Abstract
Chemoreception is a principle modality by which organisms gain information from their environment, and extensive variation in odor-mediated behavior has been documented within and among species. To examine the mechanisms by which sensory systems mediate these responses, we ask to what extent variation in Drosophila melanogaster odorant receptor genes contributes to variation in odor-mediated behavior. Significant differences in behavioral responses to structurally similar odorants, methyl hexanoate and ethyl hexanoate, were found in a natural population. Polymorphisms in 3 genomic regions (Or22a/Or22b, Or35a, and Or47a) were identified and associated with variation in behavior to these esters. Overall similarity in association profiles for both odorants was observed, except for Or47a in which polymorphisms were associated solely with variation in responses to ethyl hexanoate. Our analyses were then extended to examine polymorphisms in 3 odorant receptors previously reported to contribute to variation in olfactory behavior for the chemically distinct odorants benzaldehyde and acetophenone. Two Or10a polymorphisms were associated with variation in response to ethyl hexanoate. Finally, differences in Or35a and Or47a expression were associated with variation in responses to ethyl hexanoate. These results demonstrate that the genetic variation at the peripheral sensory stage plays a role in mediating differences in odor-mediated behavior.
Keywords: behavior, chemosensory, olfaction, single nucleotide polymorphism
Introduction
Understanding the mechanisms by which sensory systems mediate behavioral responses to stimuli is a fundamental goal in behavioral neuroscience, and numerous advances have been made uncovering both genetic mechanisms and neural circuitry involved in sensory transduction and perception. In olfaction, for instance, it is well established that chemicals are bound by a diverse family of receptor proteins expressed in olfactory receptor neurons (ORNs), whose combinatorial activation results in a spatial pattern of brain activity (Buck and Axel 1991; Vassar et al. 1994; Mombaerts et al. 1996; Laissue et al. 1999; Gao et al. 2000; Vosshall et al. 2000; Bhalerao et al. 2003; Wang et al. 2003). This neural code enables an organism to identify and discriminate among chemical signals in its environment. However, despite these advances, it remains unclear how underlying molecular polymorphisms in these receptors contribute to variation in olfaction-mediated behavior within and between species and the nature and pattern of polymorphisms contributing to receptor–ligand specificity.
The Drosophila melanogaster olfactory system affords us the opportunity to begin to address this issue because odorant receptor loci have been identified and many of their response spectra have been characterized (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999; Dobritsa et al. 2003; Hallem et al. 2004; Hallem and Carlson 2006). In Drosophila, ORNs are located within sensilla distributed over the third antennal segments and maxillary palps. Each sensillum contains up to 4 neurons with specific combinations of neurons restricted to distinct spatial antennal regions (de Bruyne et al. 1999, 2001). Each ORN typically expresses a single odorant receptor, and thus, the response profile of individual ORNs is determined by the individual odorant receptor expressed, with the majority of receptors responding to numerous and overlapping suites of ligands (Dobritsa et al. 2003; Wang et al. 2003; Hallem et al. 2004; Hallem and Carlson 2006).
The D. melanogaster odorant receptor (Or) family consists of 62 seven transmembrane proteins transcribed from 60 Or genes (Clyne et al. 1999; Vosshall et al. 2000; Robertson et al. 2003; Vosshall and Stocker 2007). The membrane topology of these receptors is opposite that of vertebrate odorant receptors, with the N-terminus in the cytoplasm of the ORN (Benton et al. 2006). Evidence also suggests that these odorant receptors dimerize with a highly conserved noncanonical receptor, Orco (formerly known as Or83b), that serves to localize odorant receptors in the sensory dendritic membrane (Jones et al. 2005; Benton et al. 2006) and form a class of ligand-gated nonselective cation channels (Sato et al. 2008; Wicher et al. 2008). Most recently, studies have also identified odorant receptor function in a group of ionotropic glutamate receptor–related proteins expressed in a subset of ORNs devoid of other previously identified odorant receptors (Benton et al. 2009; Abuin et al. 2011).
Alterations in both the central and the peripheral nervous systems contribute to extensive variation in olfaction-mediated behavioral responses within and among Drosophila species (Fuyama 1976, 1978; Alcorta and Rubio 1988, 1989; Mackay et al. 1996; Wang et al. 2007; Rollmann et al. 2010, Wang et al. 2010). In a comparative study between D. melanogaster and D. simulans, for example, differences in the specificity of particular ORNs was observed, with a shift in ligand affinity from ethyl hexanoate to methyl hexanoate (Stensmyr et al. 2003). Since the response profile of individual ORNs is determined by the odorant receptor expressed, Stensmyr et al. (2003) suggested that this shift may be due to a substitution in the ligand-binding domain of the receptor. Differences in sensitivity to these 2 odorants were also observed between D. melanogaster and D. sechellia with changes in sensitivity thought to be due to coding sequence changes in receptors as well as overexpression of methyl hexanoate-responsive neurons (Stensmyr et al. 2003; Dekker et al. 2006). Within species, polymorphisms in odorant receptor loci have also been implicated in mediating variation in olfactory behavior (Rollmann et al. 2010). In a natural D. melanogaster population, sequence variation in 3 odorant receptor loci, Or10a, Or43a, and Or67b, was associated with variation in behavioral responses to structurally similar odorants, benzaldehyde and acetophenone. Association profiles for both odorants revealed that distinct polymorphic sites in these loci contribute to differences in behavior and that some but not all of the same polymorphisms were associated with behavioral responses to both odorants (Rollmann et al. 2010).
