Summary
Similarities between speech and birdsong make songbirds advantageous for investigating the neurogenetics of learned vocal communication; a complex phenotype likely supported by ensembles of interacting genes in cortico-basal ganglia pathways of both species. To date, only FoxP2 has been identified as critical to both speech and birdsong. We performed weighted gene co-expression network analysis on microarray data from singing zebra finches to discover gene ensembles regulated during vocal behavior. We found ~2,000 singing-regulated genes comprising 3 co-expression groups unique to area X, the basal ganglia subregion dedicated to learned vocalizations. These contained known targets of human FOXP2 and potential avian targets. We validated novel biological pathways for vocalization. Higher order gene co-expression patterns, rather than expression levels, molecularly distinguish area X from the ventral striato-pallidum during singing. The previously unknown structure of singing-driven networks enables prioritization of molecular interactors that likely bear on human motor disorders, especially those affecting speech.
Introduction
Speech and birdsong are examples of the rare ability to learn new vocalizations. Both depend on hearing and are supported by analogous neural pathways through the cortex and basal ganglia1. In humans, such pathways support an array of behaviors, but songbirds like the zebra finch possess well-defined sub-circuitry specialized for song learning and production, enabling the design of experiments to uncover vocal-motor specific function (Figure 1A)2. The transcription factor FoxP2, critical for birdsong and the only molecule directly linked to speech and language dysfunction3, is expressed similarly in these pathways in both species4. The discovery of FOXP2’s link to vocal-motor dysfunction was a constructive step towards understanding the genetic basis of speech, but learned vocalization is a complex phenotype and likely depends on interactions between many genes. Methodological limitations preclude the study of gene expression in behaving humans, so the neuromolecular underpinnings of speech remain poorly understood. Zebra finches, however, are well-suited as a model system for neurogenetic investigations of learned vocal-motor behaviors including speech; bolstered by the sequencing and assembly of their genome5.
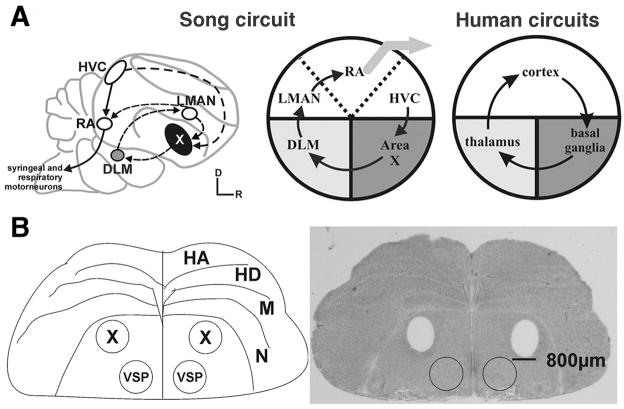
Figure 1. Neuroanatomical overview.
(A) Schematic comparison of avian and human cortico-basal ganglia loops. Left, composite sagittal view of songbird telencephalon highlights song control nuclei. Auditory input (not shown) enters the song circuit at cortical HVC, the neurons of which contribute to 2 pathways, the vocal motor pathway (plain arrows) and the anterior forebrain pathway (stippled arrows). The latter includes basal ganglia nucleus area X and rejoins the vocal motor pathway via projections from the cortical lateral magnocellular nucleus of the anterior nidopallium (LMAN) to the robust nucleus of the arcopallium (RA). Middle, songbird cortico-basal ganglia circuitry is further simplified to illustrate song-specialized sub-regions that are embedded within similar brain areas in the human brain (Right). Cortex is in white, basal ganglia dark gray and thalamus light gray. Adapted from Teramitsu et al. (2004)4.
(B) Striato-pallidal brain regions that gave rise to the oligoarray data consist of area X and VSP. Left, line drawing of a coronal section through anterior zebra finch brain shows anatomical borders and highlights area X, observable in the Nissl-stained section. Right, bilateral tissue punches of equivalent size were taken from area X (holes) and VSP (circles). Abbreviations: D-Dorsal, HA-Hyperpallium apicale, HD–hyperpallium densocellulare, M–mesopallium, N–nidopallium, R-Rostral, X–song control area X, VSP-ventral striato-pallidum. Adapted from Miller et al. (2008)9. See also Figure S1
To elucidate gene ensembles underlying learned vocalizations, we used weighted gene co-expression network analysis6 (WGCNA) to identify and investigate groups of genes co-regulated during singing. This biologically-inspired method (Supplemental Experimental Procedures) has previously yielded results that could not have been obtained using traditional microarray analyses7, with gene co-expression groups typically corresponding to functional pathways. Past uses have uncovered novel genes important for human evolution and brain development, and highlighted genes with clinical significance for pathologies such as cancer8. Our experimental design was based upon prior studies showing that FoxP2 levels within the song-specialized basal ganglia subregion, striato-pallidal area X, decrease after 2 hours of undirected singing9,10,11, a form of vocal practice12,13, with the magnitude of downregulation correlated to how much the birds sang11. In addition, we observed increased vocal variability after 2 hours of undirected singing14, and another group found abnormally variable acoustic structure in the adult song of birds that underwent knockdown of area X FoxP2 during song development15. Together, these findings imply that low FoxP2 levels in area X are coincident with increased vocal variability, and that genes normally repressed by FoxP2 become activated with increasing amounts of singing.
Using this behavioral paradigm, we performed WGCNA on microarray data arising from 2 anatomically adjacent, yet functionally distinct, regions of the songbird basal ganglia: song-dedicated area X and the ventral striato-pallidum (VSP; Figure 1B), an area important for non-vocal motor function (e.g. posture) that is also active during singing16. We then quantitatively related network structure to singing measurements (Table S1), representing the first application of WGCNA to a procedurally learned behavior. We hypothesized, and subsequently confirmed, that area X and the VSP would have distinct network structures and that FoxP2, along with its transcriptional targets, would be members of singing-regulated co-expression groups unique to area X. These results are substantiated by the identification and functional annotation of previously known singing genes in our network, and biological validation of molecular pathways not previously linked to vocal motor behavior.
Results and Discussion
Prior to network construction, we defined gene significance measures (GS, Supplemental Experimental Procedures) for each probe to relate expression variability to trait variability across all birds (n=26), e.g. to the act of singing (referred to as GS.singing.X when measured in area X and GS.singing.V when measured in VSP, see Experimental Procedures for explanation of “probe” vs. “gene”). In area X, after false discovery rate (FDR) correction, 2,659 probes representing 1,364 known genes were significantly correlated to the act of singing (q<0.05; GS.singing.X), and 3,709 probes (1,825 known genes) to the number of motifs sung (GS.motifs.X; motifs are neuroethologically relevant sequences of song notes17), with 1,132 genes common to both. In sharp contrast, 0 probes in the VSP had significant GS.singing.V or GS.motifs.V scores (Table S2). We observed small differences in probe expression values in the singing vs non-singing birds: in area X, only 177 probes (~0.9% of the total) showed >100% up- or downregulation, 65 probes >200%, 3 probes >1000%. In the VSP, only 17 probes showed >100% up- or downregulation (~0.08%), 6 probes >200%, and 0 probes >1000%. We also measured correlations to individual acoustic features such as Wiener entropy (a measure of width and uniformity of the power spectrum18; GS.entropy) that are typically used to assess song (Figures 2B and S3, Table S2). GS.age was computed for each bird as a negative control. Importantly, GS results did not influence network construction in any way.
Figure 2. Song patterns that emerged from the behavioral paradigm.
(A) Histogram shows number of song motifs produced in 600 s bins for the 18 singing birds in the microarray study.
(B) Birds who sang the most motifs exhibited greater acoustic variability. Individual bird identifier numbers are shown for the singing birds. Number of motifs sung was positively correlated with mean Wiener entropy, for which scores closer to 0 represent more disorder across the width and uniformity of the power spectrum18. The dashed line represents the linear regression of Wiener entropy on number of motifs, with the Pearson correlation coefficient r and p-value (based on Fisher’s z transformation) shown at top. See also Figure S2, Table S1.
During pre-processing, all samples were hierarchically clustered to visualize inter-array correlations and remove outliers (Supplemental Experimental Procedures). The area X vs. VSP samples segregated into 2 distinct clusters, as would be expected if tissue source influences gene expression (Figure S1A). Within area X, the singing vs non-singing birds segregated into 2 distinct sub-clusters (Figure S1B), indicating that singing is a profound regulator of gene expression in area X. Singing birds sang throughout the 2 hour recording period (Figures 2A and S2). There was a significant correlation between the number of motifs sung and Wiener entropy, replicating our prior finding of heightened vocal variability after 2 hours of singing14 (Figure 2B).
