Abstract
MicroRNAs (miRNAs) have attracted attention because of their key regulatory functions in many biological events, including differentiation and tumorigenesis. Recent studies have reported the existence of a reciprocal regulatory loop between the family of let-7 miRNAs and an RNA-binding protein, Lin28, both of which have been documented for their important roles during cell differentiation. Hence, using bipotent K562 human leukemia cells and human CD34+ hematopoietic progenitor cells as research models, we demonstrate that let-7 and Lin28 have contrary roles in megakaryocytic (MK) differentiation with a dynamic balance; expression of miR-181 is capable of effectively repressing Lin28 expression, disrupting the Lin28–let-7 reciprocal regulatory loop, upregulating let-7, and eventually promoting MK differentiation. However, miR-181 lacks a significant effect on hemin-induced erythrocyte differentiation. These results demonstrate that miR-181 can function as a ‘molecular switch' during hematopoietic lineage progression specific to MK differentiation, thus providing insight into future development of miRNA-oriented therapeutics.
Keywords: MicroRNA, megakaryocytic differentiation, Lin28, let-7
Let-7 is the second microRNA (miRNA) that was discovered after lin-4, and it has been shown to have roles in modulating the multiple stages of development in Caenorhabditis elegans and Drosophila.1 Often, multiple copies of let-7 are present in a genome, which results in the transcriptional difference between organisms, although mature let-7 is highly conserved across species from worms to humans.2 In C. elegans, temporal regulation of let-7 occurs at the transcription level,3, 4, 5 and expression of primary (pri-) and precursor (pre-) let-7s occurs concurrently with the appearance of mature let-7 at a later developmental stage.5, 6 In mammals, mature let-7s are undetectable in embryonic stem cells until differentiation, and are highly expressed in various adult tissues.7, 8, 9 Lin28 is a conserved cytoplasmic protein that regulates developmental timing in C. elegans.10 In mammals, Lin28 is a pluripotent marker that is ubiquitously expressed in embryonic stem cells. Likely, Lin28 functions in a manner opposing let-7 in cell differentiation. Studies identified that Lin28 maintains cell pluripotency partially by repressing the biogenesis of mature let-7.8, 9, 11, 12 When combining the conserved ‘GGAG' motif within the terminal loop of pre-let-7, Lin28 recruits terminal uridylyl transferase-4 (TUT4) to add uracil to the 3′ end of pre-let-7. This modification results in pre-let-7 degradation and blockade of let-7 maturation.11, 12 However, a recent report demonstrated that let-7 is also capable of promoting neural stem cell differentiation by repressing the expression of Lin28.13 These findings imply a reciprocal regulatory interaction between let-7 and Lin28 during cell differentiation.
In this study, we consider the Lin28–let-7 regulatory loop as a feedback circuit in the process of differentiation. We hypothesize that Lin28 is dominantly expressed and represses the maturation of let-7 in stem or progenitor cells. When differentiation occurs, let-7 will be upregulated and further repress Lin28 expression to maintain differentiation. Given that such a loop exists in balance, it is unknown how it would be disrupted when differentiation is initiated and whether an initial ‘molecular switch' promptly relieves the inhibitory effect of Lin28 on let-7. Therefore, we select hematopoiesis as a research model and explore the qualification of miRNAs as such a ‘switch'. Our results show that miR-181 promotes megakaryocytic (MK) differentiation by breaking down the regulatory barrier of Lin28 on the maturation of let-7, suggesting that miR-181 may have a ‘trigger' role in promoting MK hematopoiesis.
