Abstract
Hypertension affects one billion people and is a principal reversible risk factor for cardiovascular disease. A rare Mendelian syndrome, pseudohypoaldosteronism type II (PHAII), featuring hypertension, hyperkalemia, and metabolic acidosis, has revealed previously unrecognized physiology orchestrating the balance between renal salt reabsorption versus K+ and H+ excretion1. We used exome sequencing to identify mutations in Kelch-like 3 (KLHL3) or Cullin 3 (CUL3) in 41 PHAII kindreds. KLHL3 mutations are either recessive or dominant, while CUL3 mutations are dominant and predominantly de novo. CUL3 and BTB-Kelch proteins such as KLHL3 are components of Cullin/RING E3 ligase complexes (CRLs) that ubiquitinate substrates bound to Kelch propeller domains2–8. Dominant KLHL3 mutations are clustered in short segments within the Kelch propeller and BTB domains implicated in substrate9 and Cullin5 binding, respectively. Diverse CUL3 mutations all result in skipping of exon 9, producing an in-frame deletion. Because dominant KLHL3 and CUL3 mutations both phenocopy recessive loss-of-function KLHL3 mutations, they may abrogate ubiquitination of KLHL3 substrates. Disease features are reversed by thiazide diuretics, which inhibit the Na-Cl cotransporter (NCC) in the distal nephron of the kidney; KLHL3 and CUL3 are expressed in this location, suggesting a mechanistic link between KLHL3/CUL3 mutations, increased Na-Cl reabsorption, and disease pathogenesis. These findings demonstrate the utility of exome sequencing in disease gene identification despite combined complexities of locus heterogeneity, mixed models of transmission, and frequent de novo mutation, and establish a fundamental role for KLHL3/CUL3 in blood pressure, K+, and pH homeostasis.
A small number of genes causing Mendelian forms of hypertension have been identified, establishing the role of increased renal salt reabsorption in its pathogenesis10–12. The study of pseudohypoaldosteronism type II (PHAII) has identified a physiologic mechanism that orchestrates activities of diverse electrolyte flux pathways, allowing maximal salt reabsorption in response to aldosterone when angiotensin II (AII) is elevated, as in settings of reduced intravascular volume (hypovolemia), versus maximal potassium secretion in settings of hyperkalemia, in which aldosterone is elevated without changes in AII1. The role of WNK kinases in this process was revealed by discovery of their mutation in a small fraction of PHAII kindreds11. Dominant gain-of-function mutations in WNK4 or WNK1 lead to constitutively increased salt reabsorption in the distal nephron regardless of volume status, resulting in hypertension, and inhibition of K+ secretion despite marked hyperkalemia1,11,13–17.
We studied a cohort of 52 PHAII kindreds, including 126 affected subjects with renal hyperkalemia and otherwise normal renal function; hypertension and acidosis were present in 71% and 82%, respectively. There was wide variation in disease severity and age of clinical presentation (Supplementary Figs. 1 and 2). Mutations in WNK1 or WNK4 were present in only seven of these kindreds (13%). Those without WNK mutations had only 2.0 + 1.4 affected members, complicating mapping efforts.
Exome sequencing of eleven unrelated PHAII index cases without WNK mutations was performed. Index cases and affected relatives (five trios and one quartet) were also subjected to genome-wide SNP genotyping. Tabulation of high quality novel protein-altering variants revealed 124 genes with three or more variants, 50 with four or more, and 23 with five. Concurrent analysis of linkage among the multiplex families was used to prioritize loci harboring variants that co-segregated with disease; this identified 28 genes with novel protein-altering variants that co-segregated with disease in two or more multiplex families. This revealed Kelch-like 3 (KLHL3) as a strong candidate, with novel KLHL3 mutations comprising five alleles in three kindreds, all of which co-segregated with the trait. These include one kindred in which affected members are homozygous for a nonsense mutation (W470X), one in which affected members are compound heterozygotes for two missense mutations (F322C and S410L), and one segregating a heterozygous missense mutation (R528H). As a confirmation of significance, Fisher’s exact test was used to compare the prevalence of novel protein-altering variants in all genes in PHAII cases versus 699 control exomes. A single gene, KLHL3, showed a burden of mutation that surpassed genome-wide significance (p = 1.1 × 10−8; Supplementary Tables 1–3).