Here, we examine how molecular polymorphisms in odorant receptor loci contribute to variation in behavior within a natural D. melanogaster population. We first assess the extent of variation in attraction/avoidance to 2 esters, ethyl hexanoate and methyl hexanoate, which are similar in structure, differing only by a single carbon length. These esters have been shown to be physiologically active and present in fruit extracts, the natural feeding/breeding substrate of Drosophila (Stensmyr, Giordano, et al. 2003). A significant difference in olfactory sensitivity to these esters has been previously observed among Drosophila species (Stensmyr et al. 2003; Dekker et al. 2006), but within-species variation had yet to be examined. Next, we identify numerous single nucleotide polymorphisms (SNPs) and insertion and deletion polymorphisms (indels) in 3 D. melanogaster odorant receptor loci, Or22a, Or35a, and Or47a that have been shown to be electrophysiologically tuned to these esters (Dobritsa et al. 2003; Hallem et al. 2004; Hallem and Carlson 2006). We also sequence Or22b due to its close physical proximity to Or22a (540 bp downstream) on the second chromosome, its coexpression in the same cell, and the previously reported presence of a single chimeric Or22ab gene within some D. melanogaster populations (Dobritsa et al. 2003; Aguadé 2009). Subsequently, we conduct genotype–phenotype association analyses to determine which polymorphic sites (if any) in these 4 Or loci contribute to variation in olfactory behavior and examine whether lines with overall more positive versus negative behavioral response indices to ethyl hexanoate show differences in Or gene expression levels.
Finally, we extend our analyses of genotype–phenotype relationships to assess whether genetic variation in 3 previously published odorant receptor loci, Or10a, Or43a, and Or67b (Rollmann et al. 2010) also contributes to the observed behavioral differences. Polymorphisms in these loci have been previously implicated in mediating variation in behavioral responses to odorants, benzaldehyde and acetophenone (Rollmann et al. 2010). We predict that no genotype–phenotype associations will be observed with variation in Or43a and Or67b because no ligand specificity to ethyl hexanoate or methyl hexanoate has been documented. However, ethyl hexanoate has been previously demonstrated to elicit inhibitory responses in Or10a expressing neurons. Our results demonstrate that changes at the most peripheral sensory stage contribute to within-species differences in behavioral responses to environmental stimuli.
Materials and methods
Experimental animals
Isofemale lines were derived from a natural population of D. melanogaster collected in Raleigh, NC. Each line was subsequently inbred by 20 generations of full-sib mating (Ayroles et al. 2009). Flies were raised on a cornmeal/agar/molasses media at 25 °C with a 12 h light–dark cycle and behaviorally tested 3- to 7-day posteclosion.
Chemicals
Ethyl hexanoate and methyl hexanoate were purchased from Sigma-Aldrich at the highest purity available (>99% purity). Odorants were diluted in paraffin oil (Sigma-Aldrich).
T-maze olfactory assay
Behavioral assays were essentially in accordance with Helfand and Carlson (1989) with minor modification. Thirty flies were placed into the T-maze apparatus and allowed to acclimate to conditions in a holding chamber for 1 min. Flies were then given a choice between airflow (500 mL/min) from the 2 arms of the maze, one containing the diluted odorant and one solely containing the paraffin oil vehicle. Flies were allowed 1 min to choose between the odorant and the control side. The response index (RI) was calculated by the formula: RI = (No. flies choosing odor − No. flies choosing control)/(No. total flies). A positive RI represents attraction to the odor (with a maximal attractive response of +1), whereas a negative RI indicates repulsion (with a maximal repulsion response of −1). To optimally resolve variation among wild-derived lines, dose response curves were first generated for each odorant on a subset of 4 randomly chosen wild-derived lines. Subsequent behavioral assays on the remainder of the wild-derived lines were then performed at 0.1% (v/v) methyl hexanoate and 1.0% (v/v) ethyl hexanoate. Approximately 9 replicate tests were performed per line and sex. Drosophila lines were tested using a randomized design, and all assays were conducted in the morning.
Statistical analyses of behavior
Data were analyzed using JMP 8.0 (SAS Institute). Comparisons among lines were made using a 2-way factorial analysis of variance (ANOVA) model y = μ + L + S + L × S + E, where sex (S) is the fixed effect of sex, line (L) and L × S are the random effect of line and line by sex, respectively, and E indicates error. To examine if there was a correlation between the behavioral responses to ethyl hexanoate and methyl hexanoate, the mean response indices for all lines and sexes were plotted. The response index for methyl hexanoate was arbitrarily chosen as the independent variable and the response index for ethyl hexanoate was chosen as the dependent variable.
Identification of polymorphisms in odorant receptor loci
Genomic DNA was extracted using the Puregene DNA purification system (Qiagen) for the same wild-derived lines for which behavioral data were collected. Primers were designed for 4 odorant receptors (Or22a, Or22b, Or35a, and Or47a) in order to obtain overlapping fragments encompassing the full-length coding and noncoding regions and ∼700 bp of the 5′ and 3′ untranslated regions. Polymerase chain reaction (PCR) was performed, and products sequenced with original and nested PCR primers. Sequences were analyzed using Vector NTI Advance 11 software (Informax). DNA sequences for Or10a, Or43a, and Or67b were obtained previously (Rollmann et al. 2010; Or10a, N = 79; Or43a, N = 72; and Or67b, N = 60).
Genotype–phenotype association analyses
Associations between individual polymorphic sites in each odorant receptor locus and variation in behavioral responses to ethyl and methyl hexanoate were determined by 2-way factorial ANOVA, model y = μ + M + S + M × S + E, in which μ is the overall mean, the molecular marker (M) and sex (S) are fixed effects, and E represents error. A conservative Bonferroni correction was performed to account for multiple tests. Likewise, when multiple polymorphisms in a given receptor were associated with variation in behavior, haplotypes were determined, and associations between haplotype and variation in behavior was assessed according to the 2-way factorial model y = μ + H + S + H × S + E, in which μ is the overall mean, the haplotype (H) and sex (S) are fixed effects, and E represents error. Polymorphisms present in only one line were not included in the association analyses. Linkage disequilibrium (LD) between markers was determined using Tassel 2.1 software (www.maizegenetics.net/tassel) and examined using Fisher's Exact test. An R2 value of 1 was indicative of complete LD, and polymorphisms in complete LD were treated as a single linkage group in Bonferroni calculations. Thirty-eight wild-derived lines were examined for each odorant receptor locus.