Results
The interaction of Lin28 with let-7 influences MK differentiation
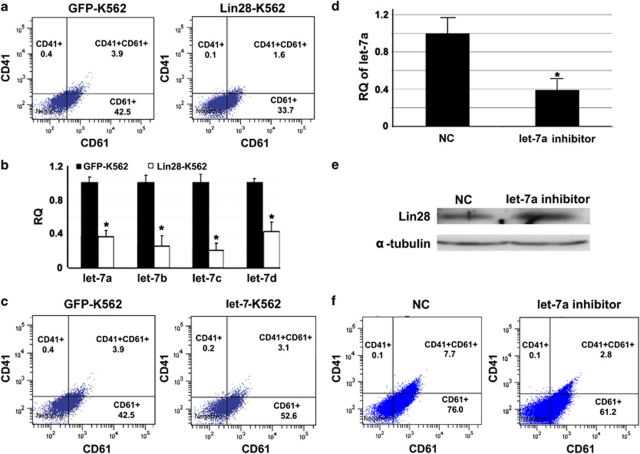
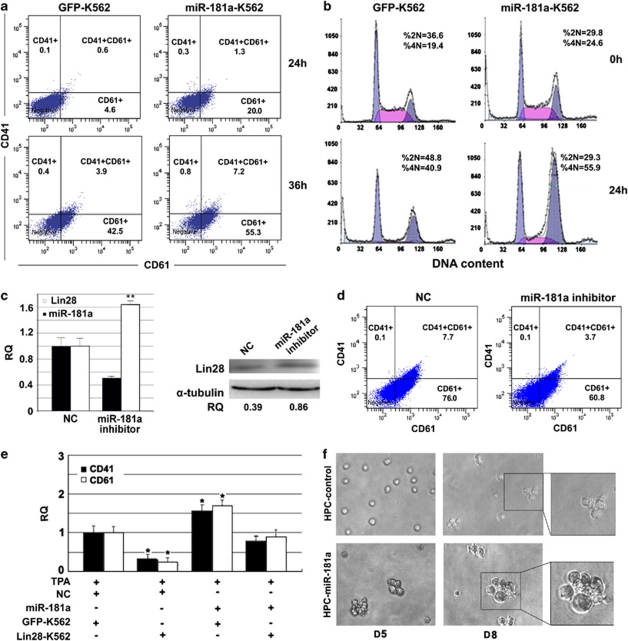
Lin28 is known to inhibit let-7 maturation, and let-7 can inversely repress Lin28 expression. Both have been shown to have key roles in cell differentiation and development,14, 15 but no reports have discussed their co-effects on hematopoiesis to date. In this study, we first examined whether Lin28 can repress MK differentiation by using a well-established model, bipotent K562 human leukemia cells induced by phorbol-12-myristate-13-acetate (TPA).16 After constructing a green fluorescent protein (GFP) fusion protein and successfully expressing Lin28 in K562 cells (Supplementary Figure S1), we induced MK differentiation by incubating the cells with TPA for 36 h. CD41 and CD61 are the integrin complexes on the cell surface, and their expressions are key signs of megakaryocytopoiesis.16 Flow cytometry results showed that Lin28-K562 cells had less CD41/61-positive cells than control cells (35.4% versus 46.8% Figure 1a). In parallel, let-7a, b, c, and d were significantly downregulated (P<0.05) in cells overexpressing Lin28 (Figure 1b). These results demonstrate that Lin28 repressed MK differentiation as well as expression of mature let-7s.
Figure 1.