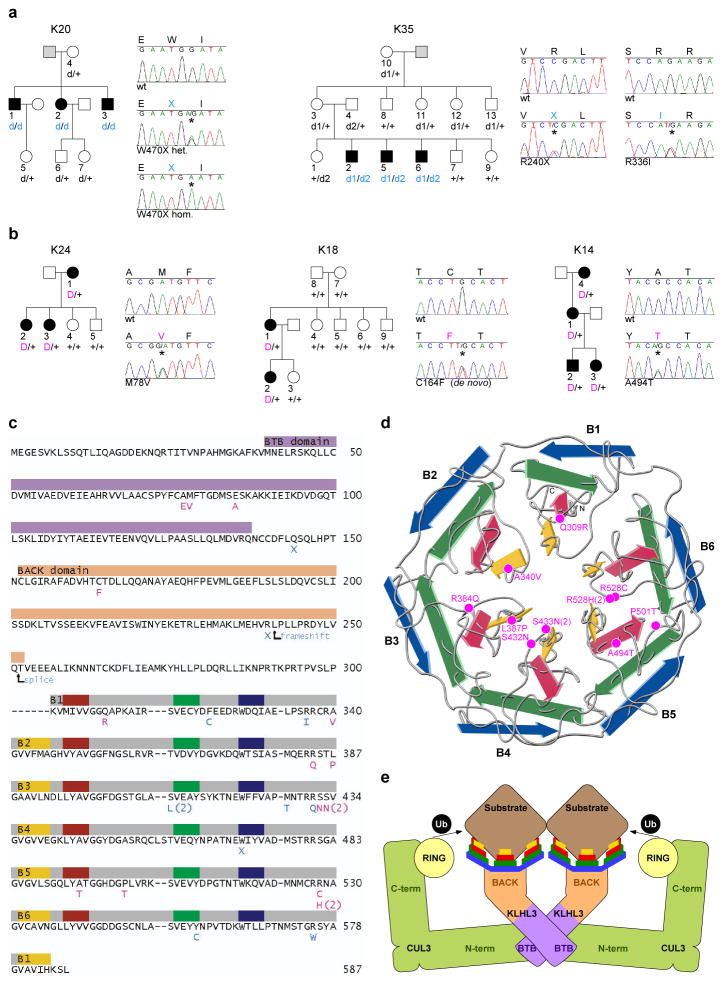
KLHL3 was sequenced in all PHAII index cases, identifying novel mutations in 24 (Fig. 1a–b, Supplementary Figs. 3 and 4). Nearly all are at positions conserved among orthologs (Supplementary Fig. 5). Sixteen kindreds have heterozygous mutations that co-segregate with the trait under a dominant model (lod score 6.9, < −2 under other models). In contrast, eight index cases inherited mutations in both KLHL3 alleles. In these kindreds, affected members are confined to siblings of index cases who inherited the same two mutations, while unaffected relatives inherited zero or one mutation (lod score 4.3 for a recessive model, < −2 for other models). Recessive transmission has not been previously described for PHAII. Consistent with two modes of transmission, subjects with dominant KLHL3 mutations had significantly higher serum K+ levels (6.2 ± 0.6 mM) than heterozgyotes for recessive mutations (4.8 ± 0.6 mM) (p < 10−4, Student’s t-test; normal range 3.5–5.0 mM). These findings establish that PHAII can be caused by either recessive or dominant KLHL3 mutations. Importantly, we infer that mutations in dominant kindreds are likely dominant-negative, because they phenocopy the features of recessive disease.
Figure 1. Recessive and dominant KLHL3 mutations in PHAII kindreds.
a–b, Representative kindreds demonstrating recessive (a) and dominant (b) KLHL3 mutations (all 24 kindreds are shown in Supplementary Figs. 3–4). Affected, unaffected, and phenotype-undetermined subjects are denoted by black, white, and gray symbols, respectively. KLHL3 alleles are denoted by ‘+’ (wild-type), ‘d’ (recessive mutation), and ‘D’ (dominant mutation). Sequence traces show wild-type (wt) and mutant (*) alleles and encoded amino acids. c, KLHL3 protein sequence. Colored bars indicate BTB domain (lavender), BACK domain (peach), and Kelch propeller blades (B1-B6, gray) with β-strands ‘a’–‘d’ in yellow, red, green, and blue respectively. Recessive (aqua) and dominant (pink) mutations are shown; recurrences indicated by numbers. d, Kelch propeller schematic, from KLHL2 crystal structure27. β-strands colored as in c; dominant mutations indicated. e, CRL schematic, comprising a BTB-Kelch protein (KLHL3), CUL3, and a ubiquitin transfer-mediating RING protein, with substrate bound via the Kelch propeller. Complex shown as a dimer7.
