SUMMARY
DNA methylation is an important epigenetic mark established by the combined actions of methylation and demethylation reactions. Plants use a base excision repair pathway for active DNA demethylation. After 5-methylcytosine removal, the Arabidopsis DNA glycosylase/lyase ROS1 incises the DNA backbone and part of the product has a single-nucleotide gap flanked by 3′- and 5′-phosphate termini. Here we show that the DNA phosphatase ZDP removes the blocking 3′-phosphate, allowing subsequent DNA polymerization and ligation steps needed to complete the repair reactions. ZDP and ROS1 interact in vitro and co-localize in vivo in nucleoplasmic foci. Extracts from zdp mutant plants are unable to complete DNA demethylation in vitro, and the mutations cause DNA hypermethylation and transcriptional silencing of a reporter gene. Genome-wide methylation analysis in zdp mutant plants identified hundreds of hypermethylated endogenous loci. Our results show that ZDP functions downstream of ROS1 in one branch of the active DNA demethylation pathway.
Keywords: epigenetics, DNA methylation, active DNA demethylation, ROS1, gene silencing
INTRODUCTION
Cytosine methylation (5-meC) is an important epigenetic mark for the establishment and maintenance of gene silencing across cellular divisions, and plays critical roles in regulation of developmental genes, genomic imprinting, X chromosome inactivation and transposon silencing (He et al., 2011; Law and Jacobsen, 2010; Suzuki and Bird, 2008; Zilberman, 2008). During the life cycle of animals and plants, active DNA demethylation processes that are still incompletely understood cause both genome-wide and local erasure of DNA methylation patterns (Feng et al., 2010; He et al., 2011). The mechanisms responsible for active DNA demethylation in mammalian cells remain debatable (Gong and Zhu, 2011; Wu and Zhang, 2010), but there is overwhelming biochemical and genetic evidence that plants possess DNA glycosylases that specifically remove 5-meC from DNA, initiating its replacement by unmethylated cytosine through a base excision repair (BER) process (Gehring et al., 2009b; Roldan-Arjona and Ariza, 2009; Zhu, 2009).
Plant 5-meC DNA glycosylases comprise a subfamily of atypical HhH-GPD enzymes whose founding members are the Arabidopsis proteins ROS1 (REPRESSOR OF SILENCING 1) (Gong et al. 2002), DME (DEMETER), DML2 and DML3 (DEMETER-LIKE proteins 2 and 3) (Choi et al., 2002; Ortega-Galisteo et al., 2008; Penterman et al., 2007). ROS1 was identified in a screen for mutants with increased silencing of the repetitive RD29A-LUC transgene (Gong et al., 2002). Together with paralogs DML2 and DML3, ROS1 is needed to counteract the robust RNA-directed DNA methylation (RdDM) pathway at hundreds of discrete regions across the plant genome (Ortega-Galisteo et al., 2008; Penterman et al., 2007; Zhu et al., 2007). DME is expressed in the central cell of the female gametophyte, and participates in demethylation of the maternal genome in the endosperm, thus contributing to gene imprinting in this nutritive, extra-embryonic part of the developing seed (Gehring et al., 2009a; Gehring et al., 2006; Hsieh et al., 2009).
ROS1 and its homologs are bifunctional DNA glycosylases/lyases that cleave the phosphodiester backbone at the 5-meC removal site by β-elimination, generating a 3′ phospho α,β-unsaturated aldehyde at the strand break (Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006; Ortega-Galisteo et al., 2008; Penterman et al., 2007). In ROS1, a significant amount of β-elimination incisions proceed to β,δ-elimination products (Agius et al., 2006; Morales-Ruiz et al., 2006). Thus, part of the final reaction product generated by ROS1 is a single-nucleotide gap flanked by 3′phosphate and 5′phosphate termini. A yet unknown DNA polymerase must fill this gap with an unmethylated cytosine before a DNA ligase can seal the remaining nick. However, all DNA polymerases characterized to date require a 3′-hydroxyl terminus to initiate synthesis. Therefore, the phosphate group present at the 3′ end of the single-nucleotide gap generated by ROS1 must be removed prior to the polymerization and ligation steps that complete the DNA demethylation pathway.
In mammalian cells, 3′-phosphate termini generated by β,δ-elimination catalysts are converted to 3′ hydroxyl by polynucleotide kinase 3′-phosphatase (PNKP) (Jilani et al., 1999), which functions not only in BER (Wiederhold et al., 2004), but also in the repair of both single-strand breaks (SSBs) (Whitehouse et al., 2001) and double-strand breaks (DBSs) (Chappell et al., 2002). The first plant enzyme acting on 3′-phosphorylated termini was identified in maize and termed ZmDP2 (Betti et al., 2001). ZmDP2 shows partial sequence similarity to mammalian PNKP and possesses 3′phosphatase activity, but it is devoid of an associated 5′-kinase activity (Betti et al., 2001). Its Arabidopsis homolog is ZDP (Arabidopsis thaliana zinc finger DNA 3′-phosphoesterase) (herein referred to as ZDP), a modular protein with a C-terminal 3′-phosphatase domain and an N-terminal DNA-binding domain containing three PARP-like zinc fingers (Petrucco et al., 2002). This enzyme has been shown to bind SSBs and DSBs and to dephosphorylate 3′-phosphate ends to generate the corresponding 3′-hydroxyl termini (Petrucco et al., 2002).
We hypothesized that 3′-phosphates generated by ROS1 are putative substrates for the 3′-phosphatase activity of ZDP, and thus ZDP may be important for epigenetic regulation through participation in active DNA demethylation. In this work we report biochemical, genetic and cell biological evidence that ZDP functions with ROS1 in active DNA demethylation and is an important player in shaping the DNA patterns of hundreds of genomic loci.
RESULTS
ZDP removes the 3′-blocking phosphate from the gapped product generated by ROS1 and increases the processivity of ROS1
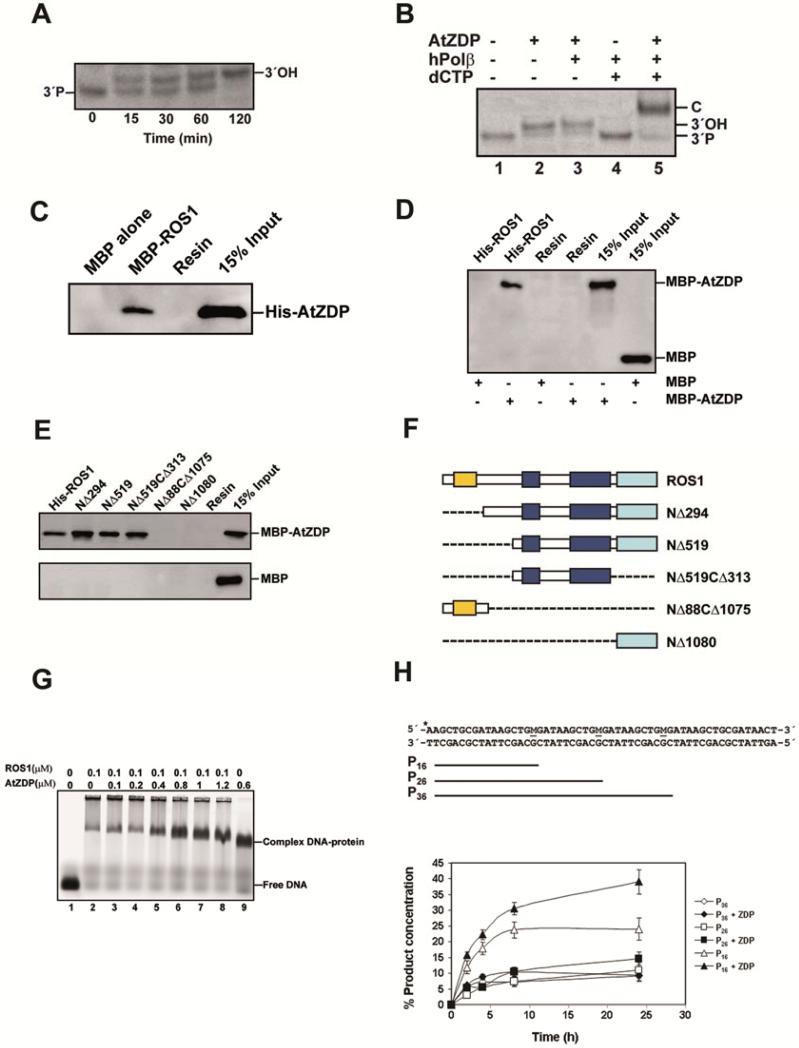
We incubated ROS1 with a 51-mer duplex oligo substrate that contained a 5-meC residue at position 29 in the upper, 5′-end labeled strand (Table S1). The products generated by ROS1 were purified and used for kinetic analysis with purified ZDP. We found that ZDP catalyzed the conversion of the 3′-phosphate end of the β,δ-elimination product into the corresponding 3′-hydroxyl species in a time-dependent manner (Figure 1A). To confirm that the 3′-terminus generated by ZDP may be used for the gap-filling step, we performed the 3′ end-cleaning reaction in the presence of human DNA polymerase β (pol β) and dCTP (Figure 1B). We found that the insertion of dCMP required the presence of both pol β and ZDP. These results indicate that ZDP is able to remove in vitro the 3′-blocking phosphate from the single-nucleotide gap generated by ROS1, producing a 3′-hydroxyl terminus that may be used by a DNA polymerase for DNA synthesis.