Essential network terminology
To identify ensembles of genes that were tightly co-regulated (modules) during singing, we performed WGCNA6 (Experimental Procedures) of the area X samples and quantitatively related the resulting modules to traits. Co-expression networks were built based exclusively on expression levels, via unsupervised hierarchical clustering on a biologically significant distance metric (topological overlap, TO; Experimental Procedures), and relationships between GS and network structure were only examined post hoc. Modules were defined as branches of the dendrogram obtained from clustering, and labeled by colors beneath the dendrogram (Figure 3A; probes outside properly defined modules were considered background and colored grey). To study module composition we defined the 1st principal component of each module as the module eigengene (ME), which can be considered a weighted average of the probe expression profiles that make up the module. Correlating MEs to traits, e.g. number of motifs sung, is an efficient way to relate expression variability within modules to trait variability. The module membership (MM) and intramodular connectivity (kIN) of each probe were defined as the correlation of its expression profile to the ME, and the sum of its network connections with other module members, respectively (Experimental Procedures). MM and kIN are closely related; high values for either indicate tight co-expression with most other module genes, signaling increased biological importance.
Figure 3. Relationships between network modules and behavioral traits.
(A) Top) Dendrogram of the subset of the area X network that includes the blue, dark green, orange, black, and salmon singing-related modules. ‘Leaves’ along ‘branches’ represent probes. The y-axis represents network distance as determined by 1 - TO, where values closer to 0 indicate greater similarity of probe expression profiles across samples. Color blocks below denote modules. Beneath, additional bands indicate positive (red) and negative (green) correlation (see scale bar in B). The top 2 bands show correlations to the number of motifs sung and the act of singing for probes in the dendrogram. The bottom 3 bands show the degree of correlation of these probes to the EGR1, FOXP2 and GAPDH probes with the most significant GS.motifs.X scores, respectively. ****passed Bonferroni for correlation to act of singing and number of motifs, and FDR for correlation to mean Wiener entropy; ***passed Bonferroni for correlation to act of singing and number of motifs, **passed Bonferroni for correlation to number of motifs and FDR for correlation to mean Wiener entropy; *passed FDR for correlation to number of motifs.
(B) Colors to the left represent the 21 proper modules in the network. For each module, the heatmap shows ME correlations to traits. Numbers in each cell report the correlation coefficients and Student asymptotic p-value (parentheses) for significant ME-trait relationships for the 5 singing-related modules as indicated by asterisks in (A). Scale bar, right, indicates the range of possible correlations from positive (red, 1) to negative (green, −1).
The Supplemental Experimental Procedures section contains further information on WGCNA methodology, definitions, and advantages.
Multiple area X co-expression modules strongly related to singing
WGCNA yielded 21 proper co-expression modules in area X (Figure 3). Correlations were computed between MEs and traits, and p-values were computed for each correlation (Experimental Procedures). After Bonferroni correction (significance threshold α=1.7e-4), the MEs of 3 modules were significantly related to the act and/or the amount of singing (Figure 3B, Table S3); the blue module (act of singing and number of motifs), the dark green module (act of singing and number of motifs) and the orange module (number of motifs). The positive correlations of the blue module (2,013 probes representing 995 known genes) indicate upregulation of its members during singing and, in general, increased expression with more singing. In contrast, the negative correlations observed for the dark green (1,417 probes representing 824 known genes) and orange (409 probes representing 234 known genes) modules indicate significant downregulation with the act of singing (dark green only) that continued in concert with increased amounts of singing (both). Since Bonferroni correction often results in false negatives19 we also performed a less conservative False Discovery Rate (FDR) procedure (Experimental Procedures), yielding 2 additional significant ME correlations to the number of motifs sung (black and salmon modules) and 2 to Wiener entropy (blue and orange modules). There were no significant correlations to age.
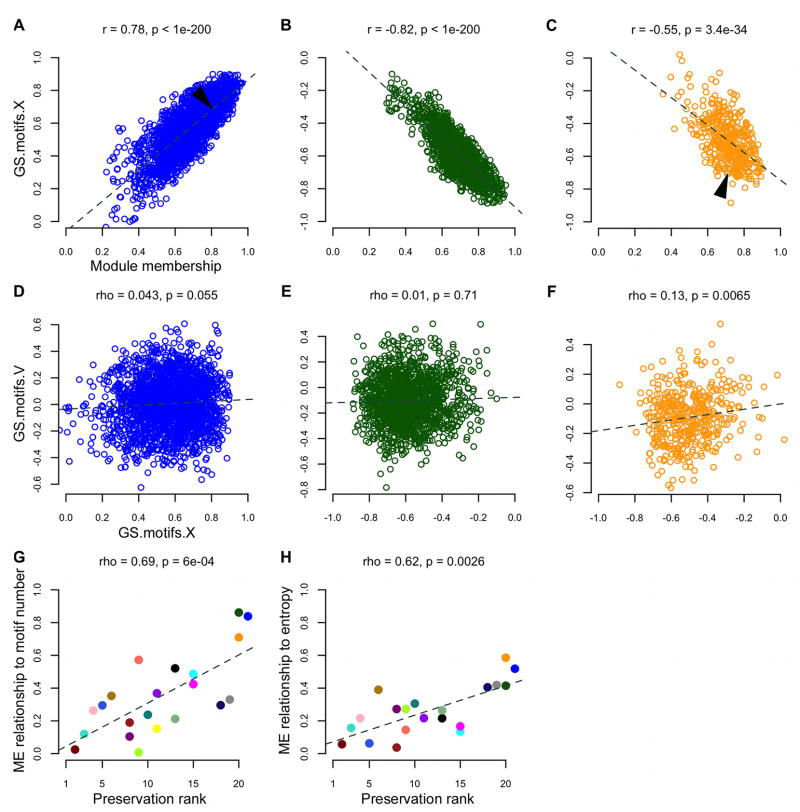
These 5 “singing-related” modules contained ~83% of the probes with significant GS.motifs.X and GS.singing.X scores. Compared to the rest of the network, genes in these modules were more strongly coupled to the act and amount of singing, and to Wiener entropy (GS.singing.X, GS.motifs.X, GS.entropy.X p<1e-200, Kruskal-Wallis ANOVA). The most interconnected probes within the singing-related modules were also the most tightly regulated by singing, as evidenced by the significant correlations of MM to GS.singing.X and GS.motifs.X in these modules (Figures 4A–C and S3A–F), indicating a strong relationship between importance in the network and behavioral relevance. MM-GS relationships such as these were not found in modules unrelated to singing, e.g. the dark red and turquoise modules, indicating that connectivity, and likely the biological functions in those modules, is relatively unspecialized with respect to vocal-motor behavior in area X, at least after 2 hours of singing.
Figure 4. Module membership predicts relationship to singing in area X.
(A–C) Area X GS scores for the number of motifs sung are plotted as a function of MM for probes in the blue (left), dark green (center) and orange (right) song modules. Each dot represents one probe. Dashed lines represent the linear regression of GS.motifs.X on MM in each module, with the Pearson correlation coefficient r and p-value (based on Fisher’s z transformation) shown at top. Arrows indicate approximate locations of the EGR1 (blue module) and FOXP2 (orange module) probes shown in Figure 3A.
(D–F) GS scores arising from the VSP (V) plotted as a function of the values in area X for the number of motifs sung. Each dot represents one probe. Dashed lines represent the linear regression of GS.motifs.V on GS.motifs.X in each module, with the Spearman rank correlation coefficient rho and p-value shown at top.
(G–H) The magnitude of ME-motifs (left) and ME-entropy (right) relationships in area X (absolute values of correlations represented in Figure 3B heatmap) plotted as a function of the degree of preservation of each module across brain regions. Each circle represents a module, colored accordingly, e.g. the blue, dark green, and orange song modules (upper right) had the strongest ME-correlations and were the least preserved in the VSP. Dashed lines represent the linear regression of ME-motifs and ME-entropy correlations on preservation rank, with Spearman’s rho and p-value shown at top. The purple and yellow modules overlap in the right panel. See also Figure S3.
Gene significance of area X song module genes is not preserved in VSP
We performed a series of comparisons between area X and the VSP to test the hypothesis that area X singing-related network structure was specific to vocal-motor function, and not due to motor function in general. We note that the region of outlying striato-pallidum selected for our analysis, the VSP, is not transcriptionally ‘muted’ during singing, rather, it exhibits immediate early gene (IEG) activation thought to reflect non-vocal movements that co-occur with singing16. To test whether single probes exhibited similar relationships to singing in both regions, we compared GS scores from area X to those measured in the VSP. As noted above, no probes had significant GS values for the amount or act of singing in the VSP, in contrast to thousands in area X. We compared GS.motifs.X and GS.singing.X within each module to GS.motifs.V and GS.singing.V for the same probes in the VSP and found weak correlations overall, especially for genes in the song modules (Figures 4D–F and S3G–L). Thus, genes whose area X expression is tightly coupled to singing have a very different relationship, or none at all, to this behavior in the VSP.
