Abstract
During development, circuits are refined by the dynamic addition and removal of synapses; however, little is known about the molecular mechanisms that dictate where and when synaptic refinement occurs. Here we describe transcriptional mechanisms that pattern remodeling of C. elegans neuromuscular junctions (NMJs). The embryonic GABAergic DD motor neurons remodel their synapses, while the later born VD neurons do not. This specificity is mediated by differential expression of a transcription factor (HBL-1), which is expressed in DD neurons but is repressed in VDs by UNC-55/COUP-TF. DD remodeling is delayed in hbl-1 mutants whereas precocious remodeling is observed in mutants lacking the microRNA mir-84, which inhibits hbl-1 expression. Mutations increasing and decreasing circuit activity cause corresponding changes in hbl-1 expression, and corresponding shifts in the timing of DD plasticity. Thus, convergent regulation of hbl-1 expression defines a genetic mechanism that patterns activity-dependent synaptic remodeling across cell types and across developmental time.
Introduction
A hallmark of all nervous systems is the dynamic addition and removal of synaptic connections. Despite its universality, synaptic remodeling has primarily been studied in vertebrates. In mammals, synaptic remodeling occurs in many, and perhaps all circuits. For example, at the neuromuscular junction (NMJ), each muscle is initially innervated by multiple axons, and the mature pattern of mono-innervation emerges following a period of synaptic elimination (Goda and Davis, 2003; Luo and O’Leary, 2005; Purves and Lichtman, 1980). Similarly, in the cerebellum, Purkinje cells eliminate exuberant climbing fibers inputs (Bosman and Konnerth, 2009). Live imaging studies in the mouse cortex also suggest that dendrites continuously extend and retract spines during development (Holtmaat et al., 2005; Trachtenberg et al., 2002; Grutzendler et al., 2002). From these and other studies, a great deal has been learned about how changes in axonal and dendritic structures are patterned during development.
Much less is known about the molecular mechanisms that pattern synaptic refinement in vertebrates. In particular, several important questions remain unanswered. Although remodeling occurs throughout the life of an animal, there is a general trend for increased plasticity earlier in development. For each circuit, plasticity often occurs during brief time intervals, which are termed critical periods (Hensch, 2004). While remodeling occurs in most, and perhaps all circuits, different cell types within a circuit exhibit the capacity for plasticity at distinct times. For example, in the visual cortex, plasticity in layer 4 ends prior to plasticity in more superficial layers (Jiang et al., 2007; Oray et al., 2004). How is plasticity restricted to specific cell types and specific developmental times? In all known cases, vertebrate synaptic refinement is highly dependent on circuit activity, which implies that plasticity is dictated by competition between cells in these circuits. A few activity-induced genes have been implicated in synaptic refinement. For example, ocular dominance plasticity is correlated with activity-induced changes in the expression of CREB and BDNF (Hensch, 2004). However, activity induces CREB and BDNF expression in many (perhaps all) neurons, including dissociated neurons in culture (Cohen and Greenberg, 2008; Lonze and Ginty, 2002). How does altered expression of general activity induced genes confer cell and temporal specificity on circuit refinement? Because circuit refinement plays a pivotal role in shaping cognitive development, there is great interest in defining the molecular and genetic mechanisms that determine how refinement is patterned.
To address these questions, we exploited an example of genetically programmed synaptic remodeling in C. elegans. During the first larval stage (L1), the DD GABAergic motor neurons undergo a dramatic remodeling whereby synapses formed with ventral body muscles in the embryo are eliminated and replaced by synapses with dorsal muscles (Park et al., 2011; White et al., 1978; Hallam and Jin, 1998). DD remodeling occurs without retraction or extension of neurite processes. Instead, the DD ventral process switches from an axonal to a dendritic fate (and vice versa for the dorsal process).
Many aspects of C. elegans larval development are controlled by cell intrinsic developmental timing genes, which are generically termed heterochronic genes (Moss, 2007). In particular, the heterochronic gene lin-14 controls the timing of hypodermal development, whereby L2 hypodermal cell fates are expressed precociously during the L1 in lin-14 mutants (Ambros and Horvitz, 1984). Similarly, lin-14 is expressed in DD neurons, and DD remodeling occurs earlier in lin-14 mutants, initiating during embryogenesis (Hallam and Jin, 1998). Thus, LIN-14 dictates when DD remodeling is initiated. This was the first study to show that heterochronic genes play a role in post-mitotic neurons to pattern synaptic plasticity. Because lin-14 orthologs are not found in other organisms, it remains unclear if control of synaptic plasticity by heterochronic genes represents a conserved mechanism. DD plasticity (like other forms of invertebrate plasticity) is generally considered to be genetically hard wired, i.e. dictated by specific cell intrinsic genetic pathways. Thus, it also remains unclear if activity-induced refinement of vertebrate circuits and DD plasticity represent fundamentally distinct processes, which are mediated by distinct molecular mechanisms.
Here we show that a second heterochronic gene, hbl-1, regulates several aspects of DD plasticity. The hbl-1 gene encodes the transcription factor HBL-1 (Hunchback like-1) (Fay et al., 1999). We show that convergent pathways regulate hbl-1 expression in D neurons, conferring cell and temporal specificity and activity dependence on D neuron plasticity. Thus, our results define a cell intrinsic genetic pathway that dictates a form of activity dependent synaptic refinement.
Results
VD neurons undergo ectopic synaptic remodeling in unc-55 mutants
The DD motor neurons are born during embryogenesis, and remodel their synapses during the L1. A second class of GABAergic motor neurons, the VD neurons, is born during the late L1 stage but does not undergo remodeling. VD neurons share many other characteristics with DD neurons, including similar cell body positions, similar axon morphologies, similar roles in controlling locomotion, and similar expression profiles (Jorgensen, 2005). Like DDs, VD neurons initially form ventral synapses; however, unlike the DDs, VD neurons retain these ventral synapses in the adult. VD and DD neurons also differ in that a transcriptional repressor (UNC-55) is expressed in the VD but not in the DD neurons, and this difference has been proposed to explain the disparity in their ability to undergo synaptic remodeling (Shan et al., 2005; Walthall, 1990; Walthall and Plunkett, 1995; Zhou and Walthall, 1998).
Prior studies suggested that VD neurons undergo ectopic remodeling in unc-55 mutants (Shan et al., 2005; Walthall and Plunkett, 1995; Zhou and Walthall, 1998). These studies showed that adult unc-55 mutant VD neurons lacked ventral axonal varicosities and ventral GFP-tagged synaptobrevin (SNB-1) puncta, consistent with the idea that ventral VD synapses in unc-55 had been eliminated due to ectopic expression of the DD neuron remodeling program (Shan et al., 2005; Walthall and Plunkett, 1995; Zhou and Walthall, 1998) (Fig. 1a). To confirm these results, we analyzed VD synapses in adult unc-55 mutants by both imaging and electrophysiology. To image these synapses, we expressed two GFP-tagged pre-synaptic proteins (UNC-57 endophilin and SNB-1 synaptobrevin) in the D neurons (using the unc-25 GAD promoter). In wild type adults, both UNC-57 and SNB-1 were expressed in a punctate pattern in the nerve cords, and these puncta were closely apposed to post-synaptic sites in body muscles (labeled with mCherry-tagged UNC-49 GABAA receptors) (Fig. S1a and data not shown). These ventral cord puncta likely correspond to VD NMJs, since the VDs are the only neurons that form ventral GABAergic synapses in adults (White et al., 1986). In unc-55 adults, the density of UNC-57 puncta in the ventral cord was significantly reduced compared to wild type controls (Fig. 1b–c). By contrast, pre-synaptic (UNC-57) and post-synaptic (UNC-49 GABAA) puncta densities were significantly increased in the dorsal cord of unc-55 adults (Fig. 1d–e, Fig. S1b–c). To assay the function of GABAergic synapses, we recorded inhibitory post-synaptic currents (IPSCs) from adult ventral and dorsal body muscles. In unc-55 mutants, ventral IPSC rates were significantly reduced (33 Hz wild type, 0.1 Hz unc-55, p <0.0001), whereas dorsal IPSC rates were significantly increased (33 Hz wild type, 65 Hz unc-55, p <0.0001 Student’s t test) (Fig. 1f–i). Thus, inactivation of unc-55 shifts GABAergic NMJs from ventral to dorsal muscles, as assessed by both imaging and electrophysiology. The rates and amplitudes of excitatory post-synaptic currents (EPSCs) were indistinguishable in wild type and unc-55 ventral body muscles (Fig. S1d–f), suggesting that cholinergic transmission was unaltered. Consequently, the loss of ventral synapses in unc-55 mutants was specific for GABAergic (i.e. VD) synapses.