Figure 1. Characterization of ZDP biochemical activity and interaction with ROS1.
(A) Purified ROS1 (37.5 nM) was incubated at 30°C for 16 h with a double-stranded oligonucleotide substrate (40 nM) containing a 5-meC:G pair (See Table S1 for the sequence of the substrates). Reaction products were incubated with purified ZDP (1.5 nM) at 30°C. Reactions were stopped at the indicated times, products were separated in a 15% denaturing polyacrylamide gel and visualized by fluorescence scanning. (B) Reaction products of ROS1 were incubated with purified ZDP (6 nM) and human DNA polymerase β (hPol β, 0.5 U) during 30 min at 37°C, in the absence (lane 3) or presence (lane 5) of dCTP (0.2 mM). Lane 1: 3′-phosphate (3′P) marker. Lane 2: control reaction without hPol β. Lane 4: control reaction without ZDP. (C) Maltose Binding Protein (MBP) either alone or fused to ROS1 full-length (MBP-ROS1) was expressed in E. coli and fixed to an amylose resin. His-tagged ZDP purified from E.coli (His-ZDP) was incubated with either MBP-ROS1 or MBP bound to the resin. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane and immunoblotted with antibodies against His6- tag. (D and E) Purified full-length ROS1 (FL-ROS1) or truncated ROS1 polypeptides fused to His6 -tag were fixed to a Ni-Sepharose column. The proteins bound to the column were incubated in the presence of either MBP-ZDP or MBP alone. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane and immunoblotted with antibodies against MBP. (F) Schematic diagrams of full-length ROS1 and the different truncated ROS1 derivatives used in (E): lysine-rich domain (yellow), DNA glycosylase domain (blue), and C-terminal domain (cyan). (G) EMSA. Full-length ROS1 (0.1 μM) and increasing concentrations of ZDP were incubated with a labeled DNA duplex containing a single-nucleotide gap flanked by 3′-P and 5′-P termini. After nondenaturing gel electrophoresis, protein-DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. See Figure S1 for the proteins used in pull-down assay. (H) Effect of ZDP on ROS1 processivity. Purified ROS1 (18 nM) was incubated with a double-stranded oligonucleotide substrate (40 nM) containing three 5-meC:G pairs (M, upper panel) in the absence (open symbols) or presence (closed symbols) of purified ZDP (144 nM) (lowe panel). Reactions were stopped at the indicated times and products were separated in a 12% denaturing polyacrylamide gel. Product concentration was quantified by fluorescence scanning: 16 nt (triangles), 26 nt (squares) and 36 nt (diamonds). Mean values of product concentration with standard errors from three independent experiments were shown. Also see Figure S1 for purified proteins used in pull-down assay and effect of ZDP on ROS1 activity.
We used in vitro pull-down assays to test for a direct interaction between ROS1 and ZDP. As shown in Figure 1C, MBP-ROS1, but not MBP alone, bound to His-ZDP. Conversely, MBP-ZDP, but not MBP alone, was bound by His-ROS1 (Figure 1D). In order to define ROS1 regions implicated in the interaction, we generated a series of N-and C-terminal His-ROS1 deletion mutants, and the purified proteins (Figure S1A) were used to analyze their capacity to bind MBP-ZDP (Figures 1E and 1F). We found that the N-terminal 519 and C-terminal 313 amino acids from ROS1 are not required for the interaction. The region needed to interact with ZDP coincides with the protein segment that contains the discontinuous DNA glycosylase domain of ROS1 (Ponferrada-Marin et al., 2011).
Since we have previously reported that ROS1 binds tightly to its reaction product (Ponferrada-Marin et al., 2010), we performed EMSA analysis to examine whether ZDP may occupy the gapped DNA in the presence of ROS1. When only ROS1 was present in the binding reaction, a band corresponding to the complex between the DNA glycosylase and the gapped DNA product was detected (Figure 1G, lane 2). Part of the labeled probe remained trapped in the wells, hinting to the formation of insoluble ROS1-DNA complexes. When the concentration of ZDP increased (Figure 1G, lanes 3-8), the amount of labeled material gradually disappeared from the wells. Moreover, the band corresponding to the ROS1-DNA complex increased in intensity, but was gradually converted into a new band that migrated at a slower rate than the complex seen when only ZDP was incubated with DNA (Figure 1G, lane 9). Importantly, no bands corresponding to single ROS1-DNA or ZDP-DNA complexes were observed when both proteins were present. These results suggest that ROS1, ZDP and the gapped DNA substrate may form a ternary complex.
We also examined whether ZDP may exert any effect on the highly distributive behavior of ROS1, which does not exhibit significant processivity in vitro, due to strong binding to its reaction product (Ponferrada-Marin et al., 2010; Ponferrada-Marin et al., 2009). We incubated ROS1, either in the absence or the presence of ZDP, with a double-stranded oligonucleotide substrate containing three 5-meC residues in the upper strand separated by 9 nt and located in the same sequence context (Figure 1H). In this assay, a processive mechanism would rapidly convert the substrate to a final reaction product represented by a 16-nt labeled fragment, whereas a distributive mechanism would lead to the accumulation of partially processed reaction intermediates represented by 26- and 36-nt labeled fragments. Consistent with our previously reported observations, we found that ROS1 processed 5-meC in a highly distributive fashion, exhibiting a steady accumulation of the 16-nt fragment but also of significant amounts of partially processed substrates, even after long incubation times. However, the addition of ZDP (which does not show detectable enzymatic activity under these conditions) significantly increased the amount of substrate converted by ROS1 to the final reaction product (P16), without apparently changing the accumulation of reaction intermediates (P26 and P36). We also found a positive effect of ZDP on ROS1 activity against a DNA substrate containing a single 5-meC residue (Figure S1B-D). Altogether, these results suggest that ZDP stimulates the enzymatic activity of ROS1.