KLHL3 contains an N-terminal BTB domain, a BACK domain, and C-terminal Kelch-like repeats that form a six-bladed β-propeller structure2,4,5 (Fig. 1c–e). There are over 50 BTB-Kelch genes in humans4; their propeller domains bind substrate proteins, promoting substrate ubiquitination via interaction of the BTB domain with Cullin 3 (CUL3), a component of a Cullin/RING E3 ubiquitin ligase (CRL)3,5,6. Ubiquitination serves diverse functions, including targeting proteins for proteasomal degradation as well as non-degradative roles such as modulation of protein activity, interaction, and localization7,8.
While recessive KLHL3 mutations are distributed throughout the encoded protein, dominant KLHL3 mutations show striking clustering (Fig. 1c). Nine of sixteen dominant mutations alter one of the last four amino acids of the six ‘d-a’ loops that connect the outermost (‘d’) β-strand of one Kelch propeller blade to the innermost (‘a’) β-strand of the next blade. Two others are in ‘b-c’ loops. These dominant PHAII mutations lie near the hub of the propeller (Fig. 1d) at or near sites implicated in substrate binding in paralogs9 (Supplementary Fig. 5). Three other dominant mutations cluster within the BTB domain, at or near sites implicated in Cullin binding in paralogs5. We infer that dominant mutations in KLHL3 likely impair binding either to specific substrates or to CUL3.
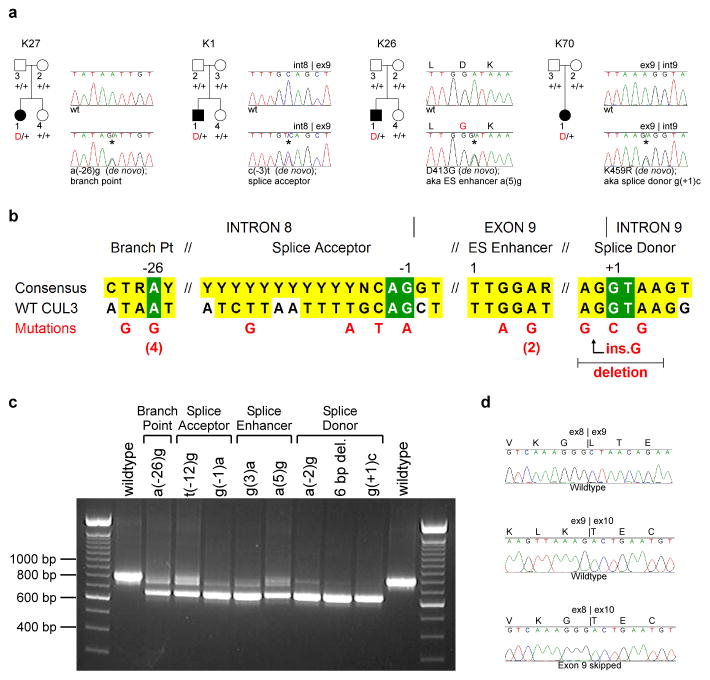
After accounting for KLHL3, WNK1, and WNK4 mutations, 21 PHAII kindreds without mutations remained. We considered KLHL3’s presumed functional partner, CUL3 (Fig. 1e), as a potential candidate. Among PHAII exomes, novel heterozygous CUL3 variants were suggested in two. Sequencing CUL3 in all index cases identified seventeen with novel heterozygous mutations, all in cases without KLHL3, WNK1 or WNK4 mutations (Fig. 2a, Supplementary Fig. 6). Eight of these mutations were documented to be de novo, providing overwhelming evidence that these mutations are disease-causing. CUL3 mutations all cluster in sites likely involved in splicing of exon 9, including the intron 8 splice acceptor (n = 4), the intron 9 splice donor (n = 5), the putative intron 8 splice branch site (n = 5), and a putative splice enhancer in exon 9 (n = 3, within a TTGGA[T/A] splice enhancer consensus sequence18) (Fig. 2b).