Fig. 1. Ectopic VD remodeling in unc-55 mutants.
(a) Schematic illustrations of VD neuron NMJs (filled ovals) in wild type and unc-55 adults are shown. Dorsal is up and posterior is to the right in both illustrations; open circles are cell bodies. In wild type adults VD neurons retain ventral NMJs, whereas in unc-55 mutants ventral NMJs are eliminated and replaced with dorsal synapses. (b–e) Imaging of GABAergic NMJs (using the UNC-57::GFP marker). Representative images and summary data for ventral (b,c) and dorsal (d,e) GABAergic NMJs are shown for the indicated genotypes. (f–i) Representative traces and summary data for endogenous IPSCs recorded from adult ventral (f,g) and dorsal (h,i) muscles are shown for the indicated genotypes. Summary data for IPSC amplitudes are shown in Fig. S3g–h. See also figure S1.
The absence of ventral GABAergic NMJs in unc-55 adults could result from decreased formation or decreased retention of ventral NMJs. To assay ventral synapse formation, we imaged ventral GABAergic synapses in L2 larvae. We observed similar patterns of closely apposed pre-synaptic (UNC-57) and post-synaptic (UNC-49 GABAA receptor) puncta in the ventral cord of unc-55 and wild type L2 larvae, indicating that inactivation of unc-55 did not disrupt ventral synapse formation by VD neurons (Fig. S1g–j). These ventral NMJs in L2 animals were detected using transgenes driving UNC-57::GFP expression in both DDs and VDs (using the unc-25 promoter; Fig. S1g–h), and those driving expression in VD and AS neurons (using the unc-55 promoter; Fig. S1i–j). The AS neurons are cholinergic neurons that form dorsal NMJs (White et al., 1986); consequently, the ventral puncta labeled by both transgenes likely correspond to ventral VD synapses. Collectively, these results suggest that VD neurons initially form ventral synapses in unc-55 mutants but that these ventral synapses are subsequently removed by ectopic expression of the DD remodeling pathway, as proposed in the prior studies (Shan et al., 2005; Walthall and Plunkett, 1995; Zhou and Walthall, 1998).
The unc-55 gene encodes an orphan nuclear hormone receptor that is expressed in the VD but not the DD motor neurons (Zhou and Walthall, 1998). Several results suggest that UNC-55 acts as a transcriptional repressor. In VD neurons, UNC-55 represses expression of the proneuropeptide gene flp-13 (Shan et al., 2005; Melkman and Sengupta, 2005). Furthermore, UNC-55 orthologs in mammals (COUP-TF) and Drosophila (Sevenup) both function as transcriptional repressors (Pereira et al., 2000; Tsai and Tsai, 1997; Zelhof et al., 1995). These results lead to the hypothesis that UNC-55 inhibits remodeling of VD synapses by repressing expression of target genes required for remodeling (Zhou and Walthall, 1998).
UNC-55 inhibits hbl-1 expression in VD neurons
In Drosophila, Sevenup represses expression of the C2H2-type Zinc finger transcription factor hunchback (Kanai et al., 2005; Mettler et al., 2006). Prompted by the Sevenup data, we considered the possibility that the C. elegans hunchback ortholog (hbl-1) is an UNC-55 target (Fay et al., 1999). Consistent with this idea, the hbl-1 promoter contains four predicted UNC-55 binding sites, and similar binding sites were found in promoters of hbl-1 orthologs in C. remanei, C. briggsae, C. brenneri, and C. japonica (Fig. S2a). Furthermore, we found that expression of the hbl-1 mRNA (as assessed by qPCR) was increased in whole worm lysates isolated from unc-55 mutants, compared to wild type controls (14±1.7% increase, p < 0.01). Based on these initial results, we did several further experiments to test the idea that hbl-1 is an UNC-55 target.
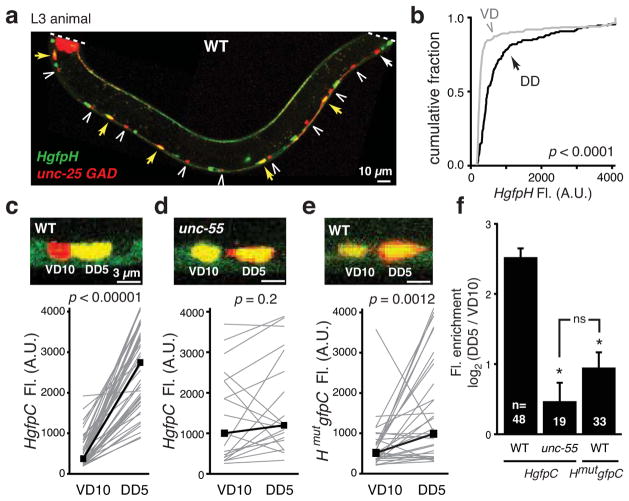
If hbl-1 is an UNC-55 target, then hbl-1 expression in DD neurons should be greater than that found in VDs. To test this idea, we analyzed expression of two GFP reporter constructs containing the hbl-1 promoter (Fig. 2). To distinguish between transcriptional and post-transcriptional regulation of hbl-1, the reporter constructs contain 3′ UTR sequences derived from either a control (unc-54 myosin) or the hbl-1 mRNA (HgfpC and HgfpH, respectively). VD and DD neurons were identified using a GABA marker (mCherry expressed by the unc-25 GAD promoter) and were distinguished based on the position and morphology of their cell bodies (detailed in the methods). We compared hbl-1 reporter expression in VD10 and DD5, which have adjacent cell bodies in the ventral cord. For both reporters, DD5 expression was significantly higher than that observed in VD10 (DD5/VD10 fluorescence ratios: HgfpC 6.6±0.8, p <0.0001 paired Student’s t test; HgfpH 3.6±0.7, p <0.05 paired Student’s t test; Fig. 2c). Similar results were observed when reporter expression was compared in all DD and VD neurons (DD/VD fluorescence ratios: HgfpC 5.6 ±0.5, p <0.0001 paired Student’s t test; HgfpH 2.6±0.4, p <0.005 paired Student’s t test; Fig. 2a–b, S2b,d). These results indicate that the hbl-1 promoter is expressed at significantly higher levels in DD neurons than in VD neurons.
Fig. 2. UNC-55 COUP-TF represses hbl-1 expression in VD neurons.