Genotyping
To assess support for the significant genotype–phenotype relationships in a larger sample, SNPs or indels identified as significantly associated with natural variation in olfactory behavior in our analyses of 38 wild-derived lines were genotyped in 82 additional lines from the same population. The genotyping approach was dictated by the number of polymorphisms significantly associated with behavior for a given locus and the pattern of LD. Either DNA sequencing of the genomic region of interest or Taqman Sample-to-SNP kit based methods (Applied Biosystems) was used. The presence or absence of the Or22ab chimera was determined by independent design of 2 primer pairs: One pair had one of the primers located within the deleted region (Or22a-F: GGTGTGAAGGTGTAGTTTGTAG, Or22a-R: TATCAAGCGGTGATCTCG). The second pair had primers spanning the deleted region (Or22a-F2: GACGCTTCACACTCTCCGTG, Or22a-R2: GATTACTACTCGTAACAAGC). Taqman primers and probes were as follows: Or35a-F: GGGAGCAAATCCTGCAGTACT, Or35a-R: GGCCAAGTTAATGAGCTTTAGTAATCG, Or35a-variant1: VIC-TCGCTGGGATTTGTAG, Or35a-variant2: FAM-CGCTGGGACTTGTAG, Or47a-F: CGCACGGCGATGTCCTA, Or47a-R:GCACAAAAATCAATCCAAAGCCTTGT, Or47a-variant1: VIC-ATGCTGAGATCATTCTCCT, and Or47a-variant2: FAM-TGCTGAGATCCTTCTCCT.
Gene expression
Total RNA was isolated from 50 D. melanogaster heads using Trizol (Invitrogen) and DNase treated (Ambion) according to manufacturers' instructions. Total RNA for 3 independent samples per sex and line was isolated. First strand cDNA was then synthesized using the AccuScript High Fidelity First Strand cDNA synthesis kit (Agilent Technologies). Quantitative real-time PCR assays were conducted with an ABI 7300 and using Taqman chemistry (Applied Biosystems). For each independent RNA isolation for each line and sex, 2 technical replicate measurements were made. Taqman gene expression assays for each gene were as follows: Or22a, Dm01814175_g1; Or22b, Dm01844971_g1; Or35a, Dm01807569_g1; Or47a, Dm01818125_g1; and Rpl32 (endogenous control), Dm02151827_g1. The ΔΔCt method was used for comparison of relative gene expression.
Results
Behavioral responses to select esters and behavioral correlations
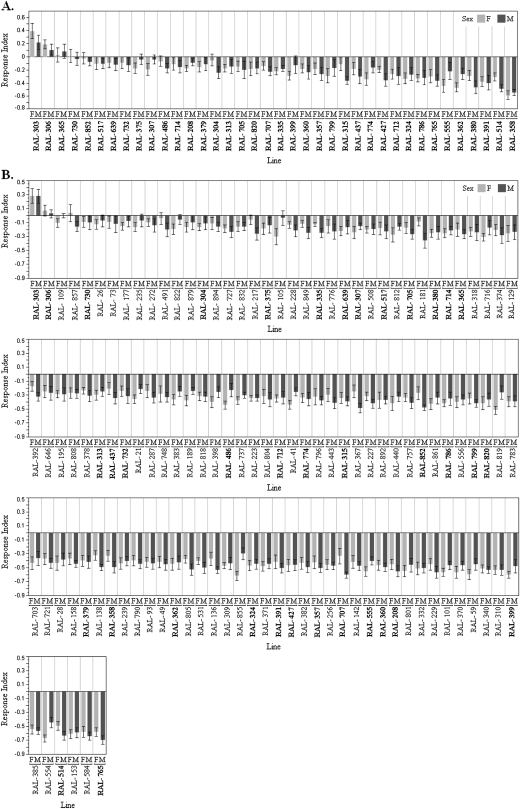
Behavioral variation was quantified among the wild-derived inbred lines to assess the extent of naturally occurring variation in olfactory behavior to 2 esters, ethyl hexanoate and methyl hexanoate. Significant phenotypic variation was detected among the wild-derived lines in response to both odorants (MetHex: degrees of freedom [df] = 37, F = 6.16, P < 0.0001; EtHex: df = 119, F = 6.24, P < 0.0001; Figure 1A and B, respectively). The behavioral response indices were broad and ranged from 0.394 to −0.589 for methyl hexanoate and 0.278 to −0.702 for ethyl hexanoate (Figure 1A and B). No significant difference was observed between the sexes for behavioral response to either odorant (MetHex: df = 1, F = 0.31, P = 0.58; EtHex: df = 1, F = 0.66, P = 0.42).
Figure 1.
Mean behavioral response indices to (A) methyl hexanoate and (B) ethyl hexanoate. Female response indices are indicated by light gray bars and male indices by dark gray bars (mean ± standard error). The wild-derived lines examined for initial genotype–phenotype associations are indicated in bold.