Area X-specific co-expression patterns correspond to singing
Next, we compared co-expression relationships within each area X module to the co-expression relationships between the same probes in the VSP, assigning each module a preservation score based on statistical comparisons of module composition and structure (Table S3)20. Area X modules were preserved to varying degrees in the VSP, with the blue, dark green, and orange song modules being the least preserved, and the modules most unrelated to singing (e.g. dark red and turquoise) being the most preserved. The song modules were effectively non-existent outside of area X, and there was a significant relationship between the strength of ME-singing correlations (Figure 3B) and module preservation ranks (Figures 4G–H), revealing a direct link between singing-relatedness and area X specific network structure in the basal ganglia.
Area X-specific co-expression patterns do not correspond to gene expression levels
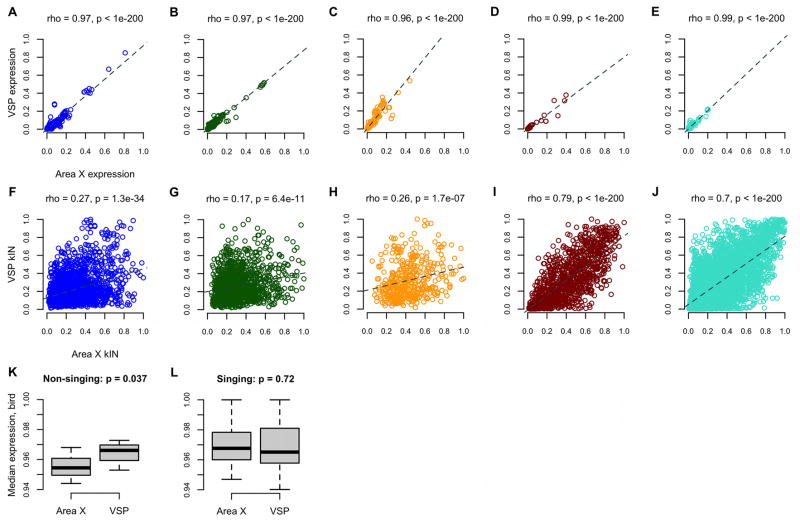
To test whether the regional differences in singing-related network structure were simply due to differences in gene expression levels, we began by computing correlations between the expression values for each probe in area X and VSP. There was remarkable similarity overall (cor=0.98, p<1e-200). Inspection of individual modules revealed a range of strong correlations between area X and VSP expression values (0.94–0.99; Figures 5A–E). In contrast, we observed a weaker overall correlation between area X and VSP network connectivity (cor=0.61, p<1e-200), especially within the 3 song modules (Figures 5F-J; blue, dark green, orange: mean cor=0.23; all other modules: mean cor=0.49).
Figure 5. Gene co-expression levels, rather than individual expression levels, distinguish area X song modules.
(A–E) Probe normalized median expression levels in the VSP are plotted as a function of levels in area X for 5 illustrative modules, revealing extremely strong correlations, whereas intramodular connectivity values (kIN, Table S2; F–J) were much less correlated, especially in the song modules. The dark red and turquoise modules were unrelated to singing and the most preserved in VSP (Table S3).
(K–L) Box and whisker plots show birds’ normalized median gene expression levels grouped by brain region for each singing state. Whiskers extend to the most extreme data points, box edges represent the 1st and 3rd quartiles, horizontal lines inside each box represent the median. Kruskal-Wallis rank sum test p-values are shown.
Activity in certain area X neurons increases during singing21. One possibility for why the song modules were observed in area X but not VSP is that this increase in neuronal firing leads to increased gene expression levels only in area X. To test this, we computed the normalized median gene expression levels in both brain regions for each bird. In non-singers, levels were higher in VSP than in area X (Figure 5K). This difference disappeared in singing birds; gene expression levels in area X and VSP became very similar (Figure 5L). These results imply that the area X-specific song modules cannot be accounted for by higher (or lower) area X gene expression levels compared to VSP during singing. Rather, as revealed here by WGCNA, the relevance of transcriptional activity in these regions to singing is determined more by region-specific co-expression relationships, which comprise ‘molecular microcircuitry’ that arises during a specific behavior (singing) within a specific brain region (area X) supporting that behavior. In line with the idea that mere neural activity levels do not account for the song-specialized gene modules, we previously found that activation of the IEG Synaptotagmin 4 (Syt4), is not achieved by overall depolarization of neurons but rather requires the patterned activation underlying singing22.
In silico validation of singing-driven co-expression networks
The new relationships we uncovered between gene co-expression patterns and singing are substantiated by the presence of previously identified area X singing-regulated genes in the song modules (e.g. EGR112 and FOS23: blue; FOXP210: dark green/orange; by convention, gene symbols are capitalized and italicized and are not meant here to denote the human form)24. Consistent with prior reports, EGR112 and FOXP210,11 were up and downregulated by song, respectively. The lack of correlation between GAPDH and singing-related probes validates its use as a control gene in area X under these conditions (Figure 3A). We compared our results to 2 prior studies which used microarrays to examine individual fold changes in gene expression in area X during singing, one of which also performed post hoc clustering5,25. Going further, we examined GS scores, MM and kIN.X for these genes in our data.
Wada et al. (2006) identified 33 genes whose expression levels differed in singing vs. non-singing birds, 31 of which were regulated in area X25. 29/31 were in our network (1 was not on the array, 1 was filtered out in pre-processing; Table S2); 19/29 were in the blue song module (p=8.9e-14, Fisher’s exact test; Table S2). In both studies, these 19 genes were upregulated by singing, as were probes representing 2 genes Wada et al. (2006) found to be regulated in other song nuclei, but not area X; BDNF and SYT4 (8/8 SYT4 and 2/4 BDNF probes had positive GS.motifs.X). Compared to the rest of the network, these 29 genes (170 probes total) had greater increases in expression in singing vs non-singing birds (p=3.5e-27, Kruskal-Wallis), and higher GS.motifs.X (p=3.5e-35) and GS.singing.X (p=3.5e-32). Wada et al. (2006) divided the genes they found into groups based on peak time of expression and regulation pattern. We found significant changes for multiple metrics across these groups in our data (Figure S4).
Warren et al. (2010) revisited singing driven gene regulation in area X and found 474 known genes (represented by 807 probes) that were regulated over the course of 0.5–7 hours of singing5. 300 of these genes were in our network, with subsets enriched in the 3 song modules (blue: 71 genes, with, e.g., SHC3, SMEK2, NTRK2 having the highest GS.motifs.X, p<4e-28; orange: 17 genes, e.g. CSRNP3, SCN3B, PLCB1, p<3e-6; dark green: 38 genes, e.g. BSDC1, VLDLR, RORA, p<5e-5; Fisher’s exact test; Table S2), and in 1 other module (yellow: 104 genes, p<5e-7; Table S2). Compared to the rest of the network, probes for all 300 genes had greater expression increases (p=1.9e-12, Kruskal-Wallis test; 882 probes total), higher GS.motifs.X (p=7.8e-11) and GS.singing.X (p=2.7e-11; Table S2). These genes were also more interconnected in their respective modules throughout the network (kIN.X, p=4.2e-4), especially in the blue song module (p=3.8e-14). A separate aspect of the study revealed enrichment for the functional annotation term “ion channel activity” in 49 genes posited to have undergone positive selection in zebra finches, which are also suppressed in the auditory forebrain during song perception. 42/49 were in our network (114 probes; Table S2), with 6 in the orange song module (p<3.3e-4, Fisher’s exact test). One of the ion channel genes, TRPV1 (dark green/salmon modules) was highly connected and strongly suppressed by singing in our data, and thus selected for validation in area X in vivo (see below and Table S2).
Singing-related modules contain human FOXP2 transcriptional targets
We previously showed that FoxP2 mRNA and protein are lower in area X following 2 hours of undirected singing compared to non-singing, with the magnitude of downregulation correlated to singing9,10,11. This finding was reproduced here; expression levels for all 12 FOXP2 probes in the network were negatively correlated with the number of motifs sung (Figure S5). Although our study used an indirect approach, i.e. a behavioral paradigm in which the birds’ natural singing behavior significantly alters FoxP2 levels within area X9,10,11, we predicted that this paradigm coupled with WGCNA would reveal FoxP2 transcriptional targets in area X singing-related modules. To test this, we screened the network for direct FOXP2 targets previously identified by 3 studies. Of 175 targets found in human fetal basal ganglia26, 56 were in our network (149 probes total; Table S2). These had relatively high MM in the orange song module (p=0.05, Kruskal-Wallis; Table S2) which contained genes that were downregulated with continued singing, including 9/12 probes for FOXP2. Of 302 targets found by a second study in SY5Y cells27, 119 were in our network (246 probes total; Table S2). Interestingly, these targets showed the opposite regulatory pattern, displaying high MM in modules upregulated with singing (blue: p=9e-4; black: p=8.6e-3; Table S2), but low MM in the orange module (p=9.6e-5; Table S2). The comparison of GS scores from these 2 groups of genes reiterated their contrary regulation during singing (GS.motifs.X scores were more negative in fetal brain targets, p<0.04; Table S2). These differences may be attributed to the different tissue types used in each study.