(a–b) A representative image and summary data of hbl-1 (HgfpH, green) and GAD (red) reporter expression in wild type L3 animals. Yellow arrows indicate DD cell bodies expressing both markers, white arrows indicate DD cell bodies lacking HgfpH expression, and carrots indicate VD cell bodies. HgfpH expression was significantly lower in VD than DD neurons (p <10−5 by Kolmogorov-Smirnov test; 150 DD (black) and 260 VD (gray) cells were analyzed). (c–f) HgfpC (c,d) or HmutgfpC (e) transcriptional reporter expression (green) is compared for adjacent VD10 and DD5 neurons in wild type (c,e) and unc-55 (d) mutant animals. Gray lines connect VD10 and DD5 cells in the same animal, black lines connect median values (p-values by paired Student’s t test). Average log2 of the ratio of DD5 to VD10 fluorescence for HgfpC and HmutgfpC was plotted in (f), and n = number of animals analyzed (*, p<10−5 difference from WT; ns, p=0.2; by Student’s t test). Error bars indicate SEM. See also figure S2.
The decreased hbl-1 reporter expression in VD neurons could result from UNC-55 mediated repression of the hbl-1 promoter. To test this possibility, we analyzed expression of the HgfpC reporter in unc-55 mutants. HgfpC expression in VD neurons was significantly increased in unc-55 mutants (197% wild type levels, p<0.001 Student’s t test), indicating increased transcription of the hbl-1 promoter in unc-55 mutant VD neurons (Fig. 2d, S2b–d). The magnitude of the increased HgfpC expression differed in individual VD neurons. For VD10, HgfpC expression in unc-55 mutants rose to the same level observed in DD5 neurons (Fig. 2d); however, in most cases, HgfpC expression in unc-55 mutant VD neurons remained significantly lower than that observed in DD neurons (DD/VD fluorescence ratio in unc-55: HgfpC 2.3±0.4, p <0.001 Student’s t test; Fig. S2c–d). By contrast, HgfpC expression in DDs did not increase in unc-55 mutants and instead was modestly decreased (Fig. 2d, S2d). This is unlikely to be a direct effect of UNC-55 on the hbl-1 promoter because unc-55 is not expressed in DD neurons (Zhou and Walthall, 1998). Taken together, these data support the idea that UNC-55 inhibits expression of the hbl-1 promoter in VD neurons and that hbl-1 expression in D neurons is likely regulated by additional factors beyond UNC-55.
In Drosophila, the UNC-55 ortholog (Sevenup) represses Hunchback (Hb) transcription (Kanai et al., 2005; Mettler et al., 2006). As in Drosophila, the C. elegans hbl-1 promoter contains four predicted UNC-55 binding sites, suggesting the hbl-1 could be a direct target for UNC-55 repression. To test this idea, we mutated the UNC-55 binding sites in the hbl-1 promoter, and assayed its expression pattern. The mutant hbl-1 promoter (HmutgfpC) had a significantly reduced DD5/VD10 expression ratio (HgfpC 6.6 ±0.8; HmutgfpC 2.7 ±0.3, p<0.0001 Student’s t test) (Fig. 2e), which was not significantly different from the ratio observed for the wild type reporter (HgfpC) in unc-55 mutants (1.8 ±0.3, p = 0.17, Student’s t test) (Fig. 2f). Thus, the UNC-55 binding sites are required for differential expression of the hbl-1 promoter in VD and DD neurons.
hbl-1 is required for ectopic remodeling of VD synapses in unc-55 mutants
If UNC-55 repression of hbl-1 prevents VD remodeling, we would expect that mutations reducing hbl-1 activity would diminish ectopic remodeling of VD synapses in unc-55 mutants. In this scenario, unc-55; hbl-1 double mutant adults would have significantly more ventral GABAergic synapses and fewer dorsal synapses than unc-55 single mutants. We did several experiments to test this idea. For these experiments, we utilized the hbl-1(mg285) mutation, which significantly reduces (but does not eliminate) hbl-1 gene function (Lin et al., 2003). It was not possible to analyze hbl-1 null mutations as these mutants are not viable (Lin et al., 2003; Roush and Slack, 2009).
We imaged both ventral and dorsal GABAergic synapses with the UNC-57::GFP pre-synaptic marker (expressed in both DD and VD neurons). The unc-55; hbl-1 double mutant adults had a significant increase in ventral UNC-57 puncta density and a corresponding decrease in dorsal UNC-57 puncta density compared to unc-55 single mutants (Fig. 3a–d). Thus, inactivation of hbl-1 in unc-55 mutants shifts GABAergic NMJs from dorsal to ventral muscles. This shift could be caused by reduced remodeling of either DD or VD synapses in unc-55; hbl-1 double mutants. We did two experiments to distinguish between these possibilities. First, ventral and dorsal UNC-57 puncta density, and ventral and dorsal IPSC rates were all unaltered in hbl-1 single mutants, suggesting that DD remodeling was successfully completed in hbl-1 adults (Fig. 3a–h). Second, we selectively labeled DD synapses with UNC-57::GFP (using the flp-13 promoter). Using this DD specific synaptic marker, we did not detect any ventral synapses in hbl-1 adults (data not shown). Consequently, defects in DD remodeling are unlikely to explain the dorsal to ventral shift of GABA synapses in unc-55; hbl-1 double mutants. Instead, these results support the idea that hbl-1 mutations decreased ectopic VD remodeling in unc-55; hbl-1 double mutants.
Fig. 3. Ectopic VD remodeling in unc-55 mutants requires HBL-1.
(a–d) Imaging of GABAergic NMJs (using the UNC-57::GFP marker). Representative images and summary data for ventral (a,b) and dorsal (c,d) GABAergic NMJs are shown for the indicated genotypes. (e–h) Representative traces and summary data for endogenous IPSCs recorded from adult ventral (e,f) and dorsal (g,h) muscles are shown for the indicated genotypes. Summary data for IPSC amplitudes are shown in Fig. S3g–h. (i) The ventral coiling phenotype of unc-55 adults during backward locomotion is shown and quantified. The unc-55 coiling defect was partially suppressed by an hbl-1 mutation, and was restored by transgenes containing either the hbl-1 cosmid or an hbl-1 cDNA expressed by a GABA promoter (unc-25 GAD) (D neuron rescue). A single transgenic line is shown for each rescue; similar results were obtained with multiple independent transgenic lines (Fig. S3e). Error bars indicate SEM. Significant differences (p <0.01) are indicated as follows: *, significantly different from WT; #, significantly different from unc-55 single mutants; ##, significantly different from unc-55; hbl-1 double mutants. The number of animals analyzed (a–h) or replicate behavioral assays (20 animals/assay; i) is indicated for each genotype. See also figure S3.
To assay the function of the ventral VD synapses, we recorded IPSCs from ventral and dorsal body muscles. We found that, compared to unc-55 single mutants, unc-55; hbl-1 double mutants had a significantly higher ventral IPSC rate and a significantly lower dorsal IPSC rate (Fig. 3e–h), both indicating decreased VD remodeling in double mutants. In both dorsal and ventral recordings, unc-55 IPSC defects were only partially suppressed in unc-55; hbl-1 double mutants. The dorsal IPSC rate observed in unc-55; hbl-1 double mutants remained significantly higher than that observed in hbl-1 single mutants (Fig. 3g–h). By contrast, the rates and amplitudes of excitatory post-synaptic currents (EPSCs) in ventral body muscles (Fig. S3b–d) were unaltered in both hbl-1 single mutants and hbl-1; unc-55 double mutants, suggesting that cholinergic transmission was unaffected. The restoration of ventral IPSCs in double mutants was partially penetrant, i.e. the increased ventral IPSC rate was only observed in a subset of the double mutant animals (14 out of 43 recordings). Double mutant recordings fell into either of two categories, having ventral IPSC rates similar to unc-55 or to wild type, while none had intermediate values (Fig. 3e–f). Incomplete penetrance of ventral remodeling in double mutants was also observed by imaging. In unc-55; hbl-1 double mutants, we observed patches of the ventral nerve cord that contained an approximately normal number of synapses, while other regions totally lacked synapses (data not shown). A transgene expressing hbl-1 in the VD and DD neurons of unc-55; hbl-1 double mutants (using the unc-25 promoter) decreased the ventral IPSC rate to that observed in unc-55 single mutants (Fig. 3f) but did not rescue the non-neuronal hbl-1 defects (Fig. S3a). These results suggest that HBL-1 acts in VD neurons to promote ectopic remodeling.