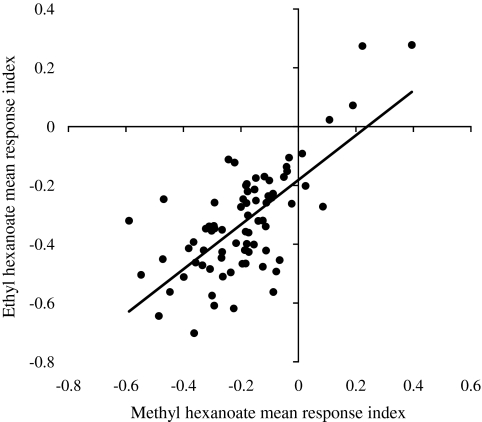
There was a significant positive correlation between responses to ethyl and methyl hexanoate (F = 69.13, P < 0.0001, R = 0.695; Figure 2). This was expected because methyl hexanoate and ethyl hexanoate have similar chemical structures (differing solely in their carbon length), and electrophysiological response profiles for several odorant receptor loci often revealed overlapping sensitivity to both odorants (Dobritsa et al. 2003; Hallem et al. 2004; Hallem and Carlson 2006). Our results demonstrated that significant intraspecific variation in olfactory behavior exists in this natural (Raleigh, NC) population and that odorants of similar chemical structure elicit comparable behavioral responses among lines.
Figure 2.
Correlation between behavioral responses to ethyl hexanoate and methyl hexanoate. The mean response indices for ethyl hexanoate for each sex and line are plotted against the behavioral response indices for methyl hexanoate.
Identification of molecular polymorphisms
Four receptors (Or22a, Or22b, Or35a, and Or47a) were sequenced for 38 inbred lines, derived from a natural population, for which behavioral responses to ethyl hexanoate and methyl hexanoate were measured. Three of these receptors (Or22a, Or35a, and Or47a) have been previously shown to respond strongly to esters (Hallem et al. 2004; Hallem and Carlson 2006), and Or22b was sequenced as it is physically located close to Or22a.
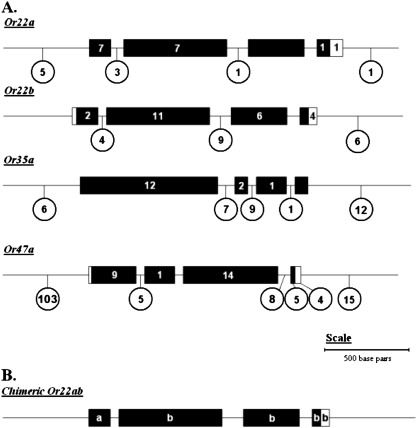
Multiple SNPs and indels were identified in each of the 4 odorant receptors (Supplementary Table 1 and Figure 3). The ∼2563-bp region sequenced for Or35a had a total of 50 polymorphisms, 4 of which result in amino acid substitutions (Lys38Ile, Leu47Val, Thr250Ile, and Asn348Ser). The ∼2549-bp sequence for Or47a had a total of 164 polymorphisms, with one resulting in an amino acid substitution (Val176Ile). Because the Or22a and Or22b regions are in close physical proximity they were considered together. This ∼4460 bp sequenced gene region contained 68 polymorphisms, which results in 11 amino acid substitutions in Or22a (His8Tyr, Val30Leu, Glu38Val, Trp51Leu, Met59Ile, Ile67Met, Met93Val, Cys100Ser, Glu196Ala, Asp201Glu, and Tyr382Ala) and 6 in Or22b (Ile25Val, Asn92Asp, Arg194Met, Asp201Ala, Val213Leu, and Leu354Ile). One polymorphism, a ∼2090-bp deletion encompassing the last 3 exons of Or22a and the first exon of Or22b, resulted in a chimeric Or22ab gene (Figure 3B) with a frequency of 0.4 in the population sample. This Or22ab chimera has been reported previously by Aguadé (2009) and has been shown to respond strongly to esters (de Bruyne et al. 2010).
Figure 3.
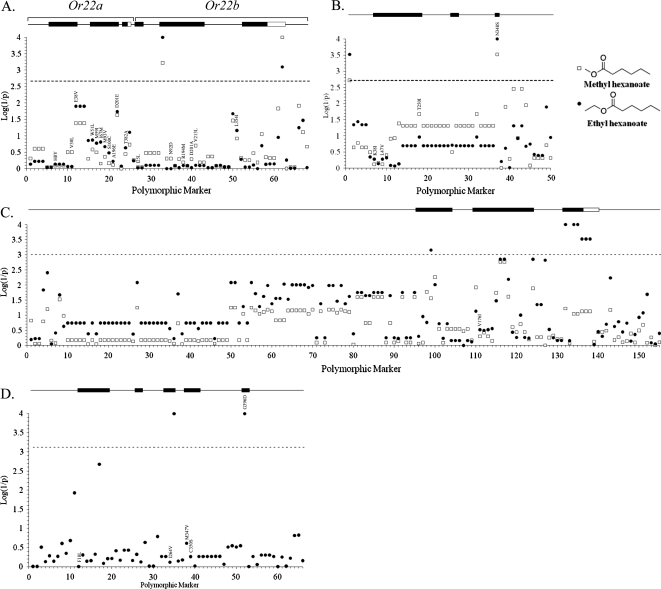
A schematic of the odorant receptor gene structure and molecular polymorphisms in each Or gene region. (A) The genomic region is depicted with a solid line with black boxes representing exons and white boxes untranslated regions. The number of polymorphic markers in each region is depicted numerically either directly within the exons or in circles underneath. (B) For the chimeric Or22ab gene, the source of each exon is indicated by an “a” for Or22a or a “b” for Or22b.