zdp mutants are deficient in DNA 3′-phosphatase activity
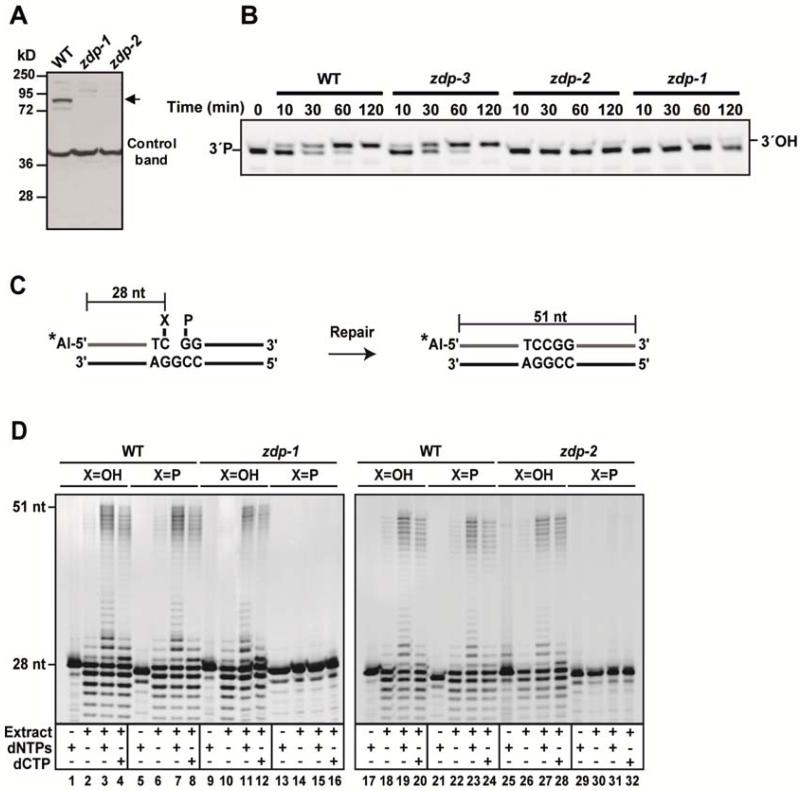
To examine the possible role of ZDP in the DNA demethylation process initiated by ROS1, we searched for zdp mutants in public T-DNA insertion line collections. We identified two Arabidopsis lines with insertions located at introns 14 (SAIL_60_C08) and 15 (SAIL_813_B03) (Sessions et al., 2002) and a third mutant line with an insertion 76 bp upstream of the translation start site (SALK_130907) (Alonso et al., 2003). We termed the corresponding mutant alleles zdp-1, zdp-2 and zdp-3, respectively (Figure S2A). RT-PCR analysis with ZDP-specific primers corresponding to the 3′-end region of the gene detected the expected product in wild type and zdp-3 plants (data not shown), but the level was very low in zdp-2 plants and undetectable in zdp-1 mutants (Figure S2B). Both zdp-1 and zdp-2 mutants were further characterized by Western blotting using an antibody raised against ZDP. A specific band of the expected size (about 80 kDa) was detected in wild-type but not in the mutant plants (Figure 2A). We tested the 3′-phosphatase activity of whole-cell extracts from WT and zdp mutant plants by measuring conversion from 3′-P to 3′-OH after incubation with a 5′-end labeled DNA substrate containing a single nucleotide gap (Figure 2B). We found that extracts from zdp-3 plants retained full 3′-end processing activity compared to WT plants. In contrast, zdp-1 and zdp-2 mutants exhibited a barely detectable phosphatase activity. We conclude that the T-DNA insertions carried by zdp-1 and zdp-2 plants inactivate the wild-type function of the ZDP gene. The homozygous zdp-1 and zdp-2 mutant plants did not exhibit obvious morphological or developmental abnormalities. However, both mutants were hypersensitive to the DNA damaging agent MMS (Figure S2C). In contrast, they displayed levels of sensitivity to H2O2 similar to those of WT plants (Figure S2D).
Figure 2. zdp mutant plants are unable to process DNA demethylation intermediates containing a gap flanked by 3′- and 5′-phosphate groups.
(A) Detection of ZDP protein levels in wild-type and mutant plants by Western blotting using an antibody against ZDP. A prominent non-specific band from the Western blot serves as a control for loading. Arrow points to the position of ZDP protein. (B) A DNA duplex containing a single-nucleotide gap flanked by 3′-P and 5′-P termini was incubated at 30°C with cell extracts from wild-type and zdp mutants. Reactions were stopped at the indicated times, products were separated in a 15% denaturing polyacrylamide gel and visualized by fluorescence scanning. (C) Schematic diagram of molecules used as DNA substrates in the repair assay. Double-stranded oligonucleotides contained a single-nucleotide gapped flanked by either a 3′P (X= P) or 3′OH (X= OH). The Alexa Fluor-labeled 5′-end of the upper strand is indicated by a star. The size of the fully repaired product is indicated. (D) DNA substrates were incubated with extract (45 μg protein) from wild-type (lanes 1-8 and 17-24), zdp-1-/- (lanes 9-16) or zdp-2-/- (lanes 25-32) mutant plants at 30°C for 2 h in a reaction mixture containing either dCTP or all four dNTPs, as indicated. Reaction products were separated in a 12% denaturing polyacrylamide gel, and detected by fluorescence scanning. Also see Figure S2 and Figure S3 for detailed T-DNA insertion mutants’ information.
zdp mutant extracts show a reduced capacity to complete DNA demethylation in vitro
To test whether ZDP is required to process DNA demethylation intermediates that contain a 3′-blocking phosphate, we incubated cell extracts prepared from WT, zdp-1 or zdp-2 plants with DNA substrates containing a single nucleotide gap flanked by a phosphate at the 5′ terminus and either by hydroxyl or phosphate at the 3′terminus (Figure 2C-D). We found that both mutants performed repair of the 3′-OH gapped substrate with similar efficiency as WT plants, but lacked any detectable repair activity on the 3′-P gapped substrate. It is worth noting that the repair of the gapped DNA demethylation intermediates takes place both via single-nucleotide insertion and long-patch DNA synthesis (Figure 2C-D).
Some of the incisions generated by ROS1 in vitro do not proceed to β,δ-elimination and remain as β-elimination products containing a 3′-α,β-unsaturaded aldehyde (3′dRP)(Agius et al., 2006). ZDP is not active on 3′-dRP (data not shown), but such blocking aldehyde might be removed by an AP endonuclease, as reported in bacteria and mammals (Demple and Harrison, 1994; Pascucci et al., 2002). The major AP endonuclease activity in Arabidopsis cell extracts is ARP (Apurinic endonuclease-Redox Protein), which is required for BER of uracil and AP sites in vitro (Cordoba-Canero et al., 2011). Thus, we examined to what extent the downstream steps required to complete the DNA demethylation process initiated by ROS1 (i.e., gap-filling and ligation) are dependent on ZDP and/or ARP (Figure S3). ROS1 activity is too low to be detectable in Arabidopsis cell extracts (our unpublished observations). Therefore, we incubated purified ROS1 with a duplex oligonucleotide substrate that contained a 5-meC residue, and the reaction products, which contained a mixture of β- and β,δ-elimination products (Figure S3C, lane 3), were purified and incubated with cell-free extracts from WT, zdp-1 and arp plants. Figure S3C shows the analysis of the incision intermediates generated on the processed strand. We found that the zdp extracts removed the unsaturated aldehyde, but not the phosphate group, from the 3′ terminus of the gap (Figure S3C, lane 5). In contrast, both WT plants and arp mutants efficiently processed both types of blocked ends (Figure S3C, lanes 4 and 6). Cleaning of the 3′ termini generated free 3′-OH ends (Figure S3C lanes 4-6), which disappeared when deoxynucleotides were added to the reaction (Figure S3C lanes 7-9), but under these conditions the non-processed 3′-P termini persisted in the reactions with zdp extracts (Figure S3C lane 8). In order to detect the fully-demethylated products, we digested the reaction products with HpaII (Figure S3D). WT and arp extracts displayed a similar demethylation activity, but the levels of fully-demethylated products in the zdp mutant extract were reduced about 2-fold compared to those of wild-type plants. In control reactions with an AP site analog (THF), we confirmed that zdp, but not arp mutants, retain full capacity to repair an AP site (Figure S3D, right panel). These results suggest that completion of DNA demethylation initiated by ROS1 is partially dependent on ZDP. Furthermore, they indicate that the β-elimination products generated by ROS1 are processed, in a ZDP- and ARP-independent manner, by an as yet unidentified enzymatic activity present in Arabidopsis cells.