To further document the functional integrity of the ventral VD synapses, we analyzed the locomotion behavior of unc-55; hbl-1 double mutants. A prior study showed that ectopic remodeling of VD synapses in unc-55 mutants was accompanied by a locomotion defect (Zhou and Walthall, 1998). During backward movement, unc-55 mutants assume a ventrally coiled body posture, presumably due to the absence of inhibitory input to the ventral body muscles (Fig. 3i). This unc-55 coiling defect was significantly reduced (but not eliminated) in unc-55; hbl-1 double mutants (Fig. 3i). The coiling defect was restored by transgenes driving hbl-1 expression in the D neurons (using either the unc-25 GAD or the unc-30 promoter) in unc-55; hbl-1 double mutants (Fig. 3i, S3e), as would be predicted if HBL-1 acts in VD neurons to promote remodeling. Thus, the imaging, electrophysiology, and behavioral assays all support the idea that hbl-1 is a functionally important UNC-55 target whose expression promotes ectopic remodeling of VD synapses in unc-55 mutants.
The partial suppression and incomplete penetrance observed in the unc-55; hbl-1 double mutants indicate that the hbl-1(mg285) mutation did not completely abolish remodeling of VD synapses. The persistent VD remodeling observed in double mutants could reflect residual hbl-1 activity in hbl-1(mg285) mutants, or the activity of other UNC-55 target genes (Lin et al., 2003). Consistent with the latter idea, transgenic expression of hbl-1 in DD and VD neurons (with the unc-25 promoter) was not sufficient to cause ectopic remodeling of VD synapses (Fig. S3f). Thus, hbl-1 is unlikely to be the only UNC-55 target involved in D neuron remodeling.
DD remodeling occurs during a precise time window and is patterned spatially
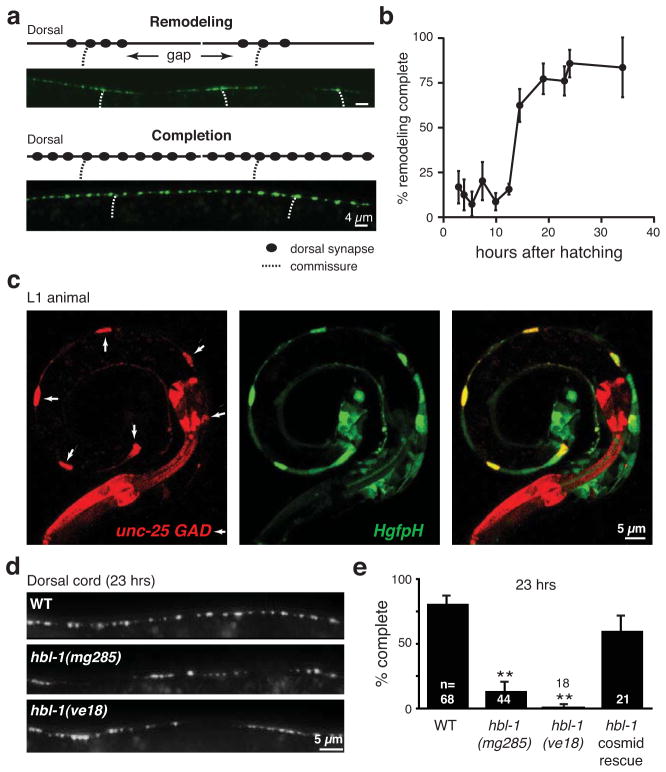
Thus far, our results show that hbl-1 promotes ectopic remodeling of unc-55 mutant VD neurons but that hbl-1 expression alone is not sufficient to cause VD remodeling. We next analyzed DD remodeling, which occurs in wild type animals (Walthall, 1990; White et al., 1978). Prior to hatching, DD neurons form ventral NMJs, which can be identified as ventral UNC-57::GFP puncta. During the L1 stage, these ventral DD synapses are eliminated and new dorsal synapses are formed (visualized as dorsal UNC-57 or RAB-3 puncta; Fig. 4a and S4a). The UNC-57::GFP transgene is expressed in both DD and VD neurons; consequently, we were unable to analyze loss of ventral DD synapses, due to the confounding signal of the nascent ventral VD synapses. For this reason, we restricted our analysis to formation of new UNC-57 puncta in dorsal cord DD axons during the L1. Using this assay, we followed the time course of DD remodeling. The entire DD remodeling process occurred in a discrete time window during the late L1 and early L2 stage (from 12–19 hours post-hatching; Fig. 4b), consistent with prior studies (Park et al., 2011; Hallam and Jin, 1998). The newly formed dorsal DD synapses occur in a stereotyped spatial pattern, where dorsal cord UNC-57 puncta adjacent to the commissures form first, while puncta in more distal axon segments form later (Fig. 4a). These results suggest that formation of dorsal DD synapses during remodeling occurs in a proximal-to-distal spatial pattern.
Fig. 4. DD remodeling is delayed in hbl-1 mutants.
(a) DD remodeling was visualized by dorsal synapse formation with the UNC-57::GFP marker. Representative images of the dorsal cord during remodeling (above) and after completion (below) are shown. During remodeling, DD neurons form en passant synapses with the dorsal muscle. DD neuron commissures are indicated by dotted lines. (b) Summary data illustrating the time course of DD neuron remodeling is shown. 15–30 animals were analyzed for all time points except 12 hrs (where n= 160 animals). (c) The HgfpH reporter (green) is expressed in DD neurons (identified with the GAD reporter, red) during the L1 when remodeling is occurring. A representative image of a wild type L1 larva is shown. Arrows indicate the six DD cell bodies. (d and e) DD remodeling is delayed in hbl-1 mutants. Representative images of dorsal DD NMJs (d), and summary data (e) for completion of DD remodeling are shown at 23 hours post-hatching. The majority of wild-type animals have completed DD remodeling, whereas significantly fewer hbl-1 mutants have finished this process (**, p <0.0001 Chi square test). This delay was rescued by a transgene containing the hbl-1 cosmid. Error bars indicate SEM. See also figure S4.
DD remodeling is delayed in hbl-1 mutants
Our analysis of unc-55 mutants suggests that hbl-1 expression promotes ectopic VD remodeling. Given these results, we wondered whether hbl-1 also plays a role in DD remodeling. Consistent with this idea, the HgfpH and HgfpC reporters were expressed in six GABAergic DD neurons of wild type L1 larvae, before the VD neurons are born (Fig. 4c, and data not shown). Thus, hbl-1 is likely to be expressed in the DD neurons during the remodeling period.