The regulatory loop of Lin28 and let-7 in the TPA-K562 cell model toward MK differentiation. (a) After 36 h of TPA induction, K562 cells with overexpressed Lin28 have a lower CD41/CD61-positive rate than the GFP control cells (35.4 versus 46.8%) as assessed by flow cytometry. (b) Overexpression of Lin28 significantly blocked the maturation of the let-7 family (let-7a, b, c, and d) as assessed by qRT-PCR. (c) After 36 h of TPA induction, K562 cells with the mutated let-7a transcript have a greater CD41/CD61-positive rate than the GFP control cells (55.9% versus 46.8%) as assessed by flow cytometry. (d) Transiently transfecting the designated inhibitor effectively downregulated the expression of let-7a when compared with the control. (e) Applying the let-7 inhibitor inversely upregulated the expression of Lin28 as shown by western blotting. (f) The let-7 inhibitor consequently retarded the transition of the MK lineage as evidenced by decline of CD41/CD61-positive rate when compared with the control (61.4% versus 83.8%). CD41 and CD61 were assessed by flow cytometry after 60 h of TPA treatment when the let-7 inhibitor was optimized to have the maximal inhibitory effect (CD41/CD61-positive rate is computed by adding the rates of CD41+, CD41+/CD61+, and CD61+). The error bars represent standard deviation (S.D.). t-tests were used for statistical analysis; * indicates P<0.05. RQ stands for relative quantitation and NC means non-target control
It is known that Lin28 can bind to the ‘GGAG' motif within the terminal loop of pre-let-7 to inhibit the maturation of let-7.12 In order to eliminate the post-transcriptional control of Lin28, we strategically mutated this binding motif on pre-let-7a-1 by substituting CCGC for GGAG (Supplementary Figure S2A). The modified transcript was transfected into K562 cells through a lentivector, and quantitative real-time PCR (qRT-PCR) results demonstrated upregulation of mature let-7a (Supplementary Figure S2B). Consequently, elevated let-7a resulted in the decline of Lin28 at mRNA and protein levels (Supplementary Figures S2C and D), which is consistent with the previous report.13 Moreover, let-7b, c, and d were sequentially upregulated resulting from repression of Lin28 by let-7a (Supplementary Figure S2E).
As stated earlier, upregulation of mature let-7 is one of the signs of cells developing to adult tissues.7, 8, 9 Here, we will investigate the function of let-7 prompting hematopoietic differentiation. After TPA treatment for 36 h, let-7a-transfected K562 cells showed higher CD41/61-positive rate than control cells (55.9% versus 46.8% Figure 1c). By adopting specific inhibitors, we successfully downregulated let-7a expression over two-fold in K562 cells (Figure 1d). As expected, western blot results showed that expression of Lin28 increased accordingly (Figure 1e). Moreover, the let-7a inhibitor significantly retarded MK differentiation in K562 cells. The positive rate of CD41/61 in K562 cells transiently transfected with the let-7a inhibitor was 61.4% compared with that in the non-target oligo-transfected control cells, which was 83.8% (Figure 1f). These results suggest that the Lin28–let-7 regulatory loop may be an important mechanism to modulate MK differentiation during hematopoiesis.
Lin28 is the direct target of miR-181
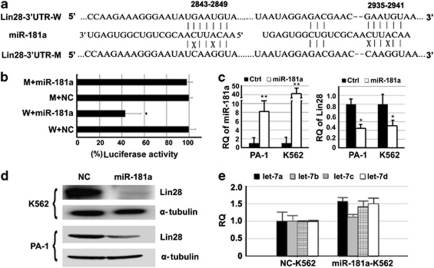
According to TargetScan, an online miRNA target prediction interface,17, 18 Lin28 is one of the putative target genes of miR-181a and b; both miRNAs have an identical seed sequence complementary to the binding site on the 3′-UTR of Lin28. Using luciferase reporter assay, we demonstrated that Lin28 is the direct target of miR-181a. First, we constructed wild-type and mutant Lin28 3′-UTR fragments (Figure 2a), and cloned them into pMIR-REPORT vectors. Second, we co-transfected Lin28 constructs and miR-181a mimics into HCT116 cells, a colorectal cancer cell line used intensively in our lab. Overexpression of miR-181a significantly decreased the luciferase activity of the reporter containing the wild-type 3′-UTR of Lin28 by approximately 60% (P<0.05) when compared with the mutant form and the controls (Figure 2b). To examine the effect of miR-181a to directly suppress expression of Lin28, we transfected miR-181a into K562 cells using the lentivector. Human ovarian cancer PA-1 cells with endogenous Lin28 were used as the second model to duplicate the interaction between miR-181a and Lin28. Figure 2c illustrates an upregulation of miR-181a and a dramatic decrease in the stability of Lin28 mRNA in both K562 and PA-1 cells, as measured by qRT-PCR assays. The suppressive effects of miR-181a on the expression of Lin28 are also confirmed by western blotting (Figure 2d). Given that Lin28 inhibits the maturation of let-7, overexpression of miR-181a should indirectly promote the expression of mature let-7 resulting from the decline of Lin28. As expected, our results show an upregulation of the let-7 family in miR-181a-K562 cells when compared with the control cells (Figure 2f), supporting that miR-181 has the potent capability of disrupting the Lin28–let-7 regulatory loop.
