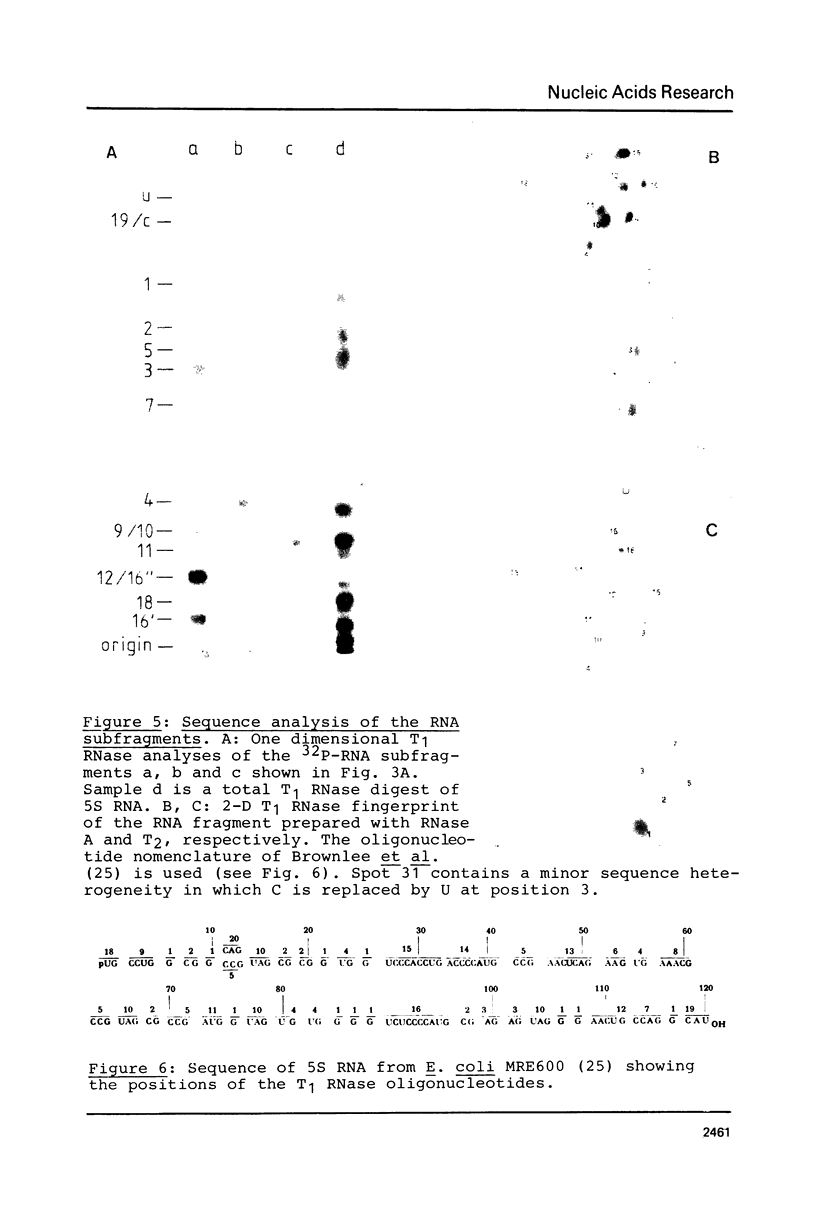
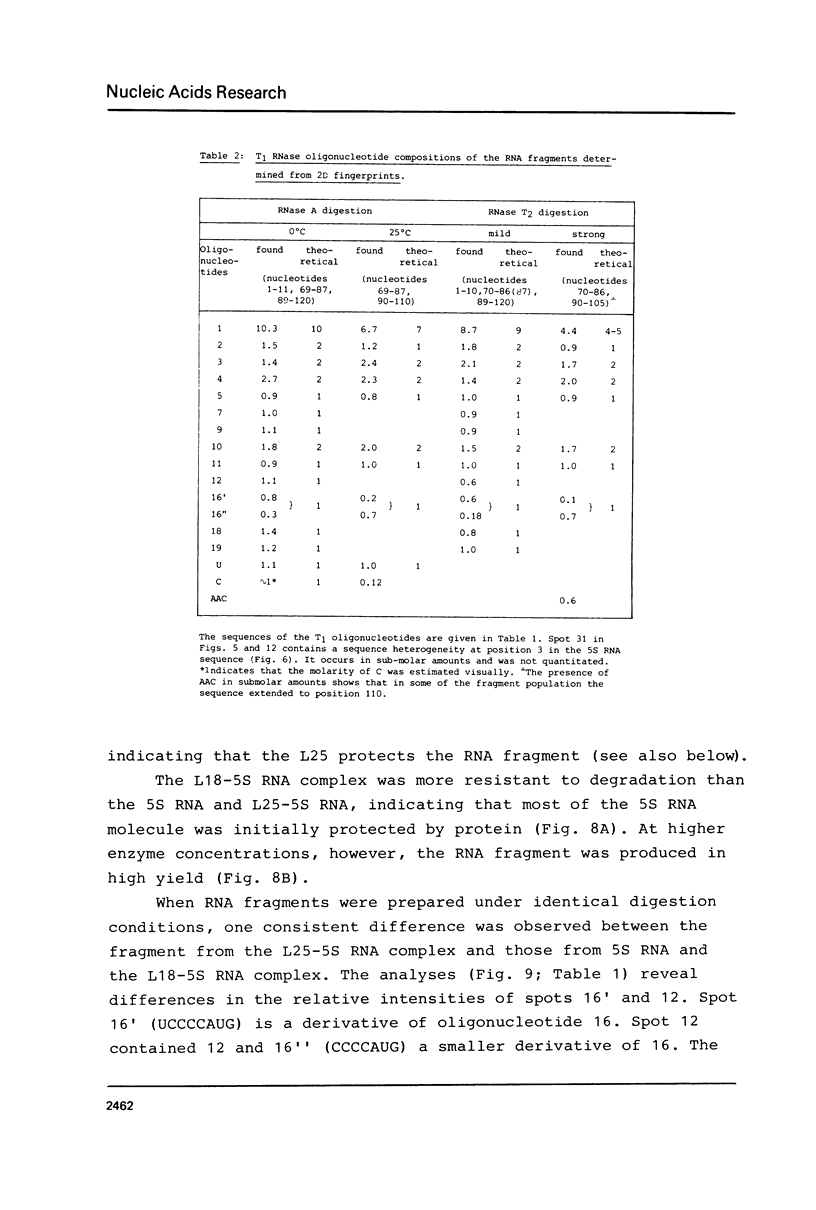
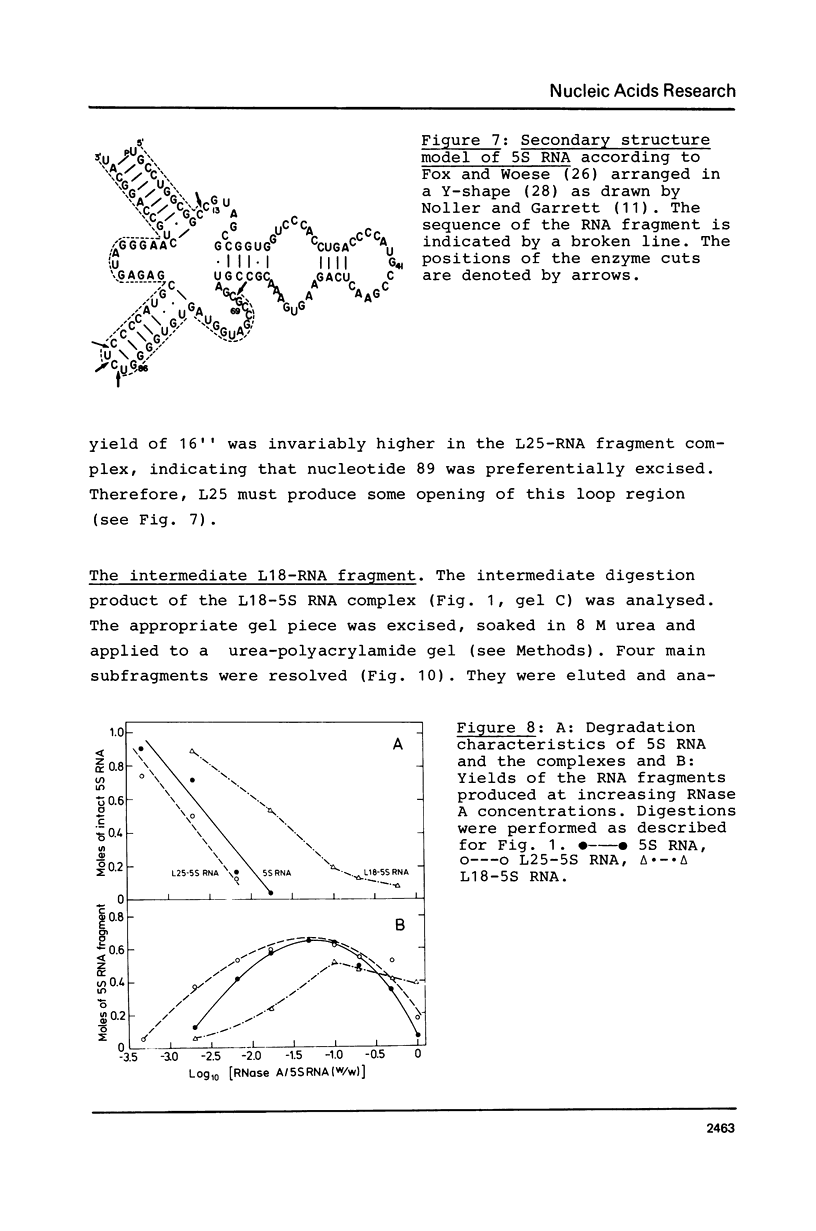
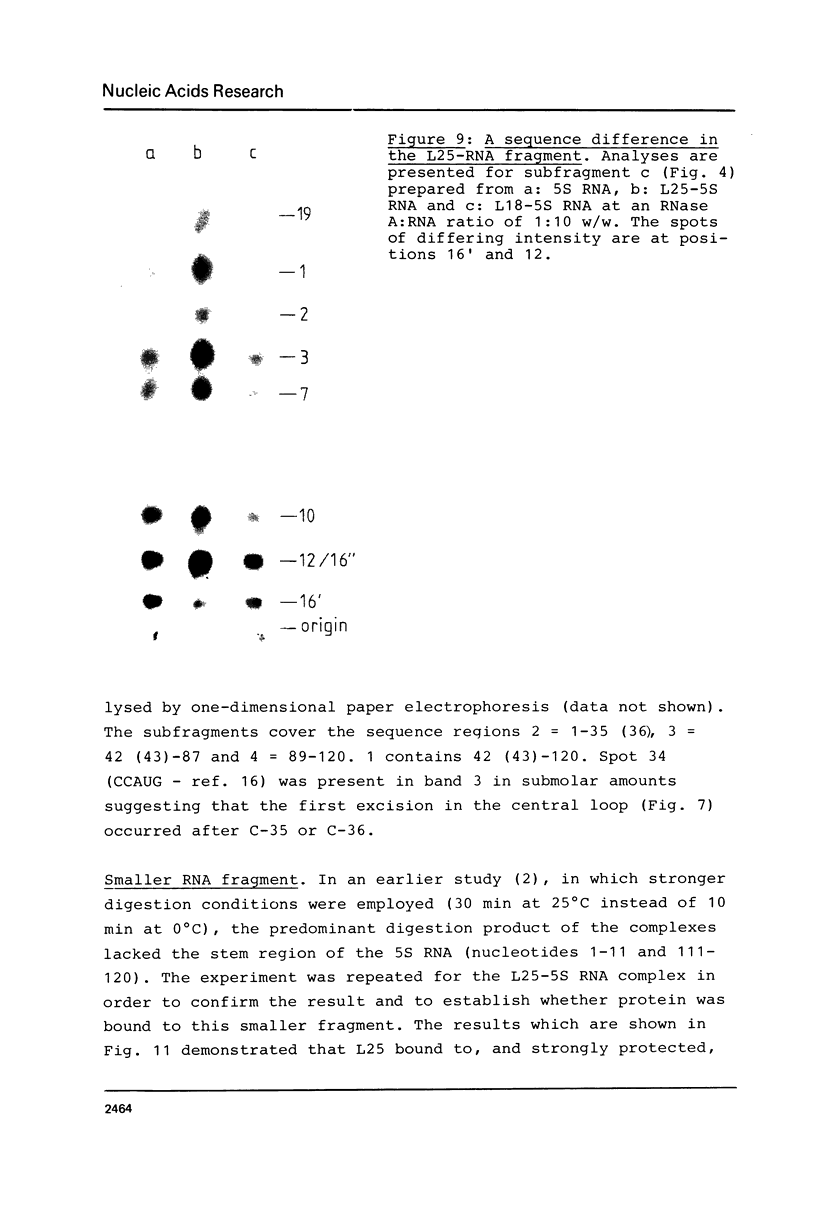
An RNA fragment, constituting three subfragments of nucleotide sequences 1-11, 69-87 and 89-120, is the most ribonuclease-resistant part of the native 5S RNA of Escherichia coli, at 0 degrees C. A smaller fragment of nucleotide sequence 69-87 and 90-110 is ribonuclease-resistant at 25 degrees. Degradation of the L25-5S RNA complex with ribonuclease A or T2 yielded RNA fragments similar to those of the free 5S RNA at 0 degrees C and 25 degrees C; moreover L25 remained strongly bound to both RNA fragments and also produced some opening of the RNA structure in at least two positions. Protein L18 initially protected most of the 5S RNA against ribonuclease digestion, at 0 degrees C, but was then gradually released prior to the formation of the larger RNA fragment. It cannot be concluded, therefore, as it was earlier (Gray et al., 1973), that this RNA fragment contains the primary binding site of L18.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Bellemare G., Monier R. Selective reaction of glyoxal with guanine residues in native and denatured Escherichia coli 5S RNA. Biochimie. 1973;55(2):135–142. doi: 10.1016/s0300-9084(73)80385-2. [DOI] [PubMed] [Google Scholar]