We next asked if HBL-1 is required for DD remodeling. At 12 hours post-hatching, DD remodeling had been initiated in both wild type and hbl-1 mutants (data not shown), implying that onset of remodeling had not been altered. By contrast, at 23 hours post-hatching, nearly all wild type animals (81 ±5%) had completed remodeling, whereas significantly fewer hbl-1 mutants (14 ±5%, p<0.0001 Student’s t test) had completed this process (Fig. 4d–e and S4a). Similar delays were observed in strains containing two independent hbl-1 alleles (mg285 and ve18), both of which reduce but do not eliminate hbl-1 gene activity (Abrahante et al., 2003; Lin et al., 2003; Roush and Slack, 2009). The hbl-1 delayed remodeling defect was rescued by a transgene containing the F13D11 cosmid (which spans the hbl-1 gene; Fig. 4e). The effect of hbl-1 was not specific to the UNC-57::GFP marker because similar delays in DD remodeling were detected using a second synaptic marker (mCherry::RAB-3; Fig. S4a). Although remodeling was delayed, hbl-1 mutants eventually completed DD remodeling, as hbl-1 adults had normal dorsal and ventral NMJs as assessed by both imaging and electrophysiology (Fig. 3a–h). This persistent remodeling activity could reflect residual gene activity in hbl-1(mg285) mutants or residual expression of other remodeling factors.
Because hbl-1 is a heterochronic gene, the delayed DD remodeling in hbl-1 mutants could be caused by a generalized delay in larval development. This seems unlikely because inactivating hbl-1 causes several aspects of hypodermal development to occur earlier (including seam cell fusions, alae formation, and division of vulva precursor cells), whereas DD remodeling is delayed (Abrahante et al., 2003; Grosshans et al., 2005; Lin et al., 2003; Nolde et al., 2007; Reinhart et al., 2000). These hbl-1 hypodermal defects occur later in development, during the L2. Therefore, we did several additional experiments to control for changes in the timing of L1 development in hbl-1 mutants. We used two developmental landmarks during the L1: the onset of expression of the mlt-10 gene (which occurs at 11–14 hours post-hatching), and the Pn.ap neuroblast (hereafter referred to as the AS/VD neuroblast) cell division (which occurs at 12.5–14 hours post-hatching) (Frand et al., 2005; Sulston, 1976). The AS/VD cell division was monitored with a GFP reporter expressed in its daughter cells (the VD and AS neurons) using the unc-55 promoter. Although completion of DD remodeling was delayed by at least 20 hours in hbl-1 mutants, corresponding delays were not observed for the onset of mlt-10 expression or for the timing of the AS/VD cell division (Fig. S4b–d). Thus, a generalized delay in the timing of L1 development is unlikely to explain the hbl-1 mutant delay in DD remodeling.
DD remodeling occurs earlier in mir-84 mutants
In the hypodermis, hbl-1 expression is negatively regulated by the let-7 family of microRNAs (Abrahante et al., 2003; Lin et al., 2003; Nolde et al., 2007; Abbott et al., 2005; Roush and Slack, 2008). The 3′ UTR of the hbl-1 mRNA contains binding sites for three let-7 paralogs (let-7, mir-48, and mir-84) (Roush and Slack, 2008). Prior studies showed that mature miR-84 is expressed in the early L1, suggesting that let-7 microRNAs could regulate hbl-1 expression in DD neurons during the remodeling process (Abbott et al., 2005; Esquela-Kerscher et al., 2005). To test this idea, we analyzed expression of the HgfpH reporter in mir-84 mutants (Fig. 5a–b). In the L1, HgfpH expression was significantly increased in mir-84 mutant DD neurons compared to wild type controls (7.5 fold increase in median, p <0.0001 Kolmogorov-Smirnov test; Fig. 5a–b). By contrast, the mir-84 mutation did not significantly change expression of the HgfpC reporter, which lacks the hbl-1 3′UTR (Fig. 5c). These results suggest that miR-84 regulates hbl-1 expression in DD neurons when remodeling is occurring.
Fig. 5. The microRNA miR-84 regulates hbl-1 expression and the timing of remodeling.
(a–b) Representative images (a) and summary data (b) are shown for HgfpH expression (green) in DD neurons (labeled with the GAD reporter, red, and indicated by arrows) of L1 larvae. In mir-84 mutants, HgfpH expression was significantly increased (** p <0.0001 by Kolmogorov-Smirnov test). (c) Summary data are shown comparing the fluorescent intensity of the HgfpC reporter in DD neurons of wild type and mir-84(tm1304) mutants; no significant difference was observed (p =0.1 Kolmogorov-Smirnov test). 54 wild-type and 118 mir-84 DD neurons were analyzed for median HgfpH expression (b); and 233 wild-type and 379 mir-84 DD neurons were analyzed for HgfpC expression (c). (d–e) DD remodeling occurs earlier in mir-84 mutants. Representative images (d) and summary data (e) are shown for dorsal DD NMJs at 11 hours post-hatching. Remodeling was completed significantly earlier in mir-84 mutants (*, p <0.01 Chi square test). (f–g) The impact of mir-84 on remodeling was eliminated in hbl-1; mir-84 double mutants. Representative images (f) and summary data (g) are shown for dorsal DD NMJs at 19 hours post-hatching. The extent of remodeling in hbl-1 single mutants and hbl-1; mir-84 double mutants were not significantly different. See also figure S5.
If miR-84 inhibits hbl-1 expression in DD neurons during the remodeling period, we would expect that the timing of remodeling would be altered in mir-84 mutants. Indeed, at 11 hours after hatching, a significantly larger fraction of mir-84 mutants had completed remodeling than was observed in wild type controls (Fig. 5d–e). These results suggest that completion of DD remodeling occurs precociously in mir-84 mutants. Corresponding changes in the timing of mlt-10 expression and of the AS/VD cell division were not observed in mir-84 mutants (Fig. S5), suggesting that global changes in the timing of L1 development are unlikely to explain the mir-84 remodeling defect. The earlier remodeling in mir-84 mutants could be caused by increased hbl-1 expression in DD neurons. Consistent with this idea, the effect of the mir-84 mutation on remodeling was eliminated in hbl-1; mir-84 double mutants (Fig. 5f–g). These results suggest that mutations increasing and decreasing HBL-1 activity (mir-84 and hbl-1, respectively) produce opposite shifts in the timing of DD plasticity.
Changes in GABA release do not alter the timing of DD plasticity
In mammals, changes in GABA transmission regulates ocular dominance plasticity as well as other aspects of synapse development (Hensch, 2004; Chattopadhyaya et al., 2007). However, GABA release is unlikely to be required for DD plasticity, as a prior study showed that DD remodeling was unaltered in unc-25 mutant adults (which lack the GABA biosynthetic enzyme GAD) (Jin et al., 1999). To confirm these results, we analyzed unc-47 mutants (which lack the vesicular GABA transporter VGAT) and unc-25 GAD mutants for DD remodeling defects in L1 and L2 larvae. We observed normal or slight changes in the timing of DD remodeling in either GABA defective mutant (Fig. S6a–b), indicating that GABA transmission does not play an important role in the timing of DD remodeling.
Circuit activity regulates hbl-1 expression and the timing of DD plasticity
Since synaptic refinement is often regulated by circuit activity, we wondered if changes in activity would also alter the timing of DD remodeling (Hua and Smith, 2004; Sanes and Lichtman, 1999). To test this idea, we analyzed mutants that have altered circuit activity. For this analysis, we used mutations that either block or exaggerate synaptic transmission. Mutants lacking UNC-13 and UNC-18 have profound defects in synaptic vesicle docking and priming, which result in dramatically reduced rates of synaptic transmission (3% and 10% of wild type rates, respectively) (Richmond et al., 1999; Weimer et al., 2003; McEwen et al., 2006). By contrast, mutations inactivating tom-1 Tomosyn and slo-1 BK channels exaggerate synaptic transmission. In tom-1 mutants, the pool of fusion competent (i.e. primed) synaptic vesicles is increased (Gracheva et al., 2006; McEwen et al., 2006). In slo-1 mutants, repolarization of nerve terminals is delayed, leading to prolonged neurotransmitter release (Wang et al., 2001).