Figure 2. Dominant CUL3 mutations in PHAII kindreds cause skipping of exon 9.
a, Representative kindreds demonstrating CUL3 mutations, depicted as in Fig. 1 (all 17 kindreds are shown in Supplementary Fig. 6). b, CUL3 mutation locations. Consensus splicing sequences18,28 and corresponding wildtype CUL3 sequences within intron 8, exon 9, and intron 9 are shown; invariant bases (green) and consensus homology (yellow) are indicated. Positions numbered relative to splice sites and first base of the exonic splice (ES) enhancer. Mutations shown in red; recurrences indicated by numbers. c, RT-PCR of spliced RNA. Wild-type CUL3 constructs produce a single product including exons 8, 9, and 10 (844 bp); all nine mutants tested produce a predominant product that skips exon 9 (673 bp). d, Representative RT-PCR sequences. Wild-type construct produces cDNA with properly spliced junctions between exons 8–9 (top) and 9–10 (middle), while mutant construct [splice donor g(+1)c] produces cDNA joining exon 8 to exon 10 (bottom).
To test the impact of these mutations on splicing, CUL3 genomic DNA spanning exon 8 to exon 10, containing either wild-type sequence or one of nine PHAII mutations, was cloned and expressed in HEK293 cells, and the spliced RNA products were analyzed. While the wild-type sequence produces a properly spliced product containing all three exons, each of the mutants produces a predominant product that skips exon 9, joining exon 8 to exon 10 (Fig. 2c–d). This results in an in-frame 57 amino acid deletion (residues 403–459) in the segment linking the BTB-binding and RING-binding domains of CUL3. The fact that CUL3 mutations phenocopy recessive KLHL3 mutations suggests that they abrogate CUL3 function at KLHL3 targets.
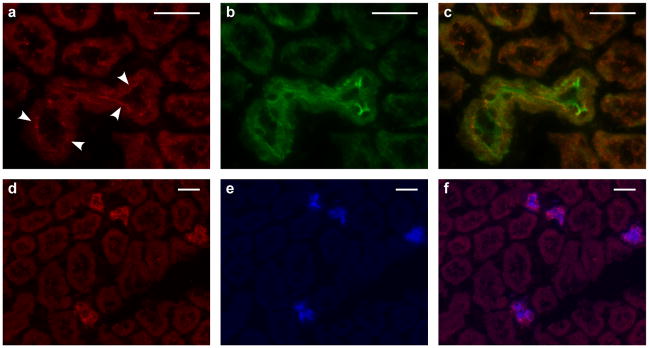
As with PHAII caused by WNK1 and WNK4 mutations1, virtually all patients with KLHL3 and CUL3 mutations have been treated with thiazide diuretics, inhibitors of the Na-Cl cotransporter NCC, with correction of phenotypic abnormalities. WNK4 regulates the activities of NCC13,14,16, the epithelial Na+ channel ENaC17, and the K+ channel ROMK15, and is co-expressed with these proteins in the renal distal convoluted tubule (DCT) and collecting duct (CD)11,19,20. Staining mouse kidney sections with specific antibodies demonstrated that KLHL3 is predominantly expressed in DCT and CD, with apical localization in DCT (Fig. 3). CUL3 is ubiquitously expressed and is in all nephron segments, with particularly high expression in the proximal tubule, but also in DCT and CD (Supplementary Fig. 7). These findings are consistent with both proteins playing a role in the regulation of salt and electrolyte homeostasis in the distal nephron.
Figure 3. KLHL3 expression in the kidney.
Mouse kidney sections stained with antibodies to KLHL3 (red), TRPM6 (a marker of the DCT29, green) and AQP2 (a marker of the CD30, blue). Scale bars 25 μm. a–c, Staining for KLHL3 (a), TRPM6 (b), and the merged image (c) demonstrates KLHL3 expression in the DCT with apical localization (arrowheads). d–f, Staining for KLHL3 (d), AQP2 (e), and the merged image (f) demonstrates KLHL3 expression in CD.
There are highly significant differences in phenotypic severity among PHAII patients with mutations in different genes (Table 1, Supplementary Figs. 1 and 2, Supplementary Table 4). Subjects with CUL3 mutations presented at much younger ages than those with mutation in KLHL3, WNK1, or WNK4, and had significantly more severe hyperkalemia and metabolic acidosis and were far more likely to have hypertension before age 18 (others commonly develop hypertension at later ages). The majority of subjects with CUL3 mutations demonstrated failure to thrive or growth impairment. These observations, in conjunction with the high rate of CUL3 de novo mutation, support impairment of reproductive fitness. Among the other mutant loci, there remain significant differences in disease severity (rank order recessive KLHL3 > dominant KLHL3 > WNK4 > WNK1).