- Bear D. G., Schleich T., Noller H. F., Garrett R. A. Alteration of 5S RNA conformation by ribosomal proteins L18 and L25. Nucleic Acids Res. 1977 Jul;4(7):2511–2526. doi: 10.1093/nar/4.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemare G., Jordan B. R., Rocca-Serra J., Monier R. Accessibility of Escherichia coli 5S RNA base residues to chemical reagents. Influence of chemical alterations on the affinity of 5S RNA for the 50S subunit structure. Biochimie. 1972;54(11):1453–1466. doi: 10.1016/s0300-9084(72)80087-7. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Sriwidada J., Fellner P., Crichton R. The identification of the RNA binding site for a 50 S ribosomal protein by a new technique. FEBS Lett. 1973 Sep 15;35(2):265–272. doi: 10.1016/0014-5793(73)80301-1. [DOI] [PubMed] [Google Scholar]
- Brosius J. Primary structure of Escherichia coli ribosomal protein L31. Biochemistry. 1978 Feb 7;17(3):501–508. doi: 10.1021/bi00596a020. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F., Barrell B. G. The sequence of 5 s ribosomal ribonucleic acid. J Mol Biol. 1968 Jun 28;34(3):379–412. doi: 10.1016/0022-2836(68)90168-x. [DOI] [PubMed] [Google Scholar]
- Chen-Schmeisser U., Garrett R. A. A new method for the isolation of a 5 S RNA complex with proteins L5, L18 and L25 from Escherichia coli ribosomes. FEBS Lett. 1977 Mar 1;74(2):287–291. doi: 10.1016/0014-5793(77)80866-1. [DOI] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Fahnestock S., Higo K., Nomura M. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Fuenteun J., Monier R., Garrett R., Le Bret M., Le Pecq J. B. Effect of 50 S subunit proteins L5, L18 and L25 on the fluorescence of 5 S RNA-bound ethidium bromide. J Mol Biol. 1975 Apr 25;93(4):535–541. doi: 10.1016/0022-2836(75)90245-4. [DOI] [PubMed] [Google Scholar]