Role of ZDP in preventing transcriptional gene silencing
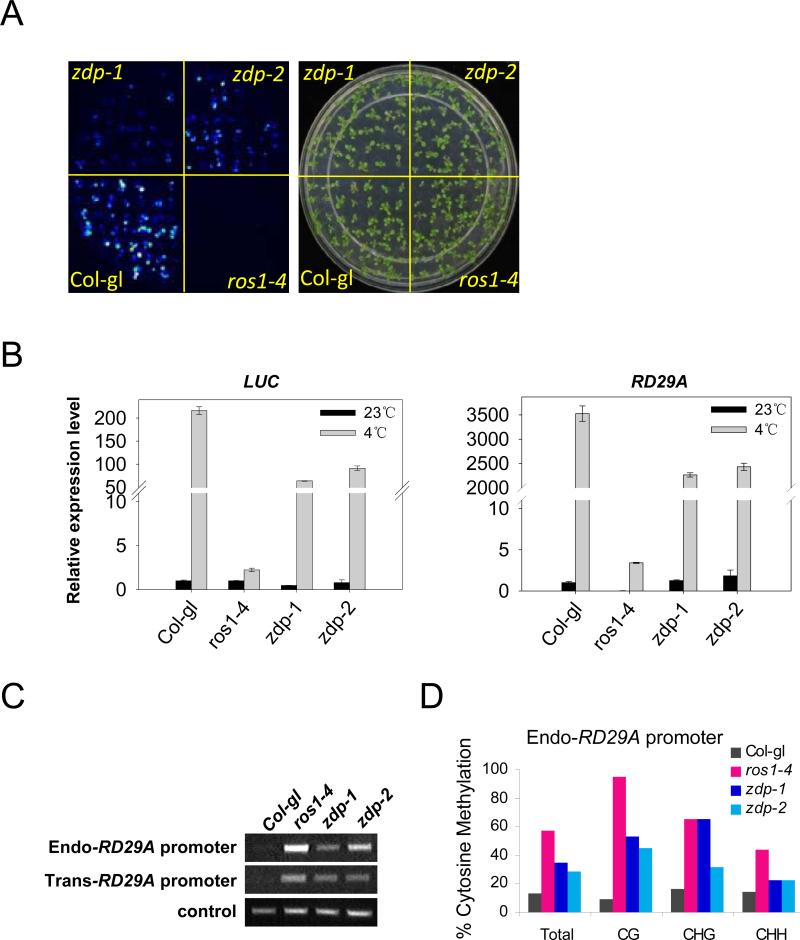
Since ROS1 is known to protect certain transgenes and endogenous genes from transcriptional silencing, we tested the potential role of ZDP in transcriptional gene silencing. We isolated zdp homozygous mutant plants with an RD29A promoter-driven LUCIFERASE reporter gene (RD29A-LUC) by crossing the zdp mutants with wild type Columbia gl1 plants expressing the reporter gene. Luminescence imaging showed that RD29A-LUC expression was reduced in the zdp-1 and zdp-2 mutants compared to the wild type (Figure 3A). Real-time PCR analysis also showed that the expression level of the LUC transgene was reduced in the mutants (Figure 3B). In addition, the analysis showed that expression of the endogenous RD29A gene was reduced in the zdp mutant plants. It is important to note that the reduction in both the transgene and endogenous gene expression in zdp mutants was not as dramatic as in the ros1-4 mutant plants (Figure 3B). The results suggest that ZDP can partially protect transgenes and endogenous genes from transcriptional silencing.
Figure 3. Partial silencing of RD29A-LUC in zdp mutant plants.
(A) Two-weeks old plants grown on MS plates were imaged for luminescence after cold treatment at 4°C for 2 days. (B) The expression levels of LUC and RD29A were determined by Real-time PCR. TUB8 was used as a control. Error bars represent s.d. (n=3). (C) Analysis of DNA methylation at transgene and endogenous RD29A promoter by CHOP-PCR (see experimental procedures for details). (D) Bisulfite sequencing analysis of promoter methylation status of endogenous RD29A promoter.
PCR analysis of the RD29A promoters that were digested with a methylation-sensitive restriction enzyme indicated that like ros1-4, both zdp-1 and zdp-2 mutations caused hypermethylation of the transgene and endogenous RD29A promoters (Figure 3C). Bisulfite sequencing confirmed the hypermethylation and revealed increased levels of methylation in not only CG but also CHG and CHH (H is A, T, or C) sequence contexts in the endogenous RD29A promoter in ros1-4 as well as zdp-1 and zdp-2 mutants (Figure 3D). These results suggest a similar genetic function of ZDP and ROS1 in preventing promoter DNA hypermethylation and transcriptional silencing of the transgenes and homologous endogenous genes.
Genome-wide effects of ZDP on DNA methylation
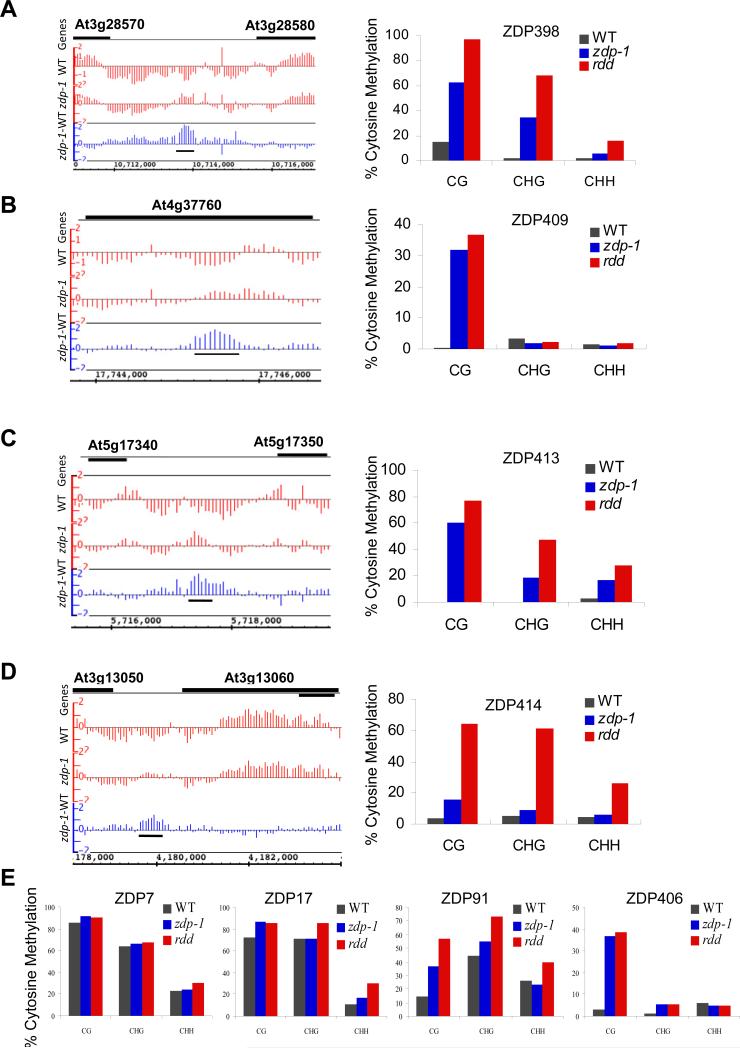
To assess the genome-wide effect of ZDP on DNA methylation, we performed whole genome methylation profiling by using methylcytosine immunoprecipitation and NimbleGen genome tiling arrays (Penterman et al., 2007). The overall DNA methylation levels in WT and zdp-1 mutant were similar (Figures S4A and S4B). However, we identified 415 loci where the methylation levels were increased in zdp-1 plants (Table S3) and 28 loci that were hypomethylated in zdp mutant plants (Table S4). The hypermethylated loci were distributed throughout the five chromosomes, but showed some enrichment around centromeric regions (Figure S4C). Transposable elements accounted for 85.8% of the affected loci. Some genic (9.4%) and intergenic regions (4.8%) were also hypermethylated in zdp-1 mutant plants (Figure S4D). We randomly chose 12 regions from the list of hypermethylated loci to verify the tiling array results by bisulfite sequencing. Eleven of these 12 loci were found to be hypermethylated in zdp-1 as well as the rdd triple mutant plants that contain T-DNA insertions in ROS1, DML2 and DML3 (Penterman et al., 2007) (Figures 4A-4E, Figure S5A and Table S5). At most of the tested loci, hypermethylation was observed in all sequence contexts (CG, CHG and CHH). The bisulfite sequencing data suggest that the tiling array analysis was quite reliable. At some of the loci examined, the increase in methylation in the zdp mutant was comparable to that seen in the rdd triple mutant (Figures 4B and 4E), whereas at some other loci the hypermethylation in zdp mutant was less dramatic than that in rdd (Figures 4A and 4C-4E). These results suggest that ZDP is important for preventing the hypermethylation of hundreds of genomic loci. The vast majority of the affected loci are transposable elements, and most if not all are also affected by ROS1 and its paralogs.