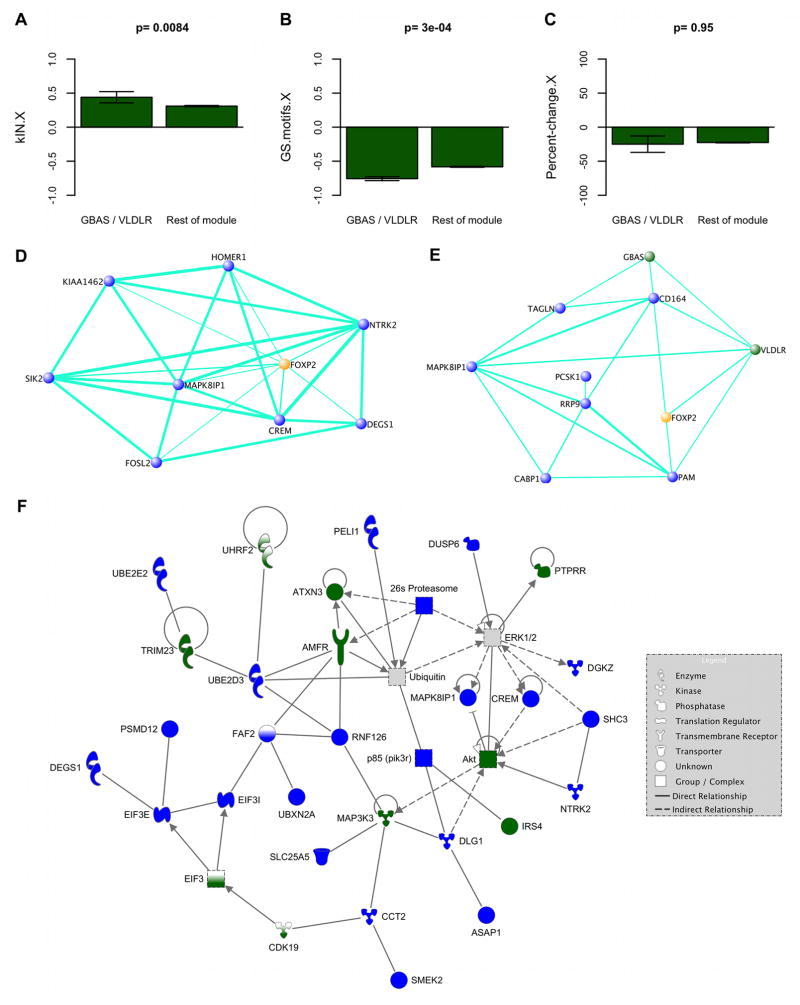
11 targets found by both studies were in our network. In line with our prediction, probes representing these 11 targets had strong relationships to singing (29 probes total; absolute values of GS.motifs.X, p=0.037; GS.singing.X, p=0.017, Kruskal-Wallis; Table S2), with a trend for greater expression increases in singing vs non-singing birds (p=0.064), compared to the rest of the network. Compared to the rest of the module, targets in the dark green song module (GBAS and VLDLR, 7 probes total) had high kIN.X and strong negative correlations to GS.motifs.X while showing no difference in expression levels (Figure 6A–C). This reinforces our finding that the connectivity of genes supersedes expression levels in dictating specification of networks for vocal behavior.
Figure 6. Behavioral regulation of gene expression coupled with WGCNA captures genes co-regulated with FOXP2.
(A–C) Barplots show intramodular connectivity (left), GS in area X for number of motifs (middle) and expression level percent-change in singing vs. non-singing birds (right), for the dark green module. Left bars in each plot represent values for 2 direct human FOXP2 targets, GBAS and VLDLR26,27, right bars represent the rest of the probes. Error bars = 95% confidence intervals. Kruskal-Wallis p-values are shown.
(D–E) VisANT51 visualizations highlight co-expression relationships among FOXP2 and subsets of its closest 300 network neighbors. TO was computed in an unsigned version of our network using the FOXP2 probe with the most significant GS.motifs.X score. D) Relationships among the most densely interconnected genes within the 20 closest FOXP2 neighbors (MM.blue>0.9 for all). E) The most densely interconnected genes within the 20 direct human FOXP226,27 targets displaying the highest TO with FOXP2. Nodes represent genes; node color, module assignment; edges, network connections; edge width, connection strength (thicker = stronger). Weak connections omitted for clarity.
(F) Canonical network involved in post-translational modification and cellular development, growth, and proliferation. All but 3 genes (CDK19, FAF2, UHRF2) were within the 300 closest FOXP2 neighbors. Connections in this graph denote biological interactions (direct = solid line; indirect = dashed) in the Ingenuity Knowledge Base (Ingenuity® Systems, www.ingenuity.com). Genes or complexes with one color had ≥1 probe assigned to a song module and are colored accordingly. Genes that are half white also reflect song module membership, but were outside the 300 closest FOXP2 neighbors. The EIF3 gene group has members in both blue and dark green modules. The ubiquitin and ERK1/2 complexes (grey) interact with song module genes and their enriched functions (Table S4). While FOXP2 does not appear here, its strong connections to these genes predicts that it interacts with them. See also Figure S5.
More recently, Vernes and colleagues (2011) performed a large scale-chromatin immunoprecipitation analysis of all known promoters and expression profiling to identify direct Foxp2 targets in embryonic mouse brain28. 557 of their putative 1,164 targets were present in our network, with 22 genes among the 300 closest network neighbors of FOXP2 (p<0.04, Fisher’s exact test). These included NTRK2 and YWHAH, which the authors validated as direct targets. In our network, NTRK2, a blue song module member, was the 3rd closest neighbor of FOXP2 (probeID=2758927) and is part of a canonical network involved in post-translational modification and cellular development, growth, and proliferation that also contains many other close network neighbors of FOXP2 (Figures 6D and 6F; Table S2). It was also found to be regulated during singing in area X by Warren et al. (2010)5. YWHAH, a gene involved in presynaptic plasticity, was in the blue song module, strongly upregulated during singing, and within the 300 closest network neighbors of FOXP2 (Table S2). 264 genes were deemed “high confidence” targets by the authors; 95 of these were in our network, including 14, 6, and 4 genes in the blue, dark green, and orange song modules, respectively. Compared to the rest of the network, these 95 genes had relatively high blue MM and low dark green and orange MM (p<1e-3, Kruskal-Wallis test), a pattern similar to what we observed for FOXP2 targets identified in SY5Y cells27 (Supplemental Experimental Procedures).
Overall, the findings by Vernes et al. (2011) indicate that in embryonic brain, Foxp2 modulates neuronal network formation by directly and indirectly regulating mRNAs involved in the development and plasticity of neuronal connections28. This is compatible with our WGCNA results emerging from adult songbird basal ganglia suggesting a role for FoxP2 in singing-related synaptic plasticity via its high interconnectedness with genes linked to MAPKK binding, NMDA receptors, actin/cytoskeleton regulation, and tyrosine phosphatase regulation (see “Biological significance of singing-related modules” below).
We also found interesting overlaps between our results and those of 2 additional studies that identified direct and/or indirect FOXP2 targets. The first study identified genes with differing expression levels in human neural progenitor cells transfected with either the human or the chimpanzee version of FOXP229. 24 such genes were in our network, and showed high kIN.X in their respective modules compared to the rest of the network (61 probes total; p=0.03, Kruskal-Wallis; Table S2). Those in the orange module had especially high kIN.X, compared to the rest of the module (CDCA7L, RUNX1T1: p=2.7e-3; Table S2). We observed a similar trend for those in the blue module (B3GNT1, HEBP2, NPTX2, TAGLN: p=0.074), but not in modules unrelated to singing that also contained many of these genes (turquoise, p=0.9; yellow, dark red, p=0.76). The second study identified 34 genes whose striatal expression levels were altered as a result of two human-specific amino-acid substitutions introduced into the endogenous Foxp2 locus of mice30. 13/34 genes were in our network (36 probes), including 3 in the song modules (ELAVL1: blue, HEXDC and YPEL5: dark green; Table S2). YPEL5 was highly connected in the dark green module and strongly suppressed by singing in our data, and was selected for validation in area X in vivo (Figure 8, Table S2). In summary, comparison of our WGCNA results with the literature identified song module genes co-regulated with FoxP2 that are common between songbird basal ganglia and mammalian tissues and, by extension, identified new genes and pathways (see below) that may be critical for speech.