First, we compared expression of the hbl-1 promoter in these activity mutants. Expression of the HgfpC reporter in DD neurons was significantly decreased in unc-13 mutants (Fig. 6a–b), whereas increased HgfpC expression was observed in tom-1 mutants (Fig. 6c–d). Thus, decreased and increased circuit activity were accompanied by corresponding changes in hbl-1 promoter expression in DD neurons.
Fig. 6. Circuit activity regulates HBL-1 expression to determine the timing of DD neuron plasticity.
(a–d) Representative images (a,c) and summary data (b,d) are shown for hbl-1 expression (HgfpC, green) in DD neurons (labeled with the GAD reporter, red, indicated by arrows). HgfpC expression significantly decreased in unc-13 mutants (a–b) and increased in tom-1 mutants (c–d; ** p <0.0001 Kolmogorov-Smirnov test; 72 wild type and 149 unc-13 L2 DD neurons; 64 wild type and 179 tom-1 L1 DD neurons). Expression of HgfpC in DD neurons was analyzed at different times in unc-13 and tom-1 mutants because the remodeling defects observed in these mutants occurred at different times. (e–h) DD remodeling in unc-13 and unc-18 mutants (e) or in tom-1 and slo-1 mutants (g). Representative images of dorsal GABAergic NMJs (e,g), and summary data for completion of remodeling (f,h) in late L3 animals (e,f) or at 11 hours post-hatching (g,h). (i) Summary data for completion of DD remodeling at 20 hours after hatching shows that the impact of slo-1 and tom-1 on remodeling was eliminated in double mutants with hbl-1. (* : significantly different than wild-type, p < 0.001, Chi squared test). Error bars indicate SEM, numbers indicate number of animals analyzed. See also figure S6.
We next asked if circuit activity alters the timing of DD plasticity. The overall rate of larval development was significantly delayed in both unc-13 and unc-18 mutants, presumably due to decreased feeding. To control for this general developmental delay, we synchronized animals at a specific stage of L3 development, defined by the dorsal turn of the gonad arms. In these late L3 larvae, unc-13 and unc-18 mutants had significantly delayed DD remodeling compared to wild type L3 larvae (Fig. 6e–f). By contrast, remodeling occurred significantly earlier in tom-1 and slo-1 mutants than in wild type controls (Fig. 6g–h). This earlier remodeling phenotype cannot be explained by a general shift in developmental timing, as neither the tom-1 nor slo-1 mutants had corresponding changes in the timing of other L1-to-L2 developmental events (Fig. S6c–d). Thus, decreased and increased synaptic activity were accompanied by corresponding changes in hbl-1 promoter expression in DD neurons, and corresponding shifts in the timing of DD plasticity. The earlier remodeling phenotypes observed in tom-1 and slo-1 single mutants were eliminated in double mutants lacking hbl-1 (Fig. 6i), suggesting that changes in hbl-1 activity are required for the activity-induced shifts in the timing of DD plasticity.
Discussion
To investigate the genetic mechanisms that pattern synaptic plasticity, we analyzed the developmentally programmed remodeling of D-type motor neuron synapses in C. elegans. Our results, together with prior studies, show that DD plasticity is extensively regulated. First, DD synapses are remodeled during a precise time window (12–19 hours post-hatching). Second, circuit activity governs the timing of remodeling. Third, plasticity is restricted to a specific cell type: the earlier born DD neurons undergo this plasticity while the later born VD neurons do not. And fourth, remodeling is patterned spatially, with new DD synapses forming in a proximal to distal order. Thus, DD plasticity shares many features with other examples of developmental plasticity (including critical period plasticity in mammals). Given these similarities, characterizing the molecular mechanisms that pattern DD remodeling may provide insights into the mechanisms underlying circuit refinement elsewhere.
A conserved role for heterochronic genes in circuit development
In both worms and flies, the timing of many aspects of development is controlled by transcriptional cascades that confer temporal cell fates. In worms, these cascades are generically referred to as heterochronic pathways. A prior study showed that LIN-14, a heterochronic transcription factor, acts cell autonomously in DD neurons, where it determines when remodeling is initiated (Hallam and Jin, 1998). Here we show that a second heterochronic gene (hbl-1) also acts cell autonomously to pattern remodeling. Several aspects of these results are significant. First, unlike lin-14, hbl-1 orthologs are found in other organisms and Drosophila Hunchback plays an analogous role in regulating temporal cell fates in neuroblast lineages (Mettler et al., 2006; Kanai et al., 2005). Thus, our results strongly suggest that heterochronic genes represent a conserved mechanism for patterning the timing of circuit development. Second, different heterochronic genes control different aspects of plasticity. LIN-14 determines when DD remodeling is initiated while HBL-1 determines when remodeling is completed. Third, a heterochronic gene can have opposite effects on developmental timing in different tissues. Inactivating hbl-1 caused delayed DD plasticity whereas hypodermal fates occurred precociously (Abrahante et al., 2003; Lin et al., 2003). By contrast, inactivating lin-14 caused precocious expression of both DD plasticity and hypodermal development (Hallam and Jin, 1998; Ambros and Horvitz, 1987). Fourth, increased and decreased HBL-1 expression produce opposite shifts in the timing of DD plasticity. Identifying genes that mutate to opposite phenotypes has historically been utilized in developmental genetics as a criterion to identify the key regulatory elements in a process. Thus, our results identify HBL-1 as a critical genetic determinant patterning DD plasticity.
The role of UNC-55 COUP-TF in circuit development
During development, maturing circuits are modified by the addition of newly born neurons, and by refinement of connectivity. We propose that the UNC-55/COUP-TF family of transcriptional repressors plays an important role in both of these aspects of circuit development. In C. elegans, synaptic remodeling is restricted to the earlier born DD neurons because UNC-55 COUP-TF represses hbl-1 expression in the later born VD neurons. Inactivating UNC-55 orthologs in other organisms alters the timing of other aspects of neural development. In Drosophila, Sevenup repression of Hunchback allows neuroblast daughters to adopt later cell fates (Mettler et al., 2006; Kanai et al., 2005). Similarly, knocking down both mouse UNC-55 orthologs (COUP-TF1 and COUP-TFII) prolongs the generation of early-born neurons at the expense of later cell types (Naka et al., 2008). Collectively, these results suggest that UNC-55 orchestrates how newly born neurons are integrated into circuits, and the capacity of developing circuits to undergo plasticity. In this respect, it is intriguing that a mouse UNC-55 ortholog (COUP-TFII) is expressed in several classes of GABAergic cortical interneurons (Armentano et al., 2007; Kanatani et al., 2008; Tripodi et al., 2004). Like UNC-55, COUP-TFII is selectively expressed in a sub-population of interneurons that have later birth dates (Zhou et al., 2001). We speculate that COUP-TFII expressing interneurons (like the VDs) will have a more limited capacity to undergo synaptic refinement compared to interneurons that are born earlier.
What role does HBL-1 play in synaptic remodeling?