Figure 4. Confirmation of randomly chosen hypermethylated regions by bisulfite sequencing.
(A-D) For each region, the left panel shows the context of the hypermethylated region (indicated by a black horizontal bar under the blue bars) and tiling array signals. The right panel shows the bisulfite sequencing results for the corresponding locus in wild type and zdp-1, as well as the rdd mutant plants. (E) Bisulfite sequencing results for four other loci which show hypermethylation in the tiling array data in zdp-1 mutant plants. See Figure S4 for the whole genome methylation profile in wild-type and zdp-1 mutant plants and Figure S5 for more loci DNA methylation level. Also see Table S2 for the primers used in bisulfite sequencing, Table S3 for hypermethylated loci in zdp-1 mutant plants, Table S4 for hypomethylated loci in zdp-1 mutant plants, and Table S5 for detailed percent methylation of loci in different plants.
The methylation level of a LINE transposon were confirmed to be increased in zdp as well as in ros1-4 mutant plants by bisulfite sequencing (Figure S5B). We examined the expression of this LINE transposon and found that its transcript levels were reduced in ros1-4 and zdp mutant plants, compared to the wild type (Figure S5C).
Genetic interaction among ZDP, ros1 and RdDM mutants
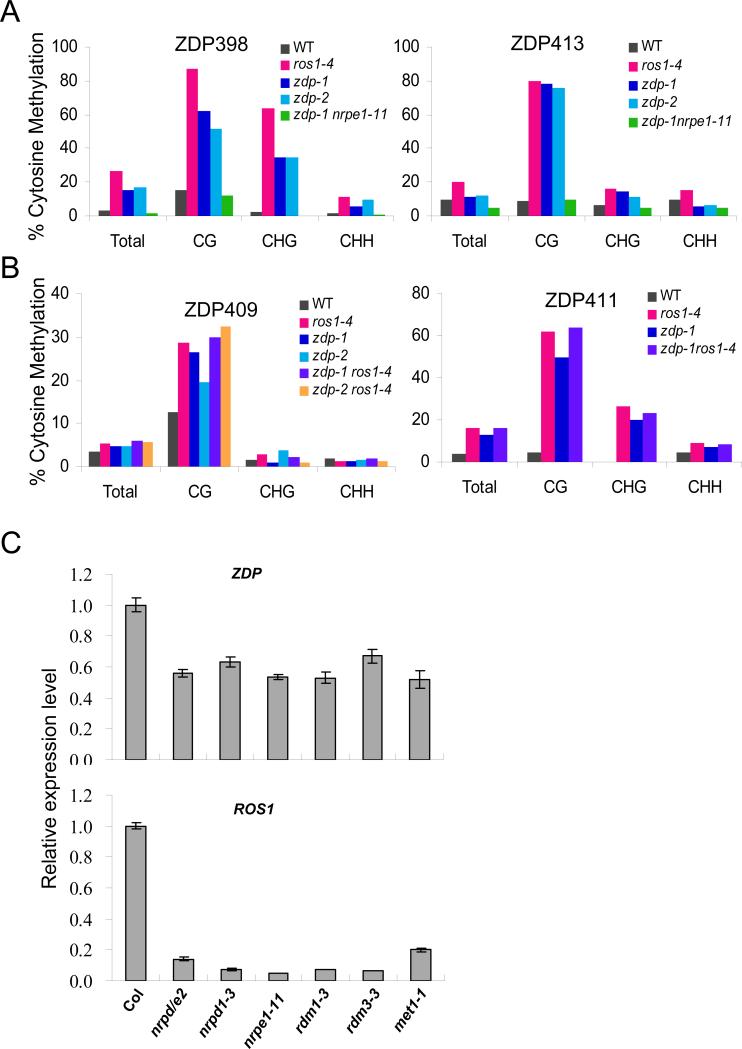
We also tested for genetic interactions among zdp, ros1 and RdDM mutants. We focused our analysis on the methylation status of ZDP386, ZDP388, ZDP398, ZDP410, ZDP413, ZDP409 and ZDP411, seven specific loci showing hypermethylation in zdp and rdd mutant plants (Figures 5A and 5B). We found that a mutation in the largest subunit of Pol V (NRPE1) suppresses the hypermethylation phenotype of the zdp-1 mutant at the ZDP386, ZDP388, ZDP398, ZDP410 and ZDP413 loci (Figure 5A and Figure S5A), thus suggesting that ZDP counteracts methylation that is dependent on the RdDM pathway. Our analysis of the zdp-1ros1-4 and zdp-2ros1-4 double mutants showed that there is little additive effect between ros1-4 and zdp-1 or zdp-2 in the hypermethylation phenotype detected at the ZDP409 and ZDP411 loci (Figure 5B). The lack of additive effect suggests that ZDP functions in the same genetic pathway as ROS1.
Figure 5. Expression and genetic interaction analysis.
(A) Suppression of the hypermethylation phenotype of the zdp-1 mutant at the ZDP398 and ZDP413 loci by the nrpe1-11 mutation. (B) Absence of additive effects between ros1-4 and zdp-1 or zdp-2 alleles on the hypermethylation phenotype at the ZDP409 and ZDP411 loci. (C) Real-time PCR analysis of ZDP and ROS1 mRNA levels in RdDM and met1-1 mutants. The transcript levels were normalized using TUB8 as an internal standard. Error bars represent s.d. (n=3).
Sub-nuclear co-localization of ZDP with ROS1 and ROS3
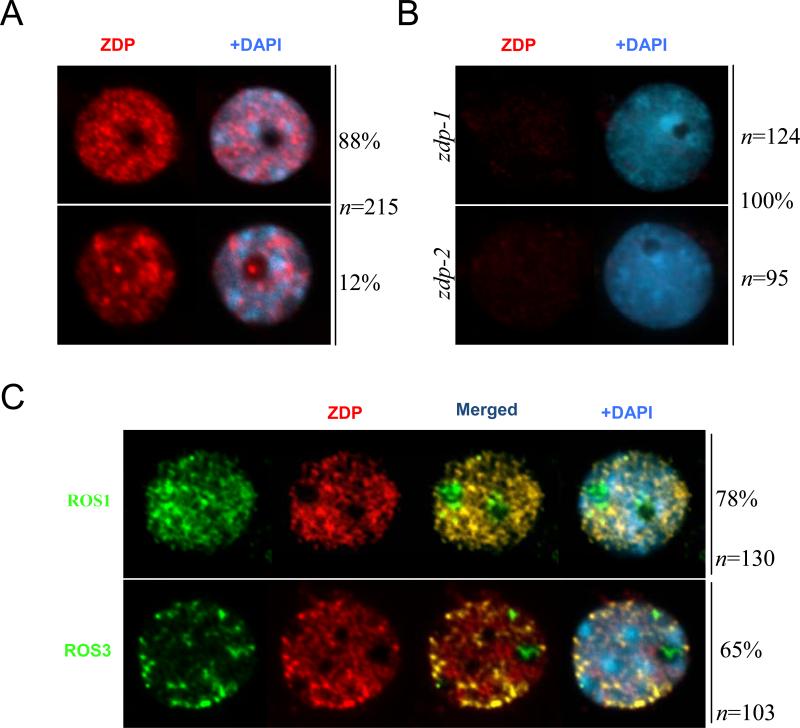
The two known active DNA demethylation factors, ROS1 and ROS3, are co-localized in the nucleolus and in discrete foci in the nucleoplasm (Zheng et al., 2008). As a component of the active DNA demethylation machinery, ZDP is expected to be in the same subnuclear sites as ROS1. To determine the subnuclear localization of ZDP protein, we generated antibodies specific to ZDP and used the antibodies for immunolocalization of ZDP in Arabidopsis leaf nuclei. As shown in Figure 6A, ZDP was broadly distributed throughout the nucleus, and was enriched in small nucleoplasmic foci. No signals were observed when the antibodies were applied to nuclei preparations of zdp-1 and zdp-2 mutant plants, indicating that the wild-type patterns reflect ZDP localization and not non-specific interactions of the antibody (Figure 6B).