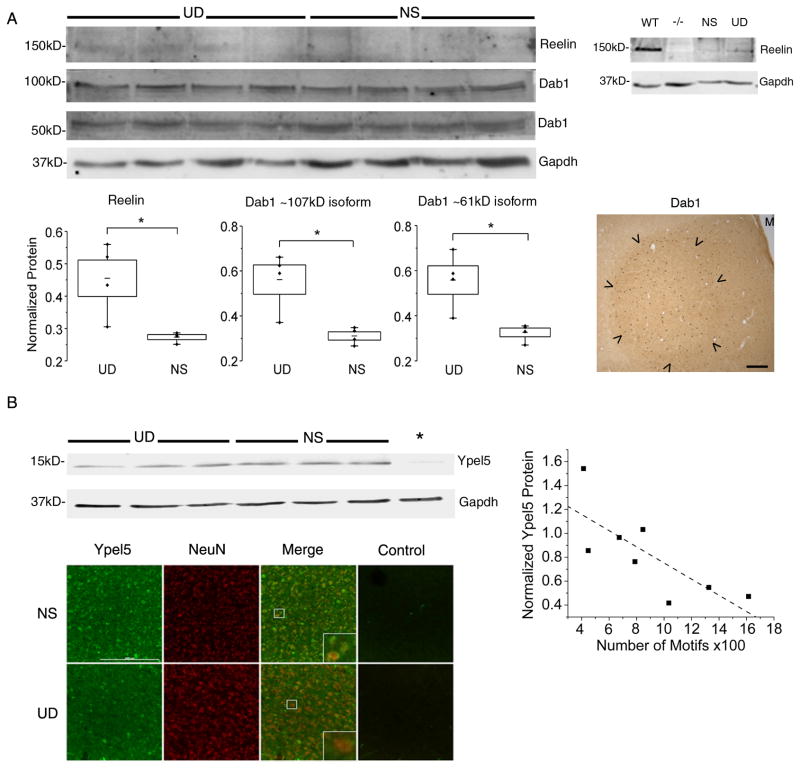
Figure 8. Behavioral regulation of hub genes and pathways in area X.
(A) Top left) Immunoblot of area X protein from 4 undirected singing (UD) and 4 non-singing (NS) birds shows bands for Reelin (~150kD) and phosphorylated forms of the Dab1 protein (~107kD, ~61kD). Top right) Reelin protein is detected in brain extracts from a wildtype mouse (WT), whereas this band is absent in a reeler mutant mouse (−/−), confirming antibody specificity. A band of similar size is observed in zebra finch area X samples from an NS and a UD bird. Bottom panels: Box and whisker plots show levels of Reelin protein (left) and of phosphorylated Dab1 isoforms (middle and right) as a function of singing. All 3 proteins are higher in area X of UD relative to NS birds (Mann-Whitney U 2-tailed test, p=0.03). Middle of each box represents the mean; top and bottom, standard error; whiskers, upper and lower 95% confidence intervals. Data from each bird is shown by individual points. At right, an immunohistochemical section at the level of area X (arrowheads) from a singing bird shows enhanced signals for Dab1 protein within the nucleus relative to outlying VSP. Scale bar = 100 m. See also Figure S7.
(B) Top left) Immunoblot of area X protein from 3 undirected singing (UD) and 3 non-singing (NS) birds shows bands at the predicted molecular weight for Ypel5 (~13kD) which are not apparent in the preadsorption control (*), indicating antibody specificity. Right) Quantification of signals from these and additional UD singers revealed a negative correlation between Ypel5 and the amount of singing (Spearman rho= −0.76; p=0.03, R2 = −0.77). Bottom, Photomicrographs of area X from a representative NS (top) and UD (bottom) bird. Immunofluorescent signals for Ypel5 (green) and the neuronal marker NeuN (red) are shown, as well as a no-primary antibody control (Control). All images were obtained at the same exposure. Qualitatively, more cell bodies appear labeled by the anti-Ypel5 antibody in the NS compared to the UD, most noticeable in the merged images where NeuN signals dominate in the UD bird. Scale bar = 200 m. Insets of boxed areas in the merged images suggest that Ypel5 and NeuN are co-expressed within area X neurons, but in different subcellular regions.
Biological significance of singing-related modules
We used the functional annotation tools available through the Database for Annotation, Visualization and Integrated Discovery (DAVID ver. 6.7)31 to characterize biological functions represented in the area X modules (Experimental Procedures). Many functional terms were enriched only in 1 of the singing-related modules, with the majority of these in the blue module; the most significant having to do with actin binding/regulation, MAP kinase activity, or proteasome activity (enrichment threshold = p<0.1). See Table S4 for all enriched terms in these modules.
To identify the most singing-relevant functions, we defined a measure of term significance (TS) as the absolute value of the product of the mean MM and GS.motifs.X for genes annotated with the term, scaled by 1 – the term’s p-value. The mean MM, GS.motifs.X, differential connectivity (kIN.diff), and clustering coefficient of genes annotated by terms with the highest TS scores were compared to the rest of the module, allowing us to hone in on particularly tight-knit, behaviorally-relevant, biological pathways/functions in the singing-related modules (Supplemental Experimental Procedures). For example, 11 genes in the blue module (ARC, CABP1, CNN3, DLG1, DLG2, DLGAP2, FREQ, HOMER1, IFNGR1, NLGN1, NTRK2) were annotated by the term “GO:0014069~postsynaptic density” (Table S4). Probes representing these genes in the blue module had high MM and GS.motifs.X (27 probes total; mean MM=0.804, GS.motifs.X=0.682), and the term “GO:0014069~postsynaptic density” had an enrichment p-value of 0.059. Thus TS for this term = 0.804 × 0.682 × (1 − 0.059) = 0.516 (7th highest of 402 enriched blue module terms; Tables S2, S4). Compared to the rest of the module, probes for the 11 genes annotated with this term had higher average MM (p=6.2e-7, Kruskal-Wallis test), GS.motifs.X (p=6.8e-5), kIN.diff (p=4.7e-6), and clustering coefficient (p=5.2e-5).
Other top ranked blue module terms included “GO:0031434~mitogen-activated protein kinase kinase binding” and “IPR019583:PDZ-associated domain of NMDA receptors”, as well as others involving actin, cytoskeleton, and tyrosine phosphatase regulation. Genes associated with these synapse related functions in the blue module were also some of FOXP2’s closest neighbors, i.e. genes with which it had high TO (Figures 6D–F, Table S2, Supplemental Experimental Procedures). This may imply a role for FoxP2 in the suppression of synaptic plasticity, since blue module genes (whose levels increased with singing in these experiments) in high TO with FOXP2 (which decreased with singing) are good candidates for repressed transcriptional targets.
Each of the song modules was enriched for astrocytic markers with developing astrocytes most enriched in the blue module (p=7.5e-6, Fisher’s exact test) and mature astrocytes in the orange module (p=4e-3)32. This observation is consistent with the recent realization that astrocytes are involved in the regulation of neuronal functions, including behavior33. We screened the modules for genes associated with Parkinson’s disease (Supplemental Experimental Procedures), since it is a basal ganglia based disorder with a vocal component and found enrichment in the black singing-related module (Figure S6). Another module that was moderately singing-related was also enriched for Parkinson’s disease associated genes, as well as autism susceptibility genes (purple module, p=2.7e-4, p=0.05, respectively, Table S2).
Biological significance of other modules
The unique presence of the song modules in area X implies that the biological pathways they represent are co-regulated in patterns specific to area X during learned vocal-motor behavior. Conversely, functions in modules found in both area X and VSP during singing may typify more general striato-pallidum-wide regulatory networks. To test this, we examined biological functions represented in the dark red, turquoise and pink modules; the 3 most preserved in VSP (Figures 4G–H, Table S3). The turquoise module was the largest in the network (4,616 probes representing 2,743 known genes; Table S2). It was the only module enriched for many functional terms related to hormone binding, morphogenesis, neurogenesis, and development, implicating it in steroid sensitivity and the ongoing neurogenesis known to occur throughout the adult songbird striatum34,35(Table S4).
The turquoise, dark red and pink modules were enriched for neuron and oligodendrocyte gene markers32 (turquoise: genes >10 fold enriched in oligodendrocytes, p=0.05, dark red: genes >20 fold enriched in neurons, p=0.03, Fisher’s exact test; Table S2), and markers of striatal and pallidal neurons (pink: p<0.02; Table S2), consistent with the mixed striatal and pallidal nature of what was formerly known as the avian ‘striatum’36,37. These findings are congruent with the idea that the preserved modules represent functions common across the striato-pallidum.