Figure 2.
Lin28 is the direct target of miR-181. (a) The design scheme for the wild-type and mutant fragments of the Lin28 3′-UTR. (b) Relative luciferase activity indicated direct binding of miR-181a to the Lin28 3′-UTR. (c) Verification of overexpression of miR-181a and its effects on the stability of Lin28 in lentivector-transfected K562 and PA-1 cells using qRT-PCR. (d) Western blotting demonstrated repression of miR-181a on Lin28 expression in both K562 and PA-1 cells. (e) Overexpression of miR-181a promoted the maturation of the let-7-family miRNAs in K562 cells. The error bars represent S.D. t-tests were used for statistical analysis; * indicates P<0.05 and ** indicates P<0.01
The expression patterns of miR-181, let-7, and Lin28 during MK differentiation in both K562 and CD34+ HPCs
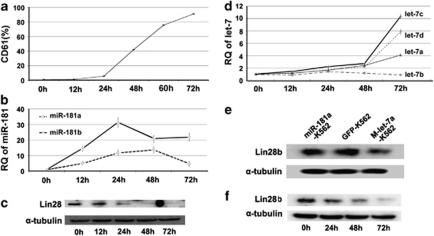
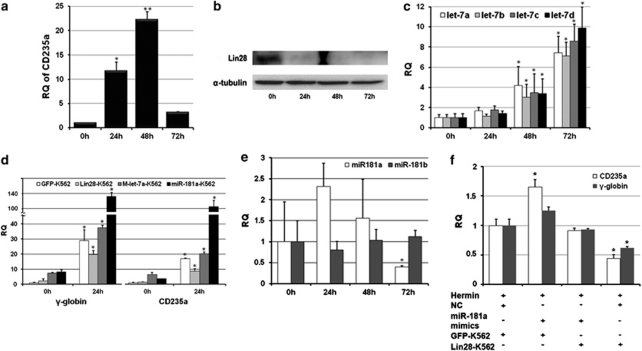
Using the K562-TPA model, we investigated the successive changes of endogenous miR-181, let-7, and Lin28 to study their interactions during MK differentiation. First, using flow cytometry, we monitored CD61 expression in K562 cells after TPA treatment and results showed that CD61 increased immediately after 0 h till peaking at 72 h (Figure 3a). Second, we examined expression of miR-181a and b by qRT-PCR and found that both miRNAs reached their peaks at 24 and 48 h, respectively (Figure 3b). As upregulation of miR-181 is capable of inhibiting the expression of Lin28, we found that Lin28 expression significantly decreased at 24 h and became undetectable by western blotting after 48 and 72 h of TPA treatment (Figure 3c). let-7a, c, and d were upregulated at 48 h, and levels peaked at 72 h, although let-7b showed minimal alteration throughout the time course (Figure 3d). These results show the successive changes of miR-181, Lin28, and let-7 during MK differentiation.
Figure 3.