HBL-1 acts cell autonomously to promote ectopic synapse remodeling of VD neurons in unc-55 mutants. We were unable to directly test if HBL-1 also acts cell autonomously for DD remodeling because the hbl-1 rescuing transgenes silence expression of the synaptic markers utilized to score remodeling (data not shown). Nonetheless, several results support the idea that HBL-1 also acts autonomously for DD remodeling. The hbl-1 promoter is expressed in DD neurons during the remodeling period. Increased hbl-1 expression in DD neurons (in mir-84 and tom-1 mutants), and decreased expression (in unc-13 mutants) cause opposite shifts in the timing of DD plasticity. These results favor the idea that HBL-1 acts autonomously in both VD and DD neurons.
HBL-1 expression could reprogram VD neurons to adopt the DD cell fate, thereby causing ectopic expression of the remodeling program. This scenario seems unlikely because bidirectional changes in hbl-1 expression produce corresponding shifts in the timing of DD plasticity. If HBL-1 were inducing the DD cell fate, we would not expect HBL-1 expression to bidirectionally alter the timing of DD remodeling. HBL-1 activity could accelerate DD remodeling by regulating expression of factors that directly mediate synapse elimination and formation. Finally, HBL-1 could be part of a timing mechanism that dictates when remodeling occurs. The effects of UNC-55 orthologs (COUP-TFs and Sevenup) and an HBL-1 ortholog (Hb) on developmental timing in flies and mice provide support for HBL-1 function as part of a conserved timing mechanism. Ultimately, identifying the relevant HBL-1 transcriptional targets will be required to distinguish between these models.
microRNA control of circuit refinement
Many aspects of early neuronal development are regulated by microRNAs (e.g. neuronal fate determination, neural tube closure, and mitotic exit) (Fineberg et al., 2009; Fiore et al., 2008). microRNAs have also been implicated in the functional plasticity of mature circuits (Fineberg et al., 2009; Fiore et al., 2008; Simon et al., 2008). Our results show that microRNAs play an important role in restricting when plasticity occurs during development. In particular, we show that miR-84 regulates the timing of DD plasticity, and that it does so by regulating hbl-1. The Drosophila microRNA Let-7 plays a similar role in dictating the timing of NMJ growth during larval development (Sokol et al., 2008; Caygill and Johnston, 2008). It is interesting that Let-7 and miR-84 are paralogs that bind to related seed sequences in target mRNAs. Thus, Let-7 microRNAs (and their targets) represent an ancient mechanism for determining the timing of circuit development.
HBL-1 mediates the effects of activity on circuit refinement
Perhaps the most surprising aspect of our results is that the timing of DD plasticity is regulated by activity. Mutations increasing and decreasing circuit activity had opposite effects on the timing of DD plasticity. These results are significant because they suggest that DD plasticity (and other forms of genetically programmed plasticity) and activity-dependent circuit refinement are not necessarily distinct processes, and may utilize similar genetic pathways. In this context, it is noteworthy that all of the genetic factors we identify (UNC-55/COUP-TF, HBL-1, and miR-84) are conserved in vertebrates, and vertebrate orthologs are all expressed in the CNS. It will be interesting to see if these molecules also play a role in refining vertebrate circuits. Several forms of plasticity are triggered by changes in the activity of the post-synaptic targets. Post-synaptic activity is unlikely to play a role in this case as mutations blocking GABA transmission had no effect on the timing DD plasticity.
Our results also identify HBL-1 as a molecular mediator of activity’s effects on DD plasticity. HBL-1 expression is restricted to a specific set of neuronal cell types, and thus could confer activity dependence in a cell and circuit specific manner. By contrast, it is unclear how the general activity-induced genes that are implicated in ocular dominance plasticity (e.g. CREB and BDNF) could mediate refinement in a cell and temporally specified manner. This result also demonstrates that the effect of hbl-1 on developmental timing is regulated by the nervous system. It will be interesting to see if the nervous system also controls other heterochronic pathways.
In summary, we show that patterning of DD plasticity is achieved by the convergence of multiple regulatory pathways on hbl-1. Convergent regulation of hbl-1 defines a cell intrinsic pathway that confers cell and temporal specificity and activity-dependence on this form of circuit refinement.
Experimental Procedures
Strains were maintained at 20°C using standard protocols, on lawns of OP50 for imaging and behavior, and on HB101 for elecrophysiology. Strains are listed in the supplementary material.
qPCR
Whole worm lysates of synchronized L3 animals were prepared by Trizol extraction (Invitrogen). Three biological replicates of wild type and unc-55(e1170) samples were collected on different days. cDNA library construction, primer validation, and quantitative RT-PCR were carried out according to standard protocols. Changes in hbl-1 mRNA levels, were normalized relative to rpl-32 levels.
HBL-1 Reporters
The hbl-1 reporters are similar to those used previously (Fay et al., 1999). These constructs contain 7.7 kb, including 6.4 kb upstream and 1.3 kb of exons 1–4. These constructs encode a protein containing the first 133 amino acids of hbl-1 fused to GFP-PEST, along with 1kb of the hbl-1 3′UTR (HgfpH) or the control unc-54 3′UTR (HgfpC). In HmutgfpC, the four 6bp UNC-55 binding sites in the hbl-1 promoter were replaced with BamHI sites. Images were collected on a laser-scanning Olympus FV1000 confocal microscope. To quantify GFP fluorescence, areas of interest were drawn around DD or VD neuron cell bodies (identified by the unc-25 GAD mCherry signal) in a single plane through the center of the cell bodies, and median GFP fluorescence was determined for that plane. DD neurons were distinguished from VD neurons based on anterior-posterior position in the ventral nerve cord, cell body size, and morphology (White et al., 1986). The ratio of GFP signal in DD5 to VD10 was determined in each animal, log2 transformed, then averaged for all animals of a genotype. To enhance our ability to detect increases in hbl-1 expression in mir-84 and tom-1 mutants, we used an HgfpH transgene (nuIs427) that has a low baseline expression level.
Electrophysiology
Electrophysiology was done on ventral and dorsal body muscles of dissected C. elegans adults as described, using 1 mM Ca2+ in the external saline solution (McEwen et al., 2006; Richmond and Jorgensen, 1999; Simon et al., 2008; Vashlishan et al., 2008). Ventral IPSC rates in unc-55; hbl-1 could not be analyzed by Student’s t test because many recordings totally lacked IPSCs; consequently, chi-squared tests were used to compare the number of recordings with and without IPSCs for unc-55 single and double mutants.
Coiling Behavior
Young adult animals were assayed for the reverse coiling behavioral phenotype as described (Walthall and Plunkett, 1995). Animals were scored as either fully coiling or not, with partial coiling or failed coiling attempts scored as not coiling.
in vivo Fluorescence Microscopy and Image Analysis
Dorsal and ventral nerve cord synapses were imaged in animals expressing GFP-tagged UNC-57/Endophilin or mCherry-tagged RAB-3 (nuIs279) using either a Zeiss Axioskop widefield epifluorescence microscope (using an Olympus PlanAPO 100x 1.4 NA objective) or an Olympus FV1000 confocal microscope (using an Olympus PlanAPO 60x 1.45 NA). Pre-synaptic markers were expressed in GABAergic neurons using the unc-25 promoter (all figures except S1i–j), or in the VD and AS neurons using the unc-55 promoter (Fig S1i–j). Animals were immobilized with 30mg/mL 2,3-butanedione monoxime (Sigma). Image stacks were captured, and maximum intensity projections were obtained using Metamorph 7.1 software (Molecular Devices). Line scans of ventral or dorsal cord fluorescence were analyzed in Igor Pro (WaveMetrics) using custom designed software as described (Burbea et al., 2002; Dittman and Kaplan, 2006).