Table 1.
PHAII phenotypes, stratified by genotype.
| Mutant Gene | # Kindreds | # Affecteds | Dx/Ref Age | K+ (mM) (nl 3.5–5.0) | HCO3− (mM) (nl 22–28) | % htn ≤age 18 |
|---|---|---|---|---|---|---|
| CUL3 | 17 | 21 | 9 ±6 | 7.5 ±0.9 | 15.5 ±2.0 | 94 |
| KLHL3 Recessive | 8 | 14 | 26 ±14 | 6.8 ±0.5 | 17.6 ±1.5 | 14 |
| KLHL3 Dominant | 16 | 40 | 24 ±18 | 6.2 ±0.6 | 17.2 ±2.5 | 17 |
| WNK4 | 5 | 15 | 28 ±18 | 6.4 ±0.7 | 20.8 ±2.3 | 10 |
| WNK1 | 2 | 23 | 36 ±20 | 5.8 ±0.8 | 22.4 ±4.6 | 13 |
| p = 0.0002 | p < 0.0001 | p < 0.0001 | p < 0.0001 |
Dx/Ref Age, age at diagnosis or referral; K+, serum potassium; HCO3−, serum bicarbonate; % htn ≤age 18, % affecteds diagnosed with hypertension by age 18; nl, normal range. Values for Dx/Ref Age, K+, and HCO3− are means ± standard deviations. Significance of differences among genotype classes was calculated by ANOVA (Dx/Ref Age, K+, HCO3−) or Fisher’s exact test (% htn ≤18).
KLHL3 and CUL3 mutations account for 79% of kindreds in our cohort. Gene identification of was complicated by the combined effects of locus heterogeneity, two modes of transmission at one locus, and few informative meioses. Many heretofore unsolved Mendelian traits may have similar complexities. Use of control exomes as comparators for analysis of mutation burden may be broadly applicable to discovery of such loci.
The most parsimonious mechanism of KLHL3 and CUL3 mutations is that they abrogate ubiquitination of targets normally bound by KLHL3, activity that is required for normal modulation of renal salt, K+, and H+ handling in response to physiologic challenge; this speculation will require biochemical verification. The fact that recessive mutations in KLHL3 cause PHAII without other diverse effects implies either that KLHL3 targets are highly restricted to the renal salt and electrolyte pathway, or that loss of KLHL3 function at other targets can be compensated by other loci. BTB-Kelch/CUL3 CRLs can act as dimers, with two substrate-binding domains capable of engaging the same target molecule7. This suggests a potential mechanism to explain dominant-negative effects of KLHL3 and CUL3 mutations.
CUL3-based CRLs participate in a wide range of critical cellular processes8 via binding diverse BTB domain-containing proteins3,5. CUL3 mutations affecting all or many of these activities would undoubtedly produce very broad phenotypes. CUL3 mutations in PHAII merely phenocopy the effects of loss of KLHL3, suggesting they selectively abrogate function at KLHL3 targets. The stereotypic consequences of CUL3 mutations, all deleting 57 amino acids in a region linking the BTB-binding and RING-binding domains, support such a specific effect. Consistent with this possibility, introduction of a flexible linker sequence to this region of CUL1 leaves substrate protein binding and ubiquitin polymerization intact, but nonetheless abolishes ubiquitination of a normal substrate21.
Thiazide diuretics correct abnormalities in virtually all PHAII subjects; similar correction is seen in a mouse model of PHAII with either thiazides or genetic ablation of NCC16. These findings suggest that increased NCC activity is likely to be a common pathogenic mechanism. Co-expression of KLHL3 and CUL3 with NCC in DCT, and evidence that NCC is ubiquitinated22 is consistent with this notion. ROMK and the H+ ATPase are respectively required for net renal K+ secretion23 and H+ secretion24 and are also likely targets, although their activity is expected to be decreased, rather than increased, in PHAII. Another thiazide-sensitive Na-Cl cotransport pathway in the collecting duct has recently been described, suggesting an additional potential target25. Whether a KLHL3/CUL3 CRL acts directly or indirectly on these targets, whether they alter delivery of NCC and other targets to, or retrieval from, the plasma membrane, and what upstream pathways regulate this activity is unknown. Similarly, whether KLHL3/CUL3 and WNKs operate within the same or different pathways is presently unknown; it is of interest that segments of NRF2 that interact with the Kelch propeller domain of KEAP1 are highly acidic9,26, akin to the domain of WNK4 that is mutated in PHAII11.