- Garrett R. A., Rak K. H., Daya L., Stöffler G. Ribosomal proteins. XXIX. Specific protein binding sites on 16S rRNA of Escherichia coli. Mol Gen Genet. 1972;114(2):112–124. doi: 10.1007/BF00332782. [DOI] [PubMed] [Google Scholar]
- Garrett R. A., Ungewickell E., Newberry V., Hunter J., Wagner R. An RNA core in the 30S ribosomal subunit of Escherichia coli and its structural and functional significance. Cell Biol Int Rep. 1977 Nov;1(6):487–502. doi: 10.1016/0309-1651(77)90086-8. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Bellemare G., Monier R. Degradation of a specific 5 S RNA - 23 S RNA - protein complex by pancreatic ribonuclease. FEBS Lett. 1972 Aug 1;24(2):156–160. doi: 10.1016/0014-5793(72)80756-7. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Bellemare G., Monier R., Garrett R. A., Stöffler G. Identification of the nucleotide sequences involved in the interaction between Escherichia coli 5 RNA and specific 50 S subunit proteins. J Mol Biol. 1973 Jun 15;77(1):133–152. doi: 10.1016/0022-2836(73)90367-7. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Garrett R. A., Stoffler G., Monier R. An attempt at the identification of the proteins involved in the incorporation of 5-S RNA during 50-S ribosomal subunit assembly. Eur J Biochem. 1972 Jul 24;28(3):412–421. doi: 10.1111/j.1432-1033.1972.tb01927.x. [DOI] [PubMed] [Google Scholar]