Hub genes and biological pathways in singing-driven co-expression networks
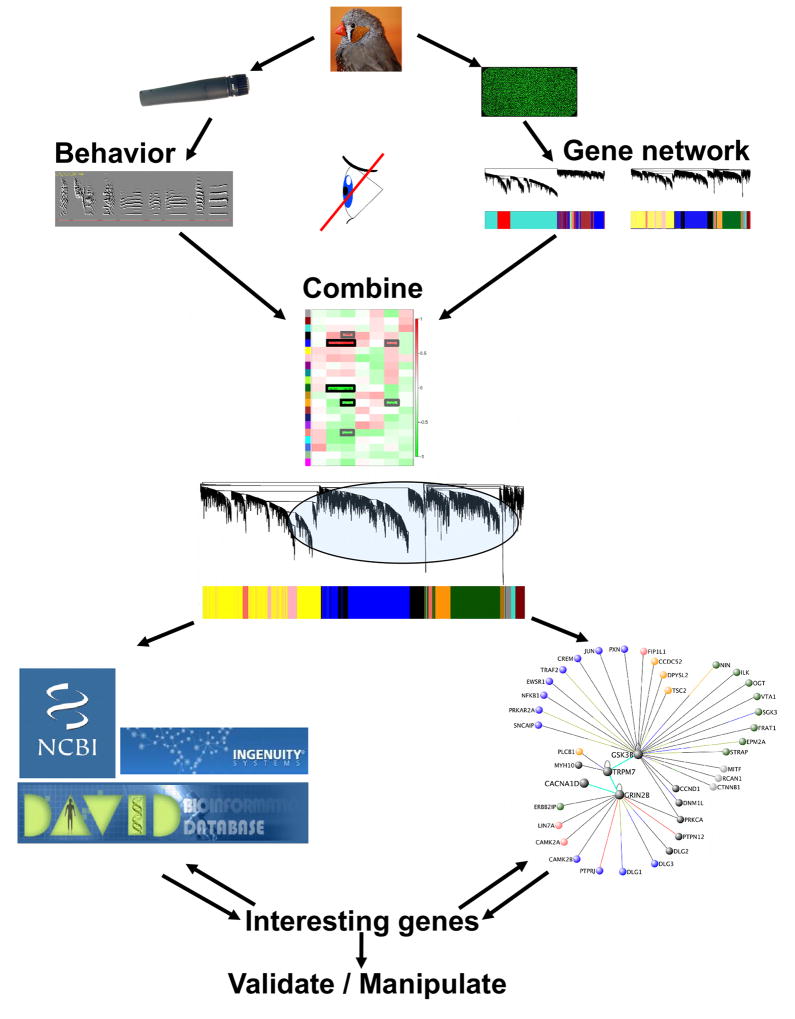
Given the large number of genes in the song modules, we sought to identify the potentially most important genes for further study. We used 2 basic approaches (Figure 7); both began by restricting further analysis to the singing-related modules. In one approach, we then focused on song module genes with high GS.motifs.X and MM, i.e. genes highly interconnected within their module (hub genes) and strongly coupled to singing, and screened them for enriched functions and biological features. The other approach is exemplified above in the “Biological significance of singing-related modules” section where we functionally annotated the singing-related modules, then prioritized enriched functional terms based on TS scores (Supplemental Experimental Procedures; Table S4), highlighting sets of tightly interconnected singing-related genes that were both important in the module and shared an enriched common feature.
Figure 7. Application of WGCNA to identify novel pathways in learned vocalization.
Schematic of the use of WGCNA to select relevant molecules and pathways for further study. Top) Singing data (left) and gene expression data (right) were gathered from the same birds. Network construction was blind to the behavioral analysis. Middle) Co-expression network structure was then related to song analysis results to identify gene modules important for the behavior. Bottom) Focusing on singing-related modules, gene ontology and functional enrichment analyses were carried out to identify functions and pathways relevant to singing (left). Concurrently, the most important molecules populating the song modules were identified via network metrics (right). The results from each of these approaches were cross-referenced to further prioritize behaviorally relevant biological pathways. Images courtesy of Maurice van Bruggen (zebra finch, http://creativecommons.org/licenses/by-sa/3.0/deed.en) and Iain Fergusson (microphone, http://creativecommons.org/licenses/by/3.0/deed.en), DAVID and Ingenuity logos used with permission.
We used these approaches to select pathways in which to test for the presence of constituent proteins in area X. The importance of studying molecules in the context of biological pathways, rather than simply validating mRNA expression, is underscored by our finding that gene co-expression relationships, rather than expression levels per se, determine molecular microcircuitry underlying vocal-motor-specific behavior. As our focus was on the protein level, area X tissue was isolated from singing and non-singing birds at 3 (rather than 2) hours following either time from the 1st motif or lights-on, respectively, to allow for potential translation of mRNA changes (see Supplemental Experimental Procedures for description of tissue processing methods).
WGCNA identified very-low density lipoprotein receptor, Vldlr, a member of the Reelin signaling pathway, as a highly connected member of the dark green song module (mean GS.motifs.X=−0.78, MM=0.82; Table S2). Vldlr was also identified in the literature as a human FOXP2 target26,27. In mammals, the Reelin pathway is critical to neuronal migration during development of the neocortex and cerebellum and to regulation of NMDA receptor-mediated synaptic plasticity in the adult hippocampus38. Reelin binds to Vldlr on migrating neurons and radial glial cells. While this pathway is well-established in cortex-containing structures, less is known about the role of these molecules in the basal ganglia of any species. In songbirds, Reelin is expressed in cortical HVC and striato-pallidal area X of adults, but behavioral regulation had not been examined39.
In line with behavioral activation of this pathway, expression of Reelin protein was significantly higher in singing vs. non-singing birds (Figure 8A). We also detected Vldlr protein expression in area X (Figure S7A). Since in mammals, binding of Reelin to Vldlr results in the activation of the cytoplasmic adapter protein disabled 1 (Dab1) by tyrosine phosphorylation, we tested for singing-driven regulation of Dab1. As expected, we detected a significant increase in phosphorylated forms of Dab1 in area X of singers relative to non-singers (Figure 8A). Dlgap2 (aka PSD95; blue module; mean GS.motifs.X=0.65, MM=0.82; Table S2) binds Vldlr to the NMDA receptor, activating downstream molecules such as the cAMP responsive element modulator (Crem). CREM (blue module; mean GS.motifs.X=0.83, MM=0.95) shares high TO with FOXP2 (Figures 6D,F; Table S2), implicating FoxP2 in regulation of synaptic plasticity through indirect connections with the Reelin signaling pathway. As noted above, tyrosine phosphorylation and NMDA receptor related functional terms stood out in the blue module, and DLGAP2 was one of 11 blue module genes annotated by “GO:0014069~postsynaptic density” (Table S4).
A second biological pathway containing yippee-like protein 5 (Ypel5) was selected for further study because of Ypel5’s identification as a putative target of the partially humanized Foxp230, its GS.motifs.X score (mean of 3 probes = −0.71) and MM in the dark green module (mean=0.86; Table S2). “PIRSF028804: protein yippee-like” and “IPR004910: Yippee-like protein” had the highest TS scores in the dark green module (Table S4). We viewed this as a rigorous test of the predictive power of WGCNA because of the relative lack of information about this molecule in vertebrates40. In immunohistochemical analyses, we observed signals for Ypel5 protein in area X (Figure 8B), as well as for its binding partner, Ran Binding Protein in the Microtubule Organizing Center40, also in the dark green module (RANBPM aka RANBP9, data not shown). In line with its strong GS.motifs.X score, Ypel5 was behaviorally regulated, with lower protein levels observed in area X of birds that sang more motifs (Figure 8B). Our results for both Reelin and Ypel5 demonstrate expression of multiple members of their respective signaling pathways in area X, with behavioral regulation of each.
As further validation, we detected protein signals within area X consistent with expression of Transient Receptor Potential Vanilloid Type 1 (Trpv1), a capsaicin receptor. We selected Trpv1 for validation because of its high MM and GS.motifs.X, and its identification as an ion channel positively selected for in the songbird lineage5 (Figure S7B). TRPV1 is in the dark green and salmon singing-related modules (1 probe in each; dark green: MM=0.85, GS.motifs.X=−0.77; salmon: MM=0.81, GS.motifs.X=−0.51; Table S2), and has been linked to endocannabinoid signaling pathways in the mammalian basal ganglia41,42. Cannabinoid exposure during zebra finch development interferes with song learning43, potentially through synaptic plasticity mechanisms such as modulation of glutamatergic synapses onto medium spiny neurons in area X44 and altered area X FoxP2 expression45. In keeping with its strong GS.motifs.X score, we observed lower levels of Trpv1 signal in birds that sang more motifs (Figure S7B). These findings provide additional biological and literature-based validation of our WGCNA.