These findings demonstrate previously unrecognized roles for KLHL3 and CUL3. Understanding the upstream regulators and downstream targets of KLHL3/CUL3 activity will provide further insight into mechanisms underlying maintenance of blood pressure and electrolyte homeostasis in response to diverse environmental challenges.
Methods summary
A cohort of 52 PHAII kindreds comprising 126 affected subjects was ascertained, characterized, and recruited for study. Index cases of eleven kindreds were chosen for whole exome capture and sequencing, and novel variants were identified. Genes were prioritized for follow-up and subjected to Sanger sequencing of index cases of each kindred. Segregation of rare variants within kindreds was analyzed. KLHL3 and CUL3 were localized in kidney by staining with specific antibodies and immunofluorescence microscopy. Effects of CUL3 mutations on RNA splicing were studied by analysis of spliced products produced in mammalian cells.
Full Methods and associated references are available in the online version of the paper at www.nature.com/nature.
Methods
Study subjects
Index cases were referred for pseudohypoaldosteronism type II (PHAII). Patients and participating family members provided consent to a study protocol approved by the Yale Human Investigation Committee. Control exomes were 699 unrelated subjects of European ancestry without hypertension, sequenced as part of diverse gene discovery projects. Genomic DNA was isolated from venous blood via standard methods.
Exome capture, sequencing and variant calling
Genomic DNA from eleven PHAII index cases and 699 controls was captured on NimbleGen 2.1M human exome arrays (Roche) and sequenced on the Illumina GenomeAnalyzer as previously described31. Reads were mapped to the reference genome (hg18) using Maq32 and genotypes of targeted bases were called with SAMtools33. Variants found in dbSNP v130 or 1000 Genomes databases were excluded from further analysis. Remaining variants were considered ‘novel’ and annotated for impact on the encoded protein, conservation, and expression31. Aligned reads were viewed with the Integrative Genomics Viewer34. Among PHAII cases, 94.2% of targeted bases were read by 8 or more independent reads; sensitivity and specificity of heterozygous calls were estimated at 93.7% and 99.9% by comparison to Illumina SNP genotyping. Among controls, 94.4% of targeted bases were read by 8 or more independent reads; sensitivity and specificity of heterozygous calls were estimated at 94.5% and 99.8%. Sanger sequencing of 212 novel variants from controls with SAMtools quality score ≥ 100 demonstrated validation in 211 and amplification failure in 1, supporting high specificity of variant calls.
SNP genotyping and linkage analysis
For the eleven PHAII index cases and their affected relatives (five trios and one quartet) genome-wide SNP genotyping was performed using Illumina Human610-Quad BeadChips and GenomeStudio software. Approximately 40K tag SNPs were extracted using Plink35. Analysis of linkage was performed using Merlin36, specifying an autosomal dominant model with no phenocopies. Variants from exome sequencing in regions of the genome that were excluded (lod score < -2) were removed from further analysis, while those that supported linkage were prioritized for further evaluation. In kindreds showing potential recessive transmission of PHAII, SNP genotypes were examined for regions of homozygosity, and linkage was performed specifying an autosomal recessive model.
Sanger sequencing of KLHL3 and CUL3
PCR amplification and Sanger sequencing from genomic DNA was performed using standard methods. Primers were designed with Primer337. Variants identified by exome sequencing were verified. All exons and flanking intronic sequences of KLHL3 and CUL3 were sequenced from all PHAII index cases. Previously unidentified mutations were discovered and verified by independent amplification and sequencing. Co-segregation of mutations with disease was determined by sequencing in all available kindred members. CUL3 exon 9 and its flanking intronic sequence was sequenced in 150 unaffected unrelated controls, none of whom were found to harbor previously unidentified variants. It is noteworthy that because of lower or absent sequence coverage at or near intron-exon junctions, splice donor and acceptor mutations in CUL3 were suggested in two of the 11 PHAII exomes (SAMtools quality scores 96 and 75) but three branch site mutations were outside the exome sequence and one splice enhancer mutation was poorly covered (SAMtools quality score 3).