- Hindennach I., Kaltschmidt E., Wittmann H. G. Ribosomal proteins. Isolation of proteins from 50S ribosomal subunits of Escherichia coli. Eur J Biochem. 1971 Nov 11;23(1):12–16. doi: 10.1111/j.1432-1033.1971.tb01585.x. [DOI] [PubMed] [Google Scholar]
- Horne J. R., Erdmann V. A. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol Gen Genet. 1972;119(4):337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry V., Brosius J., Garrett R. Fragment of protein L18 from the Escherichia coli ribosome that contains the 5S RNA binding site. Nucleic Acids Res. 1978 Jun;5(6):1753–1766. doi: 10.1093/nar/5.6.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry V., Yaguchi M., Garrett R. A. A trypsin-resistant fragment from complexes of ribosomal protein S4 with 16-S RNA of Escherichia coli and from the uncomplexed protein. Eur J Biochem. 1977 Jun 1;76(1):51–61. doi: 10.1111/j.1432-1033.1977.tb11569.x. [DOI] [PubMed] [Google Scholar]
- Osterberg R., Sjöberg B., Garrett R. A. Molecular model for 5-S RNA. A small-angle x-ray scattering study of native, denatured and aggregated 5-S RNA from Escherichia coli ribosomes. Eur J Biochem. 1976 Sep 15;68(2):481–487. doi: 10.1111/j.1432-1033.1976.tb10835.x. [DOI] [PubMed] [Google Scholar]
- Schulte C., Morrison C. A., Garrett R. A. Protein-ribonucleic acid interactions in Escherichia coli ribosomes. Solution studies on S4-16S ribonucleic acid and L24-23S ribonucleic acid binding. Biochemistry. 1974 Feb 26;13(5):1032–1037. doi: 10.1021/bi00702a031. [DOI] [PubMed] [Google Scholar]