DD Remodeling
The timing of DD remodeling was analyzed in synchronized animals. Briefly, plates containing isolated embryos were incubated at 20°C for 30 minutes and newly hatched L1 larvae were picked to fresh plates. DD remodeling was analyzed in resulting cohorts at defined times after hatching. Each time point comprises 1 hour of development (due to the time required for sample preparation and image acquisition). The extent of remodeling was quantified by counting the number of asynaptic gaps in the dorsal cord, using the GFP-tagged synaptic marker UNC-57 Endophilin expressed in the D neurons by the unc-25 GAD promoter, unless noted otherwise. Each animal can have 0 to 5 asynaptic gaps (between the 6 DD neurons). Wild type adults often have one gap (opposite the vulva opening); consequently, animals with zero or one gap were scored as completely remodeled. Images were scored in random order by an investigator unaware of the animal’s genotype.
Supplementary Material
Acknowledgments
We thank members of the Kaplan lab for helpful discussions and comments; the Caenorhabditis Genetics Center (which is funded by the NIH National Center for Research Resources (NCRR)), G. Hayes, and S. Russell for strains; and the Wellcome Trust Sanger Institute for the hbl-1 cosmid. This work was supported by a graduate research fellowship from National Science Foundation (K.T.-P.), a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research (J.B.), and grants from the National Institutes of Health (NIH) to J.M.K. (NS32196) and J.B. (K99MH085039). K.T.-P. is an Albert J. Ryan Foundation fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Developmental Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Developmental Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes & Development. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- Armentano M, Chou SJ, (null) (null), O’Leary DDM, Studer M. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci. 2007;10:1277–1286. doi: 10.1038/nn1958. [DOI] [PubMed] [Google Scholar]
- Bosman LWJ, Konnerth A. Activity-dependent plasticity of developing climbing fiber-Purkinje cell synapses. Neuroscience. 2009;162:612–623. doi: 10.1016/j.neuroscience.2009.01.032. [DOI] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Wu CZ, Knott G, Kuhlman S, Fu Y, Palmiter RD, Huang ZJ. GAD67-mediated GABA synthesis and signaling regulate inhibitory synaptic innervation in the visual cortex. Neuron. 2007;54:889–903. doi: 10.1016/j.neuron.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittman JS, Kaplan JM. Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc Natl Acad Sci USA. 2006;103:11399–11404. doi: 10.1073/pnas.0600784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. Plos Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda Y, Davis GW. Mechanisms of synapse assembly and disassembly. Neuron. 2003;40:243–264. doi: 10.1016/s0896-6273(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Gracheva EO, Burdina AO, Holgado AM, Berthelot-Grosjean M, Ackley BD, Hadwiger G, Nonet ML, Weimer RM, Richmond JE. Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. Plos Biol. 2006;4:e261. doi: 10.1371/journal.pbio.0040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Developmental Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Jin Y. Lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature. 1998;395:78–82. doi: 10.1038/25757. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period regulation. Annu Rev Neurosci. 2004;27:549–579. doi: 10.1146/annurev.neuro.27.070203.144327. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJGD, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hua JY, Smith SJ. Neural activity and the dynamics of central nervous system development. Nat Neurosci. 2004;7:327–332. doi: 10.1038/nn1218. [DOI] [PubMed] [Google Scholar]
- Jiang B, Treviño M, Kirkwood A. Sequential development of long-term potentiation and depression in different layers of the mouse visual cortex. J Neurosci. 2007;27:9648–9652. doi: 10.1523/JNEUROSCI.2655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Jorgensen E, Hartwieg E, Horvitz HR. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J Neurosci. 1999;19:539–548. doi: 10.1523/JNEUROSCI.19-02-00539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen EM. WormBook : the online review of C elegans biology. 2005. GABA; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai MI, Okabe M, Hiromi Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Developmental Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kanatani S, Yozu M, Tabata H, Nakajima K. COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J Neurosci. 2008;28:13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Developmental Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Luo L, O’Leary DDM. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- McEwen JM, Madison JM, Dybbs M, Kaplan JM. Antagonistic regulation of synaptic vesicle priming by Tomosyn and UNC-13. Neuron. 2006;51:303–315. doi: 10.1016/j.neuron.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Melkman T, Sengupta P. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development. 2005;132:1935–1949. doi: 10.1242/dev.01788. [DOI] [PubMed] [Google Scholar]
- Mettler U, Vogler G, Urban J. Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–437. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–34. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008 doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oray S, Majewska A, Sur M. Dendritic Spine Dynamics Are Regulated by Monocular Deprivation and Extracellular Matrix Degradation. Neuron. 2004;44:1021–1030. doi: 10.1016/j.neuron.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Park M, Watanabe S, Poon VYN, Ou CY, Jorgensen EM, Shen K. CYY-1/Cyclin Y and CDK-5 Differentially Regulate Synapse Elimination and Formation for Rewiring Neural Circuits. Neuron. 2011;70:742–757. doi: 10.1016/j.neuron.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FA, Tsai MJ, Tsai SY. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol Life Sci. 2000;57:1388–1398. doi: 10.1007/PL00000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Lichtman JW. Elimination of synapses in the developing nervous system. Science. 1980;210:153–157. doi: 10.1126/science.7414326. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat Neurosci. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush SF, Slack FJ. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev Biol. 2009;334:523–534. doi: 10.1016/j.ydbio.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends in Cell Biology. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Shan G, Kim K, Li C, Walthall WW. Convergent genetic programs regulate similarities and differences between related motor neuron classes in Caenorhabditis elegans. Dev Biol. 2005;280:494–503. doi: 10.1016/j.ydbio.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The MicroRNA miR-1 Regulates a MEF-2-Dependent Retrograde Signal at Neuromuscular Junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes & Development. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos Trans R Soc Lond, B, Biol Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Filosa A, Armentano M, Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- Vashlishan AB, Madison JM, Dybbs M, Bai J, Sieburth D, Ch’ng Q, Tavazoie M, Kaplan JM. An RNAi Screen Identifies Genes that Regulate GABA Synapses. Neuron. 2008;58:346–361. doi: 10.1016/j.neuron.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Walthall WW. Metamorphic-like changes in the nervous system of the nematode Caenorhabditis elegans. J Neurobiol. 1990;21:1085–1091. doi: 10.1002/neu.480210712. [DOI] [PubMed] [Google Scholar]
- Walthall WW, Plunkett JA. Genetic transformation of the synaptic pattern of a motoneuron class in Caenorhabditis elegans. J Neurosci. 1995;15:1035–1043. doi: 10.1523/JNEUROSCI.15-02-01035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Weimer RM, Richmond JE, Davis WS, Hadwiger G, Nonet ML, Jorgensen EM. Defects in synaptic vesicle docking in unc-18 mutants. Nat Neurosci. 2003;6:1023–1030. doi: 10.1038/nn1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans: the mind of a worm. Philos Trans R Soc Lond, B, Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- White JG, Albertson DG, Anness MA. Connectivity changes in a class of motoneurone during the development of a nematode. Nature. 1978;271:764–766. doi: 10.1038/271764a0. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Yao TP, Chen JD, Evans RM, McKeown M. Seven-up inhibits ultraspiracle-based signaling pathways in vitro and in vivo. Mol Cell Biol. 1995;15:6736–6745. doi: 10.1128/mcb.15.12.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tsai SY, Tsai MJ. COUP-TFI: an intrinsic factor for early regionalization of the neocortex. Genes & Development. 2001;15:2054–2059. doi: 10.1101/gad.913601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HM, Walthall WW. UNC-55, an orphan nuclear hormone receptor, orchestrates synaptic specificity among two classes of motor neurons in Caenorhabditis elegans. J Neurosci. 1998;18:10438–10444. doi: 10.1523/JNEUROSCI.18-24-10438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.