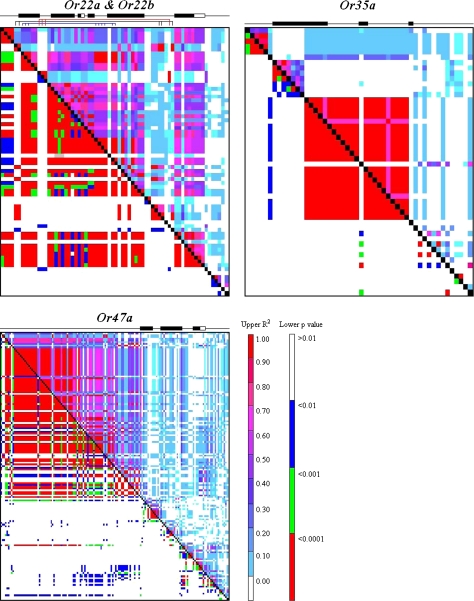
Linkage disequilibrium
LD analyses were performed for the 4 odorant receptor loci to detect nonrandom associations among polymorphic sites (Figure 4). As in previous analyses, LD analyses of the Or22a and Or22b gene regions were conducted together. For each odorant receptor gene region, LD generally decayed rapidly with physical distance in a manner consistent with mutation-drift equilibrium so that significant LD was generally observed only between polymorphisms in close physical proximity (Figure 4). However, 3 incidences of long-range LD were observed in the Or22a/Or22b complex. LD spanned a ∼3-kb region with C4T and C22T in Or22a and T860A, 909 Del (9 bp), and T960C in Or22b in complete LD. Long-range LD (across ∼2.4 kb) was also observed between A27T, C60G, and C61A in Or22a and C181G, G183A, A184C, T185C, G282A, and A334G in Or22b and between a third group consisting of A113T, A140G, C143C in Or22a and C955A in Or22b, spanning a ∼2.9-kb region.
Figure 4.
LD plots between markers for each odorant receptor. The upper diagonal represents the R2 value, whereas the lower diagonal represents the P value for all marker combinations. R2 values of 1 indicate complete LD. The location of coding and noncoding regions for each gene is shown above each LD plot with coding regions indicted by black boxes and untranslated regions by white boxes. In the case of the Or22a/Or22b gene region, colored lines below the gene region highlight distinct regions of long-range LD.
Genotype–phenotype associations
To determine the extent and nature of molecular polymorphisms in these receptors which may contribute to variation in olfaction-mediated behavior, we analyzed associations between sequence variation in 3 odorant receptor gene regions (Or22a/Or22b, Or35a, and Or47a) and variation in behavioral responses to 2 odorants, ethyl hexanoate and methyl hexanoate (Figure 5). In the Or22a/Or22b gene region, we observed 2 polymorphisms that exceeded a conservative Bonferroni correction for multiple tests and were associated with variation in responses to both methyl hexanoate and ethyl hexanoate (Figure 5A). One of these was a silent substitution in the second exon of Or22b (T240C) and the other was a 16-bp insertion (1541 Ins) in the 3′ untranslated region of Or22b, a region highly conserved among closely related Drosophila species (D. simulans, D. sechellia, and D. yakuba; Drosophila 12 Genomes Consortium 2007, http://genome.ucsc.edu/cgi-bin/hgGateway). In the case of Or35a, 2 polymorphisms (G-455A and A1195G) were significantly associated with behavioral responses to both odorants (Figure 5B). The G-455A polymorphism is located directly upstream of the initiation codon for Or35a between 2 highly conserved blocks of sequence. The A1195G is located in the third exon, which results in an amino acid substitution (Asn348Ser) near the C-terminus of the receptor. Finally, 7 polymorphic markers (G195T, T1300C, C1321T, A1324G, A1336C, 1349Del2bp, and C1357T) in Or47a were significantly associated with behavioral responses only to ethyl hexanoate (Figure 5C). G195T is a silent substitution located in the first exon of Or47a. T1300C, C1321T, A1324G, and A1336C are silent substitutions located in exon 4, with T1300C, C1321T, and A1324G in complete LD (Figure 4). An A1336C polymorphism and 2 markers (1349Del2bp and C1357T) in the untranslated region of exon 4 are in complete LD (Figure 4). The polymorphisms (1349Del2bp and C1357T) are in close proximity to the termination codon (≤12 bp away), and comparisons among closely related species (D. simulans, D. sechellia, D. yakuba, and D. erecta) indicate that they are located within a conserved region. No polymorphisms in Or47a were significantly associated with variation in behavioral responses to methyl hexanoate.
Figure 5.
Associations between each polymorphic marker and mean behavioral response index for (A) Or22a and Or22b, (B) Or35a, (C) Or47a, and (D) Or10a. Association between markers and olfactory responses to ethyl hexanoate and methyl hexanoate are represented by black circles or open boxes, respectively. Markers above the Bonferroni threshold (dashed black line) are significantly associated with behavioral variation. Above each graph is a schematic of the genomic region for each Or with coding regions indicated by a black box, nontranslated regions by a white box, and noncoding DNA segments by a line.
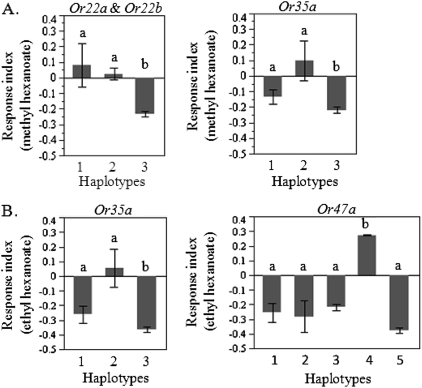
Because each odorant receptor contained multiple polymorphisms associated with variation in olfactory behavior, we also conducted haplotype analyses to determine the potential combined effects of polymorphisms on behavior. For polymorphisms associated with behavioral responses to methyl hexanoate, 3 haplotypes were identified for the Or22a/Or22b region and 3 for Or35a (Table 1 and Figure 6A). For the Or22a/Or22b region haplotypes, a significant avoidance of methyl hexanoate was observed for haplotype 3 relative to haplotypes 1 and 2 (df = 2, F = 21.14, P < 0.0001). Haplotype analysis of Or35a also revealed significant difference among haplotypes (df = 2, F = 8.99, P = 0.0003). Similarly, for sequence variants associated with variation in behavioral responses to ethyl hexanoate, significant differences in mean responses among haplotypes were observed for Or35a (df = 2, F = 14.27, P < 0.0001) and Or47a (df = 4, F = 11.11, P < 0.0001), with 3 haplotypes identified for Or35a (the same as those for methyl hexanoate) and 5 for Or47a (Table 1 and Figure 6B). Of particular note is the significantly greater mean behavioral response of haplotype 4 relative to all other haplotypes.