The expression patterns of miR-181, let-7, and Lin28 during megakaryocyte differentiation in K562 cells. (a) Expression of the MK lineage marker CD61 increased in K562 cells during the course of TPA treatment as assessed by flow cytometry. (b) Alteration of miR-181a and b in K562 cells induced to MK differentiation by TPA. (c) Lin28 protein expression declined in K562 cells accompanied by MK progression. (d) Alteration of the let-7 family in K562 cells induced to MK differentiation by TPA. (e) let-7a, but not miR-181a, targeted Lin28b, the homolog of Lin28, in K562 cells as assessed by western blotting. (f) Decline of Lin28b accompanied by upregulation of let-7 in the transition of the MK linage as assessed by western blotting. The error bars represent S.D.
We noticed that upregulation of let-7 family was obviously retarded when compared with relevant changes of miR-181 and Lin28 in K562 cells induced to MK differentiation. We think that it may be attributable to Lin28b, which is the homolog of Lin28 and also inhibits the maturation of the let-7 family. K562 cells were known to express both Lin28 and Lin28b, and knocking down Lin28b was shown to elevate the level of mature let-7 and promote K562 cell-to-MK differentiation.19 Unlike let-7, miR-181a cannot repress the expression of Lin28b (Figure 3e). Therefore, when Lin28 and Lin28b together block let-7 maturation in K562 cells, miR-181 downregulating Lin28 weakens this inhibitory effect and leads to upregulation of mature let-7. The accumulated mature let-7 gradually relieves the blockade of both Lin28 and Lin28b on its maturation, which results in increase of let-7 in a postponed manner. Figure 3f shows that the decline of Lin28b in K562 cells occurred after 48 h of TPA treatment when let-7 began to increase steeply. These results strongly support our hypothesis that elevated miR-181 is capable of acting as a ‘molecular switch' to disrupt the feedback loop between Lin28 and let-7 by first downregulating Lin28 and then upregulating mature let-7.
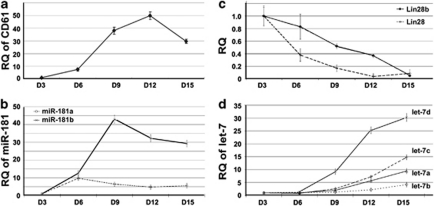
Human umbilical cord blood CD34+ progenitor cells (HPCs) treated with a CC220 cytokine cocktail were used to validate the results obtained from the K562-TPA model. CC220 contains a combination of recombinant human cytokines (thrombopoietin, stem cell factor, IL-6, and IL-9) formulated to direct the specific expansion of MK progenitors and the production of MK and platelets in cultures of human hematopoietic cells. Our results showed that CD61 expression peaked at day 12 during the time course of the CC220 treatment (Figure 4a), whereas miR-181a and b had the highest expression levels on days 6 and 9, respectively (Figure 4b). Lin28 and Lin28b reached their lowest levels on days 12 and 15 (Figure 4c); the let-7-family expression peaked by the end of day 15 (Figure 4d). Here, HPCs show a pattern similar to the successive changes and the interaction of miR-181, Lin28, and let-7 in K562 cells during MK differentiation. At day 6, although upregulation of miR-181a and b decreased Lin28 expression approximately 60%, mature let-7s were not yet obviously upregulated because of the abundance of Lin28 and/or existence of Lin28b. In the time period from day 6 to 9, steeply increased miR-181a repressed the expression of Lin28 leading to accumulation of mature let-7s, which further decreased the expression of Lin28 and Lin28b. Although miR-181a and b decreased from day 9 to 15, the elevation of let-7s continuously enhanced the steady decline of Lin28 and Lin28b. Together with the results from the TPA-K562 model, we demonstrate that miR-181 is capable of mediating MK differentiation by repressing Lin28 and consequently upregulating let-7.
Figure 4.
The expression patterns of miR-181, let-7, and Lin28 during megakaryocyte differentiation in CD34+HPCs as assessed by qRT-PCR. (a) CD61 expression increased in HPCs during the course of cytokine cocktail stimulation. (b) Alteration of miR-181a and b in HPCs toward MK differentiation. (c) Lin28 and Lin28b were differentially downregulated in HPCs accompanied by MK progression. (d) Alteration of the let-7 family in HPCs toward MK differentiation. The error bars represent S.D.