Genome-wide assessment of mutation burden
Genes show substantial variation in the prevalence of novel or rare protein-altering variants for biological reasons, including differences in gene size and variation in the proportion of bases that are under purifying selection, and for technical reasons, including difficulties in accurately mapping short sequence reads among closely related paralogs. These factors can limit the ability to directly identify disease loci by simply counting and ranking genes according to the absolute number of such variants, particularly for diseases with substantial locus or model heterogeneity. This gene-to-gene variation can be accounted for by use of control exome data. The prevalence of rare variation in each gene in case exomes was compared to the corresponding prevalence in a large set of control exomes with a Fisher’s exact test. Variants included in the analysis were protein-altering (missense, nonsense, and splice site mutations) and high quality (≥ 8 independent reads and SAMtools quality score ≥ 100). For a gene-wise test of rare variant burden in a genome with ~21,000 genes, correction for multiple testing suggests a threshold p-value of ~2.4 × 10−6, anticipated to produce a false discovery rate (FDR) of one gene per twenty experiments. The false discovery rate of the Fisher’s test was evaluated by Monte Carlo simulation, which confirmed an FDR < 1 gene per 20 experiments (Supplementary Table 1). The power to identify trait-related loci was estimated as a function of the number of variants detected in cases and the number of case exomes sequenced (Supplementary Table 2), and the test was applied comparing the eleven PHAII and 699 control exomes (Supplementary Table 3).
Ortholog and paralog comparisons
Protein sequences of orthologs and paralogs were aligned with Clustal W38. Crystal structures were examined with Cn3D39. The locations of KLHL3 propeller mutations were compared to the crystal structure of human KLHL2 (PDB ID: 2XN4)27, the closest human paralog3 (85% amino acid identity in the propeller). The location of the peptide encoded by CUL3 exon 9 was approximated by comparison to the crystal structure of human CUL1 (PDB IDs: 1LDJ, 1LDK)22,27.
Splicing assay
A 3782 bp segment of CUL3, extending from 287 bp proximal to exon 8 to 327 bp distal to exon 10, was amplified by PCR (Advantage 2 polymerase, Clontech) from genomic DNA of nine PHAII patients with different CUL3 mutations and one subject with wild-type CUL3 sequence. Products were cloned into the pcDNA6.2/GW/D-TOPO mammalian expression vector (Invitrogen), and plasmids were purified (QIAprep, Qiagen) and sequenced. HEK293 cells were transfected independently with each plasmid using Lipofectamine 2000 (Invitrogen) and harvested ~24 hours post-transfection. RNA was isolated using RNeasy with DNase on-column digestion (Qiagen). The spliced expression products were assessed by reverse transcription with oligo(dT) priming (Superscript III RT, Invitrogen) followed by PCR with vector-specific and CUL3-specific primers. Products were fractionated and visualized via agarose gel electrophoresis, and sequenced. Untransfected and water controls were negative.
Immunofluorescence
Fresh frozen mouse kidney sections were fixed with ethanol at 4°C for 30 minutes and acetone at −20°C for three minutes, washed with 1X PBS, and permeabilized with 0.1% Triton X-100 (Sigma) at room temperature for ten minutes. Sections were blocked with 10% donkey serum and 1% bovine serum albumin at room temperature for one hour, incubated with primary antibodies at room temperature for one hour or 4°C overnight, washed four times with 1X PBS, incubated with secondary antibody at room temperature for one hour, and washed four times with 1X PBS, with DAPI nuclear counterstain in the second wash. Slides were mounted with Mowiol (Polysciences) and 1% n-propyl gallate (Sigma) as an anti-fade agent. Primary antibodies included 1:100 rabbit anti-KLHL3, 1:50 rabbit anti-CUL3, 1:800 or 1:1200 guinea pig anti-TRPM6 (ab66655, ab1871, and ab47017; Abcam) and 1:400 or 1:800 goat anti-AQP2 (sc-9882, Santa Cruz Biotechnology). Secondary antibodies, diluted 1:800, included donkey Cy3 anti-rabbit, Cy2 anti-guinea pig, and 649 anti-goat IgG (AffiniPure, Jackson Immunoresearch). Staining with secondary antibodies only was consistently negative.