Table 1.
Haplotype analyses for polymorphisms significantly associated with behavioral responses to methyl hexanoate or ethyl hexanoate for each locus
| Gene region | Odorant | Number of haplotypes | Haplotypea | Frequency |
| Or22a/Or22b | Methyl hexanoate | 1 | C- | 0.053 |
| Or22a/Or22b | Methyl hexanoate | 2 | TI | 0.105 |
| Or22a/Or22b | Methyl hexanoate | 3 | T- | 0.842 |
| Or35a | Methyl hexanoate | 1 | AA | 0.158 |
| Or35a | Methyl hexanoate | 2 | GG | 0.053 |
| Or35a | Methyl hexanoate | 3 | GA | 0.789 |
| Or35a | Ethyl hexanoate | 1 | AA | 0.158 |
| Or35a | Ethyl hexanoate | 2 | GG | 0.053 |
| Or35a | Ethyl hexanoate | 3 | GA | 0.789 |
| Or47a | Ethyl hexanoate | 1 | TTCAATC | 0.132 |
| Or47a | Ethyl hexanoate | 2 | GTCACDT | 0.053 |
| Or47a | Ethyl hexanoate | 3 | GCTGCDT | 0.079 |
| Or47a | Ethyl hexanoate | 4 | TCTGCDT | 0.026 |
| Or47a | Ethyl hexanoate | 5 | GTCAATC | 0.711 |
An “I” indicates an insertion polymorphism and “D” indicates a deletion polymorphism.
Figure 6.
Haplotype analysis of polymorphisms significantly associated with behavioral responses to (A) methyl hexanoate or (B) ethyl hexanoate for each odorant receptor locus. Significant behavioral differences among haplotypes were determined by ANOVA. Differing haplotypes were subsequently identified by post hoc Tukey's test at P < 0.05 and indicated by unique letters above the bars.
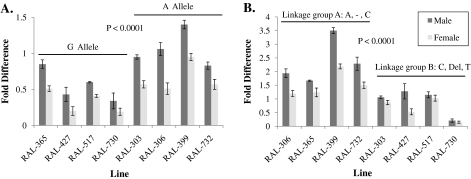
We then extended the sample to 120 wild-derived lines for which measurements of behavioral responses to ethyl hexanoate and genotypes for focal polymorphisms were obtained. With the exception of the Or47a A1336C linkage group, the statistical association between the phenotypic means for behavioral responses to ethyl hexanoate and each examined focal polymorphic marker in the sample of 120 alleles continues to support the observed significant genotype–phenotype relationships for Or35a markers G-455A (df = 1, F = 9.72, P < 0.002; frequency A allele, 0.13) and A1195G (df = 1, F = 25.09, P < 0.0001; frequency of G allele, 0.05) and variation in behavioral responses to ethyl hexanoate. Significant associations were also observed for the T1300C marker in Or47a (df = 1, F = 6.11, P < 0.01; frequency C allele, 0.15) and the T240C marker in the Or22a/Or22b region (df = 1, F = 26.88, P < 0.0001; frequency C allele, 0.05). Finally, in regard to the deletion resulting in a chimeric Or22ab gene, we found the presence/absence of this ∼2-kb deletion to be significantly associated with behavioral responses to ethyl hexanoate (df = 1, F = 4.62, P < 0.03, frequency 0.39).
Finally, we took advantage of previously published sequence data for Or10a, Or43a, and Or67b in these same wild-derived lines (Rollmann et al. 2010) to ask whether polymorphisms in these receptors were associated with variation in behavioral responses to ethyl hexanoate. Consistent with the lack of Or43a and Or67b specificity to ethyl hexanoate, no associations were found between polymorphisms in these loci and variation in responses to ethyl hexanoate. We did, however, find genotype–phenotype associations between sequence variation in Or10a, a receptor for which inhibitory electrophysiological responses to ethyl hexanoate have been observed (Hallem and Carlson 2006) and variation in behavioral responses to ethyl hexanoate (Figure 5D). More specifically, the C1131T and G1481A polymorphisms contributed to behavioral variation. These polymorphisms are distinct from those associated with variation in behavioral responses to benzaldehyde and acetophenone (Rollmann et al. 2010). The G1481A polymorphism results in a glycine to aspartic acid amino acid substitution (G396D).
Gene expression
Molecular polymorphisms in noncoding regions can result in alteration to gene expression levels. To assess whether noncoding polymorphisms associated with variation in olfactory behavior were associated with gene expression differences in the Or22a/Or22b gene region, Or35a, and Or47a, we conducted quantitative real-time PCR experiments on lines with overall more positive versus negative behavioral response indices to ethyl hexanoate. For each gene, 4 lines with more negative behavioral response indices to ethyl hexanoate and carrying a given allele and 4 lines with more positive indices carrying the alternate allele were selected. In the case of the Or22a/Or22b region, we did not observe significant differences in Or22b expression among lines with positive versus negative behavioral responses to ethyl hexanoate but confirmed the presence of the Or22ab chimera. However, for Or35a and Or47a, we did observed significant differences in gene expression levels among lines with alternate alleles and respective positive/negative behavioral response indices to ethyl hexanoate (Figure 7). Specifically, increased Or35a expression was associated with lines carrying the −455A allele that had overall positive behavioral response indices (df = 1, F = 41.40, P < 0.0001). In the case of Or47a, an A1336C polymorphism and 2 markers (1349Del2bp and C1357T) are in the noncoding region and are in complete LD. Lines carrying the linkage group consisting of 1336C, 1349 Del 2 bp, and 1357T are associated with more positive behavioral response indices and decreased levels of Or47a expression (df = 1, F = 54.72, P < 0.0001). No significant allele by sex interactions was found.