Figure 6. Sub-nuclear localization of ZDP and co-localization with ROS1 and ROS3.
(A and B) The nuclear distribution of ZDP was analyzed by immunostaining using anti-ZDP (red) in wild type (A) and zdp mutants (B). (C) Dual immunolocalization using anti-ZDP (red) in transgenic lines expressing recombinant full-length epitope-tagged MYC-ROS1 and Flag-ROS3 (green). In all panels the DNA was stained with DAPI (blue). The frequency of nuclei displaying each interphase pattern is shown on the right.
To further examine ZDP subnuclear localization, we performed coimmunolocalization analysis with ROS1 and ROS3. In our experiments, ROS1 and ROS3 were detected by making use of antibodies to their epitope tags fused to the proteins that were expressed under their respective native promoters in Arabidopsis transgenic lines (Zheng et al., 2008). As expected, we observed very similar interphase localization patterns for ROS1 and ROS3; both proteins were dispersed in the nucleoplasm and nucleolus, although the immunolocalization foci of ROS1 were generally smaller and more diffuse than ROS3, which showed a speckle-like distribution (Figure 6C). We found that ZDP co-localized with both ROS1 and ROS3 within nucleoplasmic foci, as shown by the strong yellow signals, which denoted overlapping of the green and red signals (Figure 6C). However, while the large majority of the nucleoplasmic ZDP signals colocalized with ROS1 foci, only 10-15 foci were co-localized with ROS3 (Figure 6C). Interestingly, we did not find any ZDP localization in the nucleolus (Figures 6A-6C).
ZDP is downregulated in RdDM pathway mutants
The expression of ROS1 is known to be responsive to DNA methylation levels, and exhibits a dramatic reduction both in RNA-directed DNA methylation pathway mutants and in mutants of the DNA maintenance methyltransferase MET1 (Gao et al., 2010; Huettel et al., 2006; Mathieu et al., 2007). We found that ZDP mRNA levels were also reduced in RdDM pathway mutants such as nrpd/e2, nrpd1, nrpe1, rdm1 and rdm3 (Figure 5C). The met1-1 mutation also caused a down-regulation of ZDP expression. However, the reduction of ZDP expression was not as dramatic as that of ROS1. These results suggest that, as a component of the active DNA demethylation pathway, ZDP expression is responsive to DNA methylation levels in a manner similar to ROS1 expression.
DISCUSSION
Active DNA demethylation in plants is carried out by a base excision repair pathway (Gehring et al., 2009b; Zhu, 2009). In mammals, recent studies suggest that at least some of the active DNA demethylation can also be mediated by a DNA repair pathway, although it appears that 5-meC must be processed first by Tet1 and/or deaminases before DNA glycosylases can act (Gong and Zhu, 2011; Guo et al., 2011; Popp et al., 2010; Rai et al., 2008). Thus far, the only known enzymatic components of the plant DNA demethylation pathway are the ROS1 subfamily of 5-meC DNA glycosylase/lyases (Zhu, 2009). Our study here strongly supports that ZDP is an important component in the active DNA demethylation pathway initiated by ROS1.
ZDP is needed to process part of the gapped incision intermediates generated by ROS1. Our results suggest that the dysfunction of such downstream step disrupts the demethylation pathway and alters methylation control, leading to a hypermethylated state in many loci across the genome. In fact, we have found that ZDP interacts with ROS1 and partially increases the very low processivity of the DNA glycosylase, leading to higher rates of excision in substrates containing multiple 5-meC residues. Our results show that ZDP also stimulates the enzymatic activity of ROS1 on substrates containing a single 5-meC. Two observations suggest that ZDP promotes release of ROS1 from its product, thereby liberating the enzyme for subsequent rounds of cleavage. First, in a substrate containing multiple 5-meC residues (Figure 1H), the increase in incision is higher for the final product than for the intermediate products. Second, the EMSA analysis shown in Figure 1G suggests that ZDP decreases the formation of insoluble complexes between the gapped substrate and ROS1. In summary, as in other DNA repair mechanisms, the consecutive steps of the demethylation process in vivo are strongly coordinated. The physical interaction between ZDP and ROS1 may contribute to this coordination that determines the pace of demethylation.
The zdp mutants not only show increased DNA methylation and transcriptional gene silencing (TGS) of both a siRNA (small interference RNA)-producing transgene and its endogenous counterpart, but also exhibit increased DNA methylation at more than four hundred loci throughout the genome. As in the ros1 and rdd mutants (Lister et al., 2008; Penterman et al., 2007; Zhu et al., 2007), most hypermethylated regions in zdp mutants are transposable elements, but genic regions are also represented. This result suggests that, like ROS1, ZDP may function to counteract a robust DNA methylation machinery that recognizes and silences parasitic transposable elements but also can affect normal gene expression. Similar to ROS1, ZDP expression is reduced in met1 and mutants of the RdDM pathway, thus implicating mechanisms that coordinate between DNA methylation and demethylation pathways.
Since one of the final products of ROS1 is a single-nucleotide gap flanked by 3′-P and 5′-P termini, completion of DNA demethylation requires a downstream enzymatic activity to convert the 3′-P blocking group into a free 3′-hydroxyl DNA terminus. We have found that ZDP is the major, if not the only, DNA 3′-phosphatase activity in Arabidopsis whole cell extracts, and it is strictly required for complete processing of part of the DNA demethylation intermediates generated by ROS1. An additional important finding of this work is that, after ZDP-dependent 3′-end cleaning, replacement of the excised 5-meC by an unmethylated cytosine may be carried out both via single-nucleotide insertion and long-patch DNA synthesis, as previously reported in Arabidopsis for other DNA modifications (Córdoba-Cañero et al., 2010; Córdoba-Cañero et al., 2009). The existence of two different sub-pathways for gap-filling and ligation expands the spectrum of potential proteins, particularly DNA polymerases, that may participate in the post-incision events that take place during BER demethylation.
Despite the relevance of ZDP as a downstream factor in the DNA demethylation pathway initiated by ROS1 (and potentially DME, DML2 and DML3), zdp mutants do not exhibit any dramatic phenotypic effects, at least under normal growth conditions. A partial explanation is that the T-DNA insertions in ZDP introns 14 and 15 likely create extremely hypomorphic, but not null, alleles. In fact, a very low level of WT transcript and some residual 3′-phosphatase activity was detected in homozygous zdp-2 mutants, although this residual activity was apparently not sufficient for efficient processing of DNA demethylation intermediates in vitro. In addition, although there are apparently no ZDP-like sequences present in the Arabidopsis genome, we cannot exclude the possibility that another enzyme(s) with the same activity as ZDP is specifically expressed in the central cell of the female gametophyte and functions together with DME. Another potential explanation is the existence of alternative post-incision pathways that are not dependent on a 3’ phosphatase. It should be remembered that some of the incisions generated by ROS1 in vitro do not proceed to β,δ-elimination and remain as β-elimination products containing a 3′-α,β-unsaturaded aldehyde (3′dRP). We have found that processing of the blocking 3′dRP in those ROS1 incisions that remain as β-elimination products does not require ZDP activity. It is possible that AP endonucleases (APEs) may act on 3′-dRP to generate the free 3′-OH terminus, since bacterial and mammalian APEs efficiently remove 3′-dRP termini generated after β-elimination (Demple and Harrison, 1994; Pascucci et al., 2002). The Arabidopsis genome encodes three APE homologs, and at least one of them, termed Arp, has been shown to perform incisions in DNA containing abasic sites (Babiychuk et al., 1994). However, we have found that processing of the blocking 3′dRP in those ROS1 incisions also does not require ARP, the major AP endonuclease activity detectable in Arabidopsis cells. This observation may be explained by possible functional redundancies among the three Arabidopsis APE homologs, or by the existence of an as yet unknown alternative mechanism. An enhancer mutant screening in a zdp-1 RD29ALUC background may help to identify such ZDP- and ARP-independent mechanism of 3’-end processing involved in ROS1-initiated DNA demethylation in plants. The lack of a gametophytic phenotype in zdp mutant plants suggests that ZDP-independent mechanisms have a major role in the 3’-end processing of DME reaction products in the central cell and endosperm. Based on the requirement for 3’ terminal end processing, active DNA demethylation target loci may be classified into three groups: group I (represented by ZDP409 and ZDP406 in Figures 4B and 4E) is solely or largely dependent on ZDP and exhibits similar levels of hypermethylation in the zdp and rdd mutants; group II (represented by the RD29A promoter, ZDP398 and ZDP91 in Figures 3D and 4C and 4E) relies on both ZDP and ZDP-independent mechanisms and shows less severe hypermethylation in zdp than in rdd; and group III relies on the ZDP-independent mechanism only and would show hypermethylation in rdd but not in zdp mutants (represented by Pm36 and At4g18650 promoter in Figure S5D). It would be of interest to investigate the underlying basis of the differing requirements for 3’ end processing of these different groups of demethylation target loci.