MiR-181a promotes MK differentiation
To explore the role of miR-181 in MK differentiation we transfected miR-181a into K562 cells and CD34+ HPCs using lentivectors. Supplementary Figure S3 shows successful overexpression of miR-181a in both cell types. At 24 and 36 h after TPA treatment, there were 21.6% and 63.3% CD41/61-positive cells in the miR-181a-transfected K562 cells versus 5.3% and 46.8% in the control cells, respectively (Figure 5a), which implies that miR-181 is capable of promoting MK differentiation. Another indicator of MK differentiation is polyploidization, also known as endomitosis or endoreduplication, which is a variant of mitosis without nuclear or cellular division. This event occurs in cells that contain many copies of an individual chromosome inside a single nucleus, which is recognized as a key sign of early megakaryoblasts.20 Flow cytometry results showed that miR-181a could significantly increase the number of cells undergoing endomitosis over the control (Figure 5b), which is consistent with a previous report in which nuclear DNA ploidy of K562 cells could rise to 4N (and even up to 16N) accompanied by increased cell volume in response to TPA treatment.21 Additionally, we observed the inverse effect of downregulation of miR-181a on MK differentiation. By transiently transfecting the miR-181-specific inhibitor, we successfully suppressed the expression of miR-181a over two-fold in K562 cells, and accordingly Lin28 overexpression was measured at both mRNA and protein levels (Figure 5c). Moreover, the miR-181a inhibitor significantly retarded MK differentiation as assessed by measuring CD41/61-positive rate with a flow cytometer (Figure 5d, control (83.8%) versus miR-181 inhibitor (64.6%)).
Figure 5.
MiR-181a promoted MK hematopoiesis. (a) Flow cytometry results showed that the percentage of CD41/CD61-positive cells in miR-181 stable K562 cells increased to 21.6% at 24 h and 63.3% at 36 h after TPA induction when compared with GFP control cells (5.3% and 46.8%, respectively). (b) Flow cytometry results showed that the percentage of miR-181a-K562 cells undergoing endomitosis after TPA treatment increased. (c) Transiently transfecting the designated inhibitor effectively downregulated the expression of miR-181 and upregulated Lin28 when compared with the controls as assessed by qRT-PCR and western blotting. Relative quantitation (RQ) of western blotting was computed by dividing the density values of Lin28 blots to that of α-tubulin blots, which was measured by the software GelQuantNET (http://biochemlabsolutions.com/GelQuantNET.html). The result shows that the miR-181a inhibitor is capable of upregulating the expression of the Lin28 protein 2.3-fold over the control (0.86/0.23). (d) The miR-181a inhibitor consequently retarded the transition of the MK lineage as evidenced by the decline of CD41/CD61-positive rate when compared with the control (64.6% versus 83.8%). (e) Lin28 is the direct target of miR-181a for driving K562 cells toward MK differentiation, and its overexpression abolished the effect of miR-181a on the process of MK differentiation. (f) Morphological changes of miR-181a-HPCs and control cells after CC220 induction toward MK differentiation. MiR-181a-HPCs showed early adhesion and larger nuclei than the control cells as observed by day 5 and 8. (CD41/CD61-positive rate is computed by adding the rates of CD41+, CD41+/CD61+, and CD61+). The error bars represent S.D. t-Tests were used for statistical analysis; *indicates P<0.05 and **indicates P<0.01. RQ stands for relative quantitation and NC means non-target control
In order to further study the interaction between miR-181a and Lin28 during the process of MK differentiation, we transiently transfected miR-181a mimics into characterized K562 cells induced by TPA treatment, as shown in Figure 5e. Consistent with the results from miR-181a-K562 cells shown earlier (Figure 5a), miR-181a mimics effectively induced MK differentiation as assessed by the relative quantity of CD41 and CD61, whereas the Lin28-K562 cells maintained a multipotent status with low CD41 and CD61 levels. Transfecting miR-181a mimics into Lin28-K562 cells increased CD41 and CD61 by abolishing the inhibitory effect of Lin28 on MK differentiation. Although miR-181a mimics cannot fully rescue the MK induction in Lin28-K562 cells resulting from Lin28 abundance, our results support that Lin28 is a direct target of miR-181a and silencing Lin28 is a key step for miR-181a to promote MK differentiation in K562 cells. Moreover, we confirmed that upregulation of miR-181a is not the direct response of cells to TPA treatment. Treating four cancer cell lines with TPA, namely MDA-MB-231 (breast cancer), HCT116 (colon cancer), MG63 (osteosarcoma), and U2OS (osteosarcoma), we did not find any significant changes of miR-181a expression in any of them (Supplementary Figure S4), suggesting that miR-181a upregulation is one of many sequential steps toward MK differentiation rather than a direct response to TPA treatment.