Figure 7.
Or35a and Or47a gene expression levels. For each gene, lines with overall more negative behavioral response indices to ethyl hexanoate and carrying a given allele and lines with more positive response indices carrying the alternate allele were selected. (A) Or35a expression levels across lines carrying either the −455G allele or the −455A SNP. (B) Or47a expression levels for the linkage group comprised of A1336C, 1349 Del 2 bp, and C1357T that are in complete LD. A dash indicates a lack of deletion.
Discussion
Multiple polymorphisms were identified in the Or22a/Or22b, Or35a, Or47a, and Or10a gene regions that were associated with behavioral variation in response to esters in a natural D. melanogaster population. We chose to examine these genes from the family of 60 genes that comprise this multigene Or family based on previous work elucidating their respective electrophysiological response profiles (Hallem et al. 2004; Hallem and Carlson 2006). Or22a and Or35a are broadly tuned receptors responding to a large suite of odorants including esters, with methyl hexanoate and ethyl hexanoate both eliciting excitatory responses for Or22a expressing neurons and inhibitory responses for Or35a expressing neurons. Or10a exhibits strong excitation to aromatic compounds with polymorphisms in Or10a implicated in behavioral variation in responses to the aromatic compounds, benzaldehyde and acetophenone (Hallem et al. 2004; Hallem and Carlson 2006; Rollmann et al. 2010). Or10a has also been shown to be inhibited by several odorants, including ethyl hexanoate. Or47a is narrowly tuned and responds chiefly to esters. Thus, our results identifying polymorphisms in these receptors, which were associated with variation in behavioral responses to ethyl hexanoate and methyl hexanoate, are consistent with the role of these receptors in the detection of esters. Also consistent with these results is the lack of an association between polymorphisms in Or43a and Or67b and behavioral responses to these esters as these receptors have not been shown to have ligand affinity to either ester (Hallem and Carlson 2006). Finally, our results are in line with the combinatorial nature of Ors: Multiple receptors responding to a single odorant and single receptors responding to a suite of odorants.
Given that a significant correlation was found between independent behavioral measurements of responses to ethyl hexanoate and methyl hexanoate, we predicted that the association profiles for both odorants would be globally similar. Indeed, we observed striking concordance in the association profile patterns for the 2 odorants and variation in the Or22a/Or22b region and Or35a region (Figure 5A–B). The association of these polymorphisms with independent behavioral measurements of responses to each of these structurally similar odorants serves to further validate their significant contributions to variation in odor-mediated behavior. In the case of Or47a, individual polymorphisms were associated with only one but not the other odorant raising the possibility that Or47a may be particularly adept at discriminating among these esters. The Or47a electrophysiological response profile is narrower in its tuning compared with Or22a and Or35a response profiles (Hallem and Carlson 2006). In addition, since the concentrations for optimal resolution of behavioral variation among wild-derived lines differed for ethyl hexanoate and methyl hexanoate, Or47a selectivity may be mediated by concentration-dependent changes. It should be noted that we have examined 7 receptors out of the multigene family and that variation in additional receptors may also contribute to differences in behavioral responses to these odorants. The statistical power for detecting genotype–phenotype associations increases as sample size increases. Therefore, increasing sample sizes within a population and/or surveying additional populations could potentially identify additional polymorphisms associated with variation in behavioral responses to ethyl hexanoate and methyl hexanoate.
Both coding and noncoding changes were associated with variation in behavioral responses to both odorants. Protein coding changes can result in changes in protein structure/function, and amino acid substitutions in vertebrate chemosensory receptors have been shown to change ligand-binding affinity and thereby result in differences in odorant sensitivity (Reed et al. 2004; Nie et al. 2005; Abaffy et al. 2007; Keller et al. 2007). Thus, the Asn348Ser substitution in Or35a and Gly396Asp substitution in Or10a are of particular interest. The substitutions in both genes are predicted to be located extracellularly and thus could influence ligand binding. Finally, it is possible that polymorphisms may affect dimerization with Orco, a receptor that acts to localize odorant receptors in the sensory dendritic membrane (Jones et al. 2005; Benton et al. 2006) and plays a role in the formation of ligand-gated nonselective cation channels (Sato et al. 2008; Wicher et al. 2008).
Noncoding nucleotide changes can also be primary agents of behavioral variation (Wang et al. 2007, 2010; Rollmann et al. 2010). Noncoding odorant receptor polymorphisms have been previously associated with variation in behavioral responses to benzaldehyde and acetophenone, and high behavioral responder haplotypes to benzaldehyde were associated with alterations in odorant receptor gene expression levels (Rollmann et al. 2010). SNPs in noncoding regions of odorant-binding proteins contributing to olfactory behavior also have been implicated in alterations to messenger RNA (mRNA) secondary structure and gene expression levels (Wang et al. 2007, 2010). In D. melanogaster, ∼500-bp region upstream of Or genes has been shown to be sufficient for maintaining appropriate OR expression patterns (Ray et al. 2007). Furthermore, misexpression of odorant receptors has been shown to result in altered olfactory avoidance behavior (Stortkuhl et al. 2005). In our study, the majority of polymorphisms associated with variation in olfactory behavior were noncoding changes. These polymorphisms were not located in promoter motif regions identified to date (Ray et al. 2007, 2008) nor in conserved motifs identified in 3′ untranslated gene regions. However, consistent with differences in Or gene expression levels contributing to variation in behavior, we observed significant differences in Or35a and Or47a gene expression levels among lines with more positive versus negative overall behavioral responses to ethyl hexanoate. Of particular note is the G-455A Or35a marker associated with variation in behavioral response to both methyl hexanoate and ethyl hexanoate, and in which increased expression was associated with more positive response indices to ethyl hexanoate. This polymorphism is located directly upstream of the initiation codon between 2 highly conserved blocks of sequence, suggesting it could contribute to the regulation of gene expression. Moreover, the Or47a polymorphisms (1349Del2bp and C1357T) are located within 12 bp from the termination codon and thereby could affect regulation of mRNA stability. These changes in gene expression can contribute to differences in odorant sensitivity and discrimination, with the combinatorial group of odorant receptors together influencing variation in olfactory behavior.