The co-localization of ZDP with the two known demethylation factors ROS1 and ROS3 in nucleoplasmic foci further supports its role in active DNA demethylation. The co-localization is consistent with our observation of a physical interaction between ZDP and ROS1 in vitro. Due to technical reasons, our attempts to test for their potential interaction in vivo did not yield conclusive results (data not shown). Interestingly, unlike ROS1 and ROS3, ZDP is not found in the nucleolus. This observation suggests that ZDP-dependent loci (groups I and II above) might be all demethylated in the nucleoplasm. The loci that are demethylated in the nucleolus would all have to use the ZDP-independent mechanism of 3’ end processing. The lyase activity of ROS1 at these loci probably proceeds through β−elimination rather than β,δ-elimination.
The hypersensitivity of zdp mutants to MMS indicates that, besides a role during DNA demethylation, ZDP also participates in the repair of DNA damage. As previously suggested (Petrucco et al., 2002), it is likely that ZDP functions in the repair of SSB generated either directly by DNA damaging agents or indirectly as repair intermediates. MMS does not causes DNA strand breaks directly, but generates methylated bases efficiently excised by N-methylpurine DNA glycosylases that create abasic sites and initiate a BER process (Friedberg et al., 2006). The Arabidopsis genome encodes several N-methylpurine DNA glycosylase homologs and at least one of them has been characterized and shown to restore MMS resistance to an E. coli alkA tag mutant (Santerre and Britt, 1994). The fact that zdp mutants are hypersensitive to MMS suggests that ZDP might contribute to the efficient processing of the abasic sites generated during N-methylpurine repair in Arabidopsis. A similar pathway, dependent on the DNA glycosylase/lyase NEIL and the DNA phosphatase PNK, has been proposed in mammalian cells for the repair of MMS-induced DNA damage (Wiederhold et al., 2004).
EXPERIMENTAL PROCEDURES
Plant materials and cell extract preparation
Arabidopsis mutant lines, SAIL_60_C08 (zdp-1), SAIL_813_B03 (zdp-2) and SALK_130907 (zdp-3) harboring T-DNA insertions in the ZDP gene were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). The arp mutant line was described in (Cordoba-Canero et al., 2011). Arabidopsis seedlings were grown on Murashige-Skoog (MS) nutrient agar plates at 23°C with 16 h of light and 8 h of darkness for 2 weeks, and were harvested before or after 4°C treatment for 2 days. Seedling whole cell extracts were prepared as previously described (Córdoba-Cañero et al., 2009).
Protein expression and purification
Expression and purification of ZDP, ROS1 and ROS1 deletion derivatives was performed as described in Supplemental Information.
Pull down assays
For MBP pull-down assays with ZDP His-tagged protein, 50 pmol of purified MBP alone or MBP-ROS1 in 100 μl Column Buffer (20 mM Tris pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM β- mercaptoethanol) was added to 100 μl of amylose resin (New Englands Biolabs) and incubated for 1 h at 4°C. The resin was washed twice with 600 μl of MBP Buffer (20 mM Tris pH 7.4, 1 mM EDTA, 10 mM β- mercaptoethanol, 0.5% Tritón X-100). Purified His-tagged ZDP (12 pmol) was incubated at 4°C for 1 h with either MBP or MBP-ROS1 bound to resin. The resin was washed twice with MBP Buffer. Bound proteins were separated by SDS-PAGE (7.5%), transferred to PVDF membranes and analyzed by Western blot using antibodies against His6-tag (Novagen).
For His-tag pull-down assays with MBP-ZDP protein, 50 pmol of purified FL-ROS1 or truncated ROS1 polypeptides fused to His6-tag in 400 μl of Sonication Buffer 3 (SB3: 20 mM Tris pH 8.0, 500 mM NaCl) was added to 100 μl of Ni-Sepharose resin (Amersham) and incubated for 1 h at 4°C. The resin was washed twice with MBP Buffer supplemented with 60 mM imidazole. Purified MBP or MBP-ZDP (12 pmol) was incubated at 4°C for 1 h with FL-ROS1 or truncated ROS1 polypeptides bound to resin. The resin was washed twice with MBP buffer supplemented with 60 mM imidazole. Bound proteins were separated by SDS-PAGE (7.5%), transferred to PVDF membranes and analyzed by Western blot using antibodies against MBP (Sigma).
DNA substrates
Oligonucleotides used as DNA substrates (Table S1) were synthesized by Operon and purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5 μM solution of a 5′-fluorescein-labeled or 5′-Alexa Fluor-labeled oligonucleotide (upper strand) with a 10 μM solution of an unlabeled oligomer (lower strand), heating to 95 °C for 5 min, and slowly cooling to room temperature. Annealing reactions for the preparation of the single-nucleotide gapped duplex were carried out at 95 °C for 5 min in the presence of a 2-fold molar excess of unlabeled oligonucleotides with respect to the corresponding labeled oligonucleotides followed by cooling to room temperature.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using an Alexa Fluor-labeled duplex oligonucleotide containing a single-nucleotide gapped, prepared as described above. DNA-binding reaction mixtures (15 μl) contained 100 nM of labeled duplex substrate, 0.1 μM of ROS1 and different amounts of ZDP (0, 0.1, 0.2, 0.4, 0.8, 1 and 1.2 μM) in 10 mM Tris-HCl pH 8.0, 1 mM DTT, 10 μg/ml BSA, 1 mM EDTA. After 60 min incubation at 25°C, reactions were immediately loaded onto 0.2% agarose gels in 1x TAE. Electrophoresis was carried out in 1x TAE for 40 min at 80V at room temperature. Fluorescein-labelled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
Real-time PCR
Five micrograms of total RNA from the indicated genotypes was reversely transcribed to synthesize the first-strand cDNA with Superscript III System (Invitrogen). Real-time PCR was performed using SYBR Green mix on Biorad iQ5. TUB8 was used as an internal control. The primer sequences are listed in Table S2.
Western blotting
Western blot analyses were performed as previously described (Gao et al., 2010). Anti-ZDP antibodies were generated by injecting rabbits with a recombinant ZDP C-terminal portion (amino acids 468-694) that was purified by affinity chromatography.
3′-end cleaning assay
Double-stranded oligodeoxynucleotides (40 nM) were incubated at 30°C for 16 h in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, and 37.5 nM of ROS1 protein in a total volume of 50 μl. Reactions were stopped by adding 20 mM EDTA, 0.6% sodium dodecyl sulphate, and 0.5 mg/ml proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform: isoamyl alcohol (25:24:1) and ethanol precipitated at –20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. Samples were resuspended in a reaction mixture containing 50 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM DTT, 10% glycerol and 1.5 nM of ZDP protein in a total volume of 15 μl at 30°C. Reactions were stopped at the indicated times. When testing 3′-end use by DNA polymerase, reactions products of ROS1 were incubated in a reaction mixture containing 50 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM DTT, 10% glycerol, 10 mM MgCl2, 6 nM of ZDP protein, 0.5 U of human DNA polymerase β (Trevigen) and 0.2 mM of dCTP (except where indicated) in a total volume of 15 μl at 37°C for 30 min. DNA was extracted and precipitated as described above. Samples were resuspended in 10 μl of 90% formamide and heated at 95°C for 5 min. Reaction products were separated in a 15% denaturing polyacrylamide gel containing 7 M urea. Fluorescein-labelled DNA was visualized using the blue fluorescence mode of the FLA-5100 imager and analysed using Multigauge software (Fujifilm).