Supplementary Material
Acknowledgments
We thank the PHAII subjects, their families, and health care professionals whose participation made this study possible; S. Umlauf and the staff of the Yale Center for Genome Analysis; J. Santosuosso; H. Tirrell and the staff of Beckman Coulter Genomics; V. Klump, Y. Lu, U. Scholl, and J. Zhou for providing reagents; W. Hill for artistic assistance with Fig. 1d; and E. Boyden, S. Boyden, L. Cooley, and M. Hochstrasser for helpful discussions. Supported in part by the Leducq Transatlantic Network on Hypertension.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions L.M.B., M.C., K.A.C., and R.P.L. designed experiments and analyzed data. L.M.B., C.N.W., and I.R.T. performed experiments. A.F., H.R.T., G.C., M.L., R.D.G., B.A.S., A.P., M.J.V., M.E.D.F., S.A.S., M.G., F.E.K., J.R.T., J.R.S., K.M.K.N., C.C.P., S.K.A., M.L.W., I.D.D., S.B.D., A.B., J.J.F., C.W.B., T.E.H., R.D.N., H.T., T.R.P.C., M.P., D.B., M.S., P.V., J.W.F., M.R., F.T., H.Z.A.S., J.R., A.G.G., and B.G. recruited PHAII subjects and families. R.B. and S.M.M. directed the IT and DNA sequencing infrastructure. L.M.B. and R.P.L. wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Accession numbers: KLHL3: NCBI accessions NM_017415.2, NP_059111.2; CUL 3: NCBI accessions NM_003590.3, NP_003581.1
References
- 1.Kahle KT, Ring AM, Lifton RP. Molecular physiology of the WNK kinases. Annu Rev Physiol. 2008;70:329–355. doi: 10.1146/annurev.physiol.70.113006.100651. [DOI] [PubMed] [Google Scholar]
- 2.Lai F, et al. Molecular characterization of KLHL3, a human homologue of the Drosophila kelch gene. Genomics. 2000;66:65–75. doi: 10.1006/geno.2000.6181. [DOI] [PubMed] [Google Scholar]
- 3.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 4.Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson AM, Cooley L. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell Biochem. 2008;48:6–19. doi: 10.1007/978-0-387-09595-0_2. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 11.Wilson FH, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 12.Choi M, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson FH, et al. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci U S A. 2003;100:680–684. doi: 10.1073/pnas.242735399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest. 2003;111:1039–1045. doi: 10.1172/jci17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahle KT, et al. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet. 2003;35:372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 16.Lalioti MD, et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet. 2006;38:1124–1132. doi: 10.1038/ng1877. [DOI] [PubMed] [Google Scholar]
- 17.Ring AM, et al. WNK4 regulates activity of the epithelial Na+ channel in vitro and in vivo. Proc Natl Acad Sci U S A. 2007;104:4020–4024. doi: 10.1073/pnas.0611727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 19.Bachmann S, Bostanjoglo M, Schmitt R, Ellison DH. Sodium transport-related proteins in the mammalian distal nephron - distribution, ontogeny and functional aspects. Anat Embryol (Berl) 1999;200:447–468. doi: 10.1007/s004290050294. [DOI] [PubMed] [Google Scholar]
- 20.Welling PA, Ho K. A comprehensive guide to the ROMK potassium channel: form and function in health and disease. Am J Physiol Renal Physiol. 2009;297:F849–863. doi: 10.1152/ajprenal.00181.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 22.Ko B, et al. RasGRP1 stimulation enhances ubiquitination and endocytosis of the sodium-chloride cotransporter. Am J Physiol Renal Physiol. 2010;299:F300–309. doi: 10.1152/ajprenal.00441.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon DB, et al. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996;14:152–156. doi: 10.1038/ng1096-152. [DOI] [PubMed] [Google Scholar]
- 24.Karet FE, et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- 25.Leviel F, et al. The Na+-dependent chloride-bicarbonate exchanger SLC4A8 mediates an electroneutral Na+ reabsorption process in the renal cortical collecting ducts of mice. J Clin Invest. 2010;120:1627–1635. doi: 10.1172/jci40145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong KI, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/mcb.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, et al. MMDB: annotating protein sequences with Entrez’s 3D-structure database. Nucleic Acids Res. 2007;35:D298–300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang MQ. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 29.Voets T, et al. TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J Biol Chem. 2004;279:19–25. doi: 10.1074/jbc.M311201200. [DOI] [PubMed] [Google Scholar]
- 30.Fushimi K, et al. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature. 1993;361:549–552. doi: 10.1038/361549a0. [DOI] [PubMed] [Google Scholar]
- 31.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.