Finally, a significant difference in olfactory sensitivity to these esters has been previously observed among Drosophila species (Stensmyr et al. 2003; Dekker et al. 2006), but within-species variation had yet to be examined. Our results show that significant variation in sensory perception exists within a natural D. melanogaster population for ethyl hexanoate and methyl hexanoate and that variation in the Or22a/Or22b gene region as well as in Or10a, Or35a, and Or47a contributes to this variability in behavioral response. Thus, changes at the most peripheral sensory stage can contribute to within-species differences in behavioral responses to environmental stimuli.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was supported by the National Institutes of Health [GM080592 to S.M.R.].
Supplementary Material
Acknowledgments
We thank Tiffany Cook, Richard Kirby, John Layne, Ken Petren, and Roger Ruff for comments on the manuscript or technical assistance.
References
- Abaffy T, Malhotra A, Leutje CW. The molecular basis for ligand specificity in a mouse olfactory receptor. J Biol Chem. 2007;282:1216–1224. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé M. Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol Biol Evol. 2009;26:61–70. doi: 10.1093/molbev/msn227. [DOI] [PubMed] [Google Scholar]
- Alcorta E, Rubio J. Genetical analysis of intrapopulational variation in olfactory response in Drosophila melanogaster. Heredity. 1988;60:7–14. doi: 10.1038/hdy.1988.2. [DOI] [PubMed] [Google Scholar]
- Alcorta E, Rubio J. Intrapopulational variation of olfactory responses in Drosophila melanogaster. Behav Genet. 1989;19:285–299. doi: 10.1007/BF01065911. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:240–257. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao S, Sen A, Stocker R, Rodrigues V. Olfactory neurons expressing identified receptor genes project to subsets of glomeruli within the antennal lobe of Drosophila melanogaster. J Neurobiol. 2003;54:577–592. doi: 10.1002/neu.10175. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Smart R, Zammit E, Warr CG. Functional and molecular evolution of olfactory neurons and receptors for aliphatic esters across the Drosophila genus. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2010;196:97–109. doi: 10.1007/s00359-009-0496-6. [DOI] [PubMed] [Google Scholar]
- Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Van der Goes van Naters W, Warr CG, Steinbrect RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Fuyama Y. Behavior genetics of olfactory responses in Drosophila. I. Olfactometry and strain differences in Drosophila melanogaster. Behav Genet. 1976;6:407–420. doi: 10.1007/BF01065698. [DOI] [PubMed] [Google Scholar]
- Fuyama Y. Behavior genetics of olfactory responses in Drosophila. II. An odorant specific variant in a natural population of Drosophila melanogaster. Behav Genet. 1978;8:399–414. doi: 10.1007/BF01067937. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Hallem E, Carlson J. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem E, Ho M, Carlson J. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Helfand SL, Carlson JR. Isolation and characterization of an olfactory mutant in Drosophila with a chemically specific defect. Proc Natl Acad Sci U S A. 1989;86:2908–2912. doi: 10.1073/pnas.86.8.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Nguyen TA, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr Biol. 2005;15:R119–R121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Mackay TFC, Hackett JB, Lyman RF, Wayne ML, Anholt RRH. Quantitative genetic variation of odor-guided behavior in a natural population of Drosophila melanogaster. Genetics. 1996;144:727–735. doi: 10.1093/genetics/144.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr Biol. 2005;15:1948–1952. doi: 10.1016/j.cub.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Ray A, Van der Goes van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 2008;6:1069–1083. doi: 10.1371/journal.pbio.0060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Li S, Li X, Huang L, Tordoff MG, Starling-Roney R, Taniguchi K, West DB, Ohmen JD, Beauchamp GK, et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci. 2004;24:938–946. doi: 10.1523/JNEUROSCI.1374-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollmann SM, Wang P, Date P, West SA, Mackay TFC, Anholt RRH. Odorant receptor polymorphisms and natural variation in olfactory behavior in Drosophila melanogaster. Genetics. 2010;186:687–697. doi: 10.1534/genetics.110.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1007. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Dekker T, Hansson BS. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc R Soc Lond B Biol Sci. 2003;270:2333–2340. doi: 10.1098/rspb.2003.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Giordano E, Balloi A, Angioy A, Hansson BS. Novel natural ligands for Drosophila olfactory receptor neurons. J Exp Biol. 2003;206:715–724. doi: 10.1242/jeb.00143. [DOI] [PubMed] [Google Scholar]
- Stortkuhl KF, Kettler R, Fischer S, Hovemann BT. An increased receptive field of olfactory receptor Or43a in the antennal lobe of Drosophila reduces benzaldehyde-driven avoidance behavior. Chem Senses. 2005;30:81–87. doi: 10.1093/chemse/bji003. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran JM, Nuñez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–992. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong A, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Mackay TFC, Anholt RRH. Natural variation in odorant recognition among odorant-binding proteins in Drosophila melanogaster. Genetics. 2010;184:759–767. doi: 10.1534/genetics.109.113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Shabalina SA, Mackay TFC, Anholt RRH. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics. 2007;177:1655–1665. doi: 10.1534/genetics.107.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic nucleotide activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.