DNA 3′-phosphatase assay in whole cell extracts
Phosphatase reactions (50 μl) containing 50 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM DTT, 10% glycerol, substrate DNA (20 nM) and 45 μg of extract from wild-type and mutant plants. After incubation at 30°C at the indicated times (0, 10, 30, 60 and 120 min), reactions were stopped by adding 20 mM EDTA, 0.6% SDS and 0.5 mg/ml proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol-precipitated at -20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. Samples were resuspended in 10 μl of 90% formamide, and were heated at 95°C for 5 min. Reaction products were separated in a 15% denaturing polyacrylamide gel containing 7 M urea. Alexa-labeled DNA was visualized in an FLA-5100 imager, and data were analyzed using Multigauge (Fujifilm).
DNA repair/demethylation assay in whole cell extracts
The DNA repair assay used to monitor gap-filling and ligation during DNA demethylation was performed as previously described (Córdoba-Cañero et al., 2009).
MMS and H2O2 sensitivity analysis
Seedlings grown in MS nutrient agar plates for 10 days were transferred to plates containing increasing concentrations of methyl methanesulfonate (MMS) or H2O2 and scored after 2 weeks of further growth.
DNA methylation analysis by CHOP-PCR
DNA methylation was analyzed using methylation-sensitive restriction enzyme BstUI (for transgene RD29A promoter) and BsmAI (for endogenous RD29A promoter). One μg of genomic DNA was digested with BstUI or BsmAI in 20 μl reaction mixture (no enzyme for control). After digestion, PCR was performed using 1 μl of the digested DNA as template in 10 μl PCR system. Primers used are shown in Table S2.
DNA methylation tiling array and analysis of methylated DNA
Genomic DNA was extracted from two-week seedlings using DNeasy Plant Maxi kit (Qiagen), and 2 mg of genomic DNA were used for 5-Methylcytosine immunoprecipitation (mCIP) analysis. The mCIP and amplification of immunoprecipitated products were performed according to published protocols (Zilberman et al., 2007). The hybridization to tiling array chip was performed by NimbleGen (http://www.nimblegen.com/). To identify loci in the zdp-1 mutant where the methylation level was higher than that of wild type, the updated chromosome locations of Nimblegen array probes were found by remapping probes to the TAIR9 genome assembly using SOAP2 (Li et al., 2009). Probe intensities were normalized using the Tukey-biweight scaling procedure implemented in Ringo (method= “nimblegen”) (Toedling et al., 2007). Normalized probe signal differences between the zdp mutant and the wild type were calculated and ordered along each chromosome according to probe positions. The TAMALg algorithm (Bieda et al., 2006) was used to find the hypermethylated regions (by combining L2 and L3 peaks). To remove regions that showed positive signal difference between mutant and wild type samples, but IP DNA was not enriched in either the zdp-1 mutant or wild type plants, the trimmed mean (remove the smallest and largest values) of log2 ratio of IP and input probe signals over each region in the zdp-1 sample were calculated and those with positive trimmed means were selected.
Bisulfite sequencing
One hundred ng of extracted total DNA was analyzed by sodium bisulfite genomic sequencing using BisulFlash DNA Modification Kit (Epigentek; http://www.epigentek.com/catalog/index.php) following the manufacturer's protocol. A 1 μl aliquot of bisulfite-treated DNA was used for each PCR reaction. PCR was performed in 20 μl total volume using ExTaq (Takara; http://www.takarabiousa.com/). Fragments for sequencing were amplified with primers specific for each region (Table S2). Amplified PCR products were subcloned into a pCRII-TOPO vector (Invitrogen; http://www.invitrogen.com/site/us/en/home.html) following the supplier's instructions. For each region, more than eight independent top-strand clones were sequenced from each sample. Sequenced results were calculated by CyMATE v2 (http://www.gmi.oeaw.ac.at/en/cymate-index/cymate-v2/) (Hetzl et al., 2007).
Immunolocalization
Immunofluorescence localization was performed in 2-3 weeks-old leaves as described by Pontes et al., 2006. Nuclei preparations were incubated overnight at room temperature with rabbit anti-ZDP, mouse anti-cMyc (Millipore) and mouse anti-Flag (Sigma). Primary antibodies were visualized using mouse Alexa 488-conjugated and rabbit Alexa-594 secondary antibody at a 1:200 dilution (Molecular Probes) for 2 h at 37°C. DNA was counterstained using DAPI in Prolong Gold (Invitrogen). Nuclei were examined using a Nikon Eclipse E800i epifluorescence microscope equipped with a Photometrics Coolsnap ES.
Supplementary Material
Highlights.
ZDP and ROS1 interact in vitro and co-localize in vivo
zdp mutants show increased promoter DNA methylation and TGS of a reporter gene
Hundreds of endogenous loci are hypermethylated in zdp mutant plants
ZDP functions downstream of ROS1 in one branch of the active DNA demethylation pathway
ACKNOWLEDGMENTS
We thank Carmen Haro Mariscal for help with the MMS-sensitivity assays. We thank Stefania Petrucco (University of Parma, Italy) for the kind gift of the pET-ZDP construct. The work was supported by National Institutes of Health grant R01GM070795 (J-K. Z.), the Spanish Ministry of Science and Innovation and the European Regional Development Fund grant BFU2010-18838 (T.R-A.), and by the Junta de Andalucía grant P07-CVI-02770 (T.R-A.). M-I. M-M. was the recipient of a Ph.D. Fellowship from the Spanish Ministry of Science and Innovation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
We used NimbleGen tiling array to analysis the whole genome methylation status in wild type and zdp mutant plants. The data set was deposited into NCBI (GSE33719).
REFERENCES
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Kushnir S, Van Montagu M, Inze D. The Arabidopsis thaliana apurinic endonuclease Arp reduces human transcription factors Fos and Jun. Proc Natl Acad Sci USA. 1994;91:3299–3303. doi: 10.1073/pnas.91.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti M, Petrucco S, Bolchi A, Dieci G, Ottonello S. A plant 3'-phosphoesterase involved in the repair of DNA strand breaks generated by oxidative damage. J Biol Chem. 2001;276:18038–18045. doi: 10.1074/jbc.M010648200. [DOI] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Córdoba-Cañero D, Dubois E, Ariza RR, Doutriaux MP, Roldan-Arjona T. Arabidopsis uracil DNA glycosylase (UNG) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J Biol Chem. 2010;285:7475–7483. doi: 10.1074/jbc.M109.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba-Cañero D, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009:716–728. doi: 10.1111/j.1365-313X.2009.03994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba-Canero D, Roldan-Arjona T, Ariza RR. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04720.x. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn ASM Press; Washington, D.C.: 2006. [Google Scholar]
- Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009a;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends in genetics : TIG. 2009b;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell research. 2011 doi: 10.1038/cr.2011.140. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell research. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3'-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- Pascucci B, Maga G, Hubscher U, Bjoras M, Seeberg E, Hickson ID, Villani G, Giordano C, Cellai L, Dogliotti E. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucco S, Volpi G, Bolchi A, Rivetti C, Ottonello S. A nick-sensing DNA 3'-repair enzyme from Arabidopsis. J Biol Chem. 2002;277:23675–23683. doi: 10.1074/jbc.M201411200. [DOI] [PubMed] [Google Scholar]
- Ponferrada-Marin MI, Martinez-Macias MI, Morales-Ruiz T, Roldan-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J Biol Chem. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marin MI, Parrilla-Doblas JT, Roldan-Arjona T, Ariza RR. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 2011;39:1473–1484. doi: 10.1093/nar/gkq982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marin MI, Roldan-Arjona T, Ariza RR. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic acids research. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Ariza RR. DNA demethylation. In: Grosjean H, editor. DNA and RNA modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Landes Bioscience; Austin, TX: 2009. pp. 149–161. [Google Scholar]
- Santerre A, Britt AB. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1994;91:2240–2244. doi: 10.1073/pnas.91.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Current biology : CB. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D. The evolving functions of DNA methylation. Curr Opin Plant Biol. 2008;11:554–559. doi: 10.1016/j.pbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.