Discussion
This study represents the first identification of basal ganglia gene co-expression networks specialized for vocal behavior, and the first use of WGCNA to link co-expression modules to a naturally occurring, procedurally learned behavior. We found ~2,000 genes within the song-specialized striato-pallidal area X, but not in VSP, that were significantly coupled to singing, most of which were members of one of 5 distinct singing-related modules. The 3 song modules (blue, dark green, orange; Figure 3) were unique to area X, and a given module’s singing-relatedness was highly predictive of its preservation outside of area X, i.e. the more related to singing, the less preserved (Figure 4). The VSP is active during singing, as indicated by IEG expression16, and we found gene expression levels in VSP and area X to be remarkably similar during singing (Figure 5). Thus, the regional differences we observed in network structure are likely not due to differences in expression levels, and the singing-related modules in area X are likely not a general product of neural activity, but instead reflect area X specific singing-driven gene regulation patterns.
We predict that WGCNA-type approaches applied to expression data from other song nuclei would likewise reveal song-regulated gene ensembles not found in neighboring tissue, e.g. HVC vs. surrounding cortex. The degree to which such hypothetical song modules would conform with the area X co-expression patterns described here, or whether they would represent the same biological pathways, is an open question. Since the different song nuclei apparently support distinct aspects of singing behavior, one might predict that singing-related co-expression patterns would also be distinct, or would at least relate to different song features, e.g. HVC modules might relate to measures of syllable sequencing17.
Prior microarray studies of area X gene regulation were based on singling out differentially expressed genes in singing versus non-singing birds, then placing them in groups based on the timing of their expression changes. Our approach differed in that we arranged genes into groups based only on their expression patterns, then related them to singing post-hoc. This resulted in modules that contained >1,000 genes previously unknown to be regulated by vocal behavior. The overlap of our findings with those of prior studies is dominated by genes in the blue module, which contained genes with the largest singing-driven increases in expression. This may imply that differential expression approaches are less effective at identifying gene ensembles, especially downregulated ones, with more nuanced regulation patterns. We predict WGCNA-type approaches will be more effective at uncovering biological functions vital to vocal-motor behavior that do not contain genes with massive expression perturbations. We verified our hypothesis that targets of FOXP2 in human tissue and cell lines would be important members of area X specific singing-related modules (Figure 6). Future studies could narrow the search for genes that interact with FoxP2 in a vocal-motor context using our results as a guide, beginning by screening for genes with high TO with FOXP2 that also have high singing-related GS and connectivity. We also found enriched functional categories that were unique to the singing-related modules, and described a method for prioritizing biological functions and pathways for future investigation, based on testing metrics of network importance and behavioral significance for genes annotated with significantly enriched terms. Combining this method of ranking enriched biological functions by their importance in singing-related co-expression networks with screens for FoxP2 targets, as described above, could prove fruitful for elucidating the molecular underpinnings of learned vocal-motor behavior in songbirds and humans.
We used the WGCNA area X network results and literature sources to identify novel pathways regulated by vocal behavior in area X, and demonstrated behaviorally-driven changes in protein levels in the Reelin signaling pathway and additional molecules (Figures 8, S7). Finally, enrichment for Parkinson’s disease and autism genes in the song and non-song modules (Figure S6) supports the use of songbirds not just as a model for speech, but as a model for exploring pathways in motor disorders with a vocal component.
Experimental Procedures
Behavior
Animal use was in accordance with NIH guidelines for experiments involving vertebrate animals and approved by the University of California at Los Angeles Chancellor’s Institutional Animal Care & Use Committee. For the microarrays, experiments were conducted in the morning from the time of light onset to death, 2 hours later, according to Miller et al. (2008)9. During this time, 18 adult male birds sang undirected song of varying amounts. An additional 9 males were designated ‘non-singers’ (Table S1). If any potential non-singing bird sang >10 motifs, it was excluded from the study. Males performing to a female were not included because FOXP2 mRNA levels in such directed singers are similar to non-singers and are not correlated to the amount of song10. For biological validation, 18 non-singers and 19 undirected singers were collected 3 hours following lights-on or from their 1st song motif, respectively. Songs were recorded using Shure SM57 microphones, digitized with a PreSonus Firepod (44.1 kHz sampling rate, 24 bit depth), and acquired using Sound Analysis Pro 2.091 (SAP2)18. Acoustic features of song were computed for each bird using the Feature Batch module in SAP2, and the mean values of each feature were obtained to provide 1 representative number for each bird. Motifs were counted independently by 2 experimenters via visual inspection of spectrograms in Audacity (version 1.3; http://audacity.sourceforge.net/).
Antibodies and Assays
Tissue was processed for immunoblotting or immunohistochemistry following conventional methodologies using primary antibodies to detect the following proteins: Reelin, Vldlr, phosphorylated Dab 1, Dab1, Ypel5, RanBPM, Trpv1, NeuN and Gapdh. See Supplemental Experimental Procedures for details.
Microarrays
Agilent zebra finch oligoarrays (ver. 1) containing 42,921 60-mer cDNA probes were constructed through a collaboration between the Jarvis Laboratory of Duke University, Duke Bioinformatics, and The Genomics group of RIKEN, under the direction of Drs. Erich Jarvis and Jason Howard (http://songbirdtranscriptome.net; Duke University). These arrays represent cDNA libraries obtained from Michigan State University (Dr. Juli Wade), Rockefeller University (Dr. Fernando Nottebohm), the Keck Center of the University of Illinois (Dr. David Clayton) and Duke25,46,47. Area X and VSP tissue samples were extracted from all birds (n=27). Each RNA sample was hybridized to a single array, totaling 54 arrays, 2 per bird. Each slide, containing 4 arrays, had 4 samples hybridized: bilateral area X and VSP samples from 2 different birds. Birds were selected per slide such that low or non-singers were paired with high singers to minimize possible inter-slide bias or batch effects (Table S1). During data pre-processing, 1 area X sample and 2 VSP samples, all from non-singing birds, were removed as outliers. See Supplemental Experimental Procedures for details on tissue collection, RNA isolation, array hybridization and pre-processing.
Nomenclature: Probes vs Genes
“Probe” refers to a single probe on the array. GS measurements were computed for each probe. In many cases, multiple probes for a single “gene”, e.g. FOXP2, were present on the array (Figure S5, Table S2). There were 20,104 probes in the network, 16,448 of which were annotated with a gene symbol at the time of analysis (February 2011, see http://songbirdtranscriptome.net for up-to-date annotations). Since many genes were represented by >1 probe, only 8,015 annotations were unique. Of these 8,015 unique genes, there were 2,496 unique annotations in the 5 singing-related modules. When we report GS.motifs.X for a gene, that value is the average GS.motifs.X score of all probes for that gene unless otherwise noted. The area X co-expression network was constructed using probes, thus when we report the number of genes in a module we are referring to the number of unique gene annotations found for probes in that module. Due to sources of natural and experimental variability, different probes to the same gene were sometimes assigned to different, though usually similar, modules during network construction, e.g. probes made to different regions of the same gene may bind to alternatively-spliced transcript variants with varying levels of efficiency.
Network Construction
Many methods exist for analyzing gene expression microarray data. We chose WGCNA because of its biological relevance, and other advantages (Supplemental Experimental Procedures). All WGCNA computations were done in the free statistical software R (http://www.r-project.org/) using functions in the WGCNA library48, available via R’s package installer. After pre-processing the raw microarray data to remove outliers, normalize, and filter the data from 42,921 to 20,104 probes (Supplemental Experimental Procedures), the correlation matrix was obtained by computing the signed pairwise Pearson correlations between all probes across all birds. The correlation matrix was transformed using a power function ((1 + correlation)/2)β) to form the adjacency matrix, a matrix of network connection strengths. β was determined empirically using the scale-free topology criterion6 (signed network: β=14; unsigned: β=6). The network is “weighted” because connection strengths can take on any value between 0 and 1, in contrast to “unweighted” networks where connections are binary. Connectivity (k) is defined for each probe as the sum of its connections to all other probes. The intramodular connectivity (kIN, Table S2) of each probe is the sum of its connections to other probes in its module. Intramodular connectivity in VSP (kIN.V) was computed based on the co-expression relationships in VSP of probes grouped by their area X module assignments. See Supplemental Experimental Procedures for details on the scale-free topology criterion and its biological relevance, differential connectivity, signed vs unsigned networks, and FOXP2 neighborhood analysis.
Module Definition
WGCNA identifies modules of densely interconnected probes by correlating probes with high topological overlap (TO), a biologically meaningful measure of similarity that is highly effective at filtering spurious or isolated connections49. The TO matrix was computed based on the adjacency matrix (Supplemental Experimental Procedures) and average linkage hierarchical clustering was performed using 1 – TO as the distance metric. Modules were defined using a dynamic tree cutting algorithm to prune the resulting dendrogram (Supplemental Experimental Procedures)50.