- Spierer P., Bogdanov A. A., Zimmermann R. A. Parameters for the interaction of ribosomal proteins L5, L18, and L25 with 5S RNA from Escherichia coli. Biochemistry. 1978 Dec 12;17(25):5394–5398. doi: 10.1021/bi00618a012. [DOI] [PubMed] [Google Scholar]
- Spierer P., Zimmermann R. A. Stoichiometry, cooperativity, and stability of interactions between 5S RNA and proteins L5, L18, and L25 from the 50S ribosomal subunit of Escherichia coli. Biochemistry. 1978 Jun 27;17(13):2474–2479. doi: 10.1021/bi00606a002. [DOI] [PubMed] [Google Scholar]
- Tischendorf G. W., Zeichhardt H., Stöffler G. Architecture of the Escherichia coli ribosome as determined by immune electron microscopy. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4820–4824. doi: 10.1073/pnas.72.12.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Jordan B. R. Partial enzyme digestion studies on Escherichia coli, Pseudomonas, Chlorella, Drosophila, HeLa and yeast 5S RNAs support a general class of 5S RNA models. J Mol Evol. 1977 Sep 20;10(1):77–86. doi: 10.1007/BF01796136. [DOI] [PubMed] [Google Scholar]
- Zimmermann J., Erdmann V. A. Binding sites of E. coli and B. stearothermophilus ribosomal proteins on B stearothermophilus 5S RNA. Nucleic Acids Res. 1978 Jul;5(7):2267–2288. doi: 10.1093/nar/5.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J., Erdmann V. A. Identification of Escherichia coli and Bacillus stearothermophilus ribosomal protein binding sites on Escherichia coli 5S RNA. Mol Gen Genet. 1978 Apr 17;160(3):247–257. doi: 10.1007/BF00332968. [DOI] [PubMed] [Google Scholar]