Relating Network Structure to Singing
Expression values within each module were summarized by computing module “eigengenes” (MEs): the 1st principal component of each module obtained via singular value decomposition. We defined the module membership (MM) of individual probes as their correlations to the MEs, such that every probe had a MM value in each module. To discover any significant relationships between gene expression perturbations within modules and traits, we computed the correlations between MEs and phenotypic measures, including age, acoustic features, number of motifs sung, and whether the bird sang or not (Figure 3B). P-values were obtained via the Fisher transformation of each correlation; modules with correlations to singing traits that had p-values below the Bonferroni corrected significance threshold (α=1.7e-4) are referred to as the 3 “song modules” throughout the text. We also performed the less conservative Benjamini and Hochberg FDR procedure19 and found significant correlations to singing for the black and salmon modules. P-value corrections were performed using the results from all phenotypic measures listed above, not just those highlighted in Figure 3B.
Visualization and Functional Annotation
Lists of unique gene annotations from each module were used for all module enrichment calculations using Fisher’s exact test, functional annotation studies in DAVID and Ingenuity, and when generating VisANT51 visualizations (Figures 6D-F and S6, Supplemental Experimental Procedures).
Data Accessibility
Raw and processed microarray data, and behavioral data for each bird are available at http://www.ncbi.nlm.nih.gov/geo (Accession GSE34819).
Supplementary Material
Highlights.
Singing-driven co-expression modules contain ~2,000 genes in songbird nucleus area X
High-order relationships of genes in these modules confer vocal-motor specificity
Speech-related gene FoxP2 and its transcriptional targets in singing-driven modules
Biological validation of previously unknown pathways underlying vocal behavior
Acknowledgments
We thank Peter Langfelder and Michael Oldham for advice on microarray pre-processing and network analysis, Jason Howard and Erich Jarvis for the arrays through a partnership with Agilent Technologies, Patty Phelps, Sarah Bottjer and Erica Sloan for material support, Felix Schweizer and Grace Xiao for statistical advice, and 4 anonymous reviewers for insightful commentary. Supported by NIH grants F31 MH082533 (ATH) and R01 MH070712 (SAW).
Footnotes
AUTHOR CONTRIBUTIONS: JEM, ATH and SAW designed the experiments. JEM collected the animals, tissue punches, analyzed the song, and, together with EF performed the biological validation. ATH performed the RNA isolation, array pre-processing, song analysis, and WGCNA, with guidance from SH. ATH, JEM, EF, SH, and SAW wrote/edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lieberman P. Toward an evolutionary biology of language. Cambridge: Harvard University Press; 2006. [Google Scholar]
- 2.Jarvis ED. Learned birdsong and the neurobiology of human language. Ann NY Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White SA. Genes and vocal learning. Brain and Language. 2010;115:21–28. doi: 10.1016/j.bandl.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–63. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical Applications in Genetics and Molecular Biology. 2005:4. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 7.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted Gene Coexpression Network Analysis: State of the Art. J Biopharml Stats. 2010;20(2):281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- 9.Miller JE, Spiteri E, Condro MC, Dosumu-Johnson RT, Geschwind DH, White SA. Birdsong decreases protein levels of FoxP2, a molecule required for human speech. J Neurophysiol. 2008;100:2015–2025. doi: 10.1152/jn.90415.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J Neurosci. 2006;26:7390–4. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teramitsu I, Poopatanapong A, Torrisi S, White SA. Striatal FoxP2 is actively regulated during songbird sensorimotor learning. PloS One. 2010;5:e8548. doi: 10.1371/journal.pone.0008548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci USA. 1997;94:4097–102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 14.Miller JE, Hilliard AT, White SA. Song practice promotes acute vocal variability at a key stage of sensorimotor learning. PloS One. 2010;5:e8592. doi: 10.1371/journal.pone.0008592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PloS Biol. 2007;5:2885–2897. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. Molecular mapping of movement-associated areas in the avian brain: A motor theory for vocal learning origin. PloS One. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahnloser RHR, Kozhevnikov AA, Fee MS. An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature. 2002;419:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- 18.Tchernichovski O, Nottebohm F, Ho CE, Pesaran B, Mitra PP. A procedure for an automated measurement of song similarity. Anim Behav. 2000;59:1167–1176. doi: 10.1006/anbe.1999.1416. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 20.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PloS Comp Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessler NA, Doupe AJ. Singing-related neural activity in a dorsal forebrain-basal ganglia circuit of adult zebra finches. J Neurosci. 1999;19:10461–81. doi: 10.1523/JNEUROSCI.19-23-10461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poopatanapong A, Teramitsu I, Byun JS, Vician LJ, Herschman HR, White SA. Singing, but not seizure, induces synaptotagmin IV in zebra finch song circuit nuclei. J Neurobiol. 2006;66:1613–29. doi: 10.1002/neu.20329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–25. doi: 10.1016/s0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 24.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Devel. 2000;14:142–6. [PubMed] [Google Scholar]
- 25.Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci USA. 2006;103:15212–7. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Gen. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Gen. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genetics. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao FY, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–U289. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enard W, Gehre S, Hammerschmidt K, Holter SM, Blass T, Somel M, Bruckner MK, Schreiweis C, Winter C, Sohr R, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halassa MM, Haydon PG. Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Ann RevPhysiol. 2010;72:335–55. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nottebohm F. The road we travelled – Discovery, choreography, and significance of brain replaceable neurons. Ann NY Acad Sci. 2004;1016:628–658. doi: 10.1196/annals.1298.027. [DOI] [PubMed] [Google Scholar]
- 35.Kim YH, Perlman WR, Arnold AP. Expression of androgen receptor mRNA in zebra finch song system: developmental regulation by estrogen. J Comp Neurol. 2004;469:535–47. doi: 10.1002/cne.11033. [DOI] [PubMed] [Google Scholar]
- 36.Farries MA, Perkel DJ. A telencephalic nucleus essential for song learning contains neurons with physiological characteristics of both striatum and globus pallidus. J Neurosci. 2002;22:3776–3787. doi: 10.1523/JNEUROSCI.22-09-03776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herz J, Chen Y. Reelin, lipoprotein receptors and synaptic plasticity. Nat Rev Neurosci. 2006;7:850–859. doi: 10.1038/nrn2009. [DOI] [PubMed] [Google Scholar]
- 39.Balthazart J, Voigt C, Boseret G, Ball GF. Expression of reelin, its receptors and its intracellular signaling protein, Disabled1 in the canary brain: Relationships with the song control system. Neuroscience. 2008;153:944–962. doi: 10.1016/j.neuroscience.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hosono K, Noda S, Shimizu A, Nakanishi N, Ohtsubo M, Shimizu N, Minoshima S. YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with 2 distinct proteins RanBPM and RanBP10. Genomics. 2010;96:102–111. doi: 10.1016/j.ygeno.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Musella A, De Chiara V, Rossi S, Prosperetti C, Bernardi G, Maccarrone M, Centonze D. TRPV1 channels facilitate glutamate transmission in the striatum. Mol Cell Neurosci. 2009;40:89–97. doi: 10.1016/j.mcn.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Maccarrone M, Rossi S, Bari M, De Chiara V, Fezza F, Musella A, Gasperi V, Prosperetti C, Bernardi G, Finazzi-Agrò A, et al. Anandamide inhibits metabolism and physiological actions of 2-arachidonoylglycerol in the striatum. Nat Neurosci. 2008;11:152–9. doi: 10.1038/nn2042. [DOI] [PubMed] [Google Scholar]
- 43.Soderstrom K, Tian Q. Distinct periods of cannabinoid sensitivity during zebra finch vocal development. Br Res Devel Br Res. 2004;153:225–232. doi: 10.1016/j.devbrainres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Thompson JA, Perkel DJ. Endocannabinoids mediate synaptic plasticity at glutamatergic synapses on spiny neurons within a basla ganglia nucleus necessary for song learning. J Neurophysiol. 2010;105:1159–69. doi: 10.1152/jn.00676.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soderstrom K, Luo B. Late-postnatal cannabinoid exposure persistently increases FoxP2 expression within zebra finch striatum. Dev Neurobiol. 2010;70:195–203. doi: 10.1002/dneu.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, et al. The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Wang XJ, Tannenhauser J, Podell S, Mukherjee P, Hertel M, Biane J, Masuda S, Nottebohm F, Gaasterland T. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci USA. 2007;104:6834–9. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2007;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 51.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed microarray data, and behavioral data for each bird are available at http://www.ncbi.nlm.nih.gov/geo (Accession GSE34819).