We also found that miR-181a overexpression promoted the progression of MK differentiation in HPCs. After CC220 treatment, HPCs that normally grow in suspension gave rise to adherent cells with enlarged nuclei and spreading potential similar to normal megakaryocytes.22, 23, 24 Upon observation on day 5 and 8, HPCs with miR-181a show these MK morphological phenotypes earlier than the control HPCs (Figure 5f), which demonstrates that miR-181 is capable of accelerating MK differentiation. Taking advantage of CD34+ HPCs containing committed progenitors and pluripotent stem cells, we further investigated the function of miR-181a in the transition of the MK linage. Flow cytometry results showed that, when induced to MK differentiation, miR-181a-HPCs have a higher percentage of CD34−CD61+ cells than control HPCs (Table 1). The CD34+CD61+ subsets are regarded as MK progenitors and CD34−CD61+ are the major characteristics of megakaryocytes.25, 26, 27 Our results showed that, on day 6, the ratio of CD34−CD61+/CD34+CD61+ cells is 4.07 in miR-181a-HPCs versus 1.96 in control HPCs, which implies that miR-181a may promote the development of MK progenitors (CD34+CD61+) to megakaryocytes (CD34−CD61+) instead of initiating MK differentiation, as shown in Table 1.
Table 1. Characterization of CD61+ cells in HPCs toward MK differentiation by flow cytometry.
|
Day-3 |
Day-6 |
Day-12 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
CD61+ (%) |
CD61+ (%) |
CD61+ (%) |
|||||||
| HPC | CD34+ | CD34− | Total | CD34+ | CD34− | Total | CD34+ | CD34− | Total |
| Control | 13.0 | 3.0 | 16.0 | 23.0 | 45.0 | 68.0 | 12.0 | 87.0 | 99.0 |
| miR-181a | 11.0 | 6.0 | 17.0 | 14.0 | 57.0 | 71.0 | 5.0 | 94.0 | 99.0 |
MiR-181a is a non-specific factor for erythroid differentiation in K562 cells
MiR-181a can mediate MK differentiation in K562 cells by directly targeting Lin28 and preventing the reciprocal regulatory loop between Lin28 and let-7. We further elucidated this regulatory mechanism in erythroid differentiation. After inducing K562 cells with hemin, the erythroid marker CD235a was increased and reached its peak at 48 h (Figure 6a), which indicated successful induction of erythroid differentiation. In parallel, Lin28 accordingly decreased (Figure 6b) and the let-7 family steadily increased in number (Figure 6c). When Lin28 was overexpressed, expression of CD235a and γ-globin was decreased (Figures 6d and f), but let-7 inversely promoted the expression of these two erythroid markers (Figure 6d). These results are similar to those seen with MK differentiation and provide further evidence that the feedback loop between Lin28 and let-7 is also involved in erythroid differentiation.
Figure 6.
MiR-181a is a non-specific factor for erythroid differentiation. (a) The expression time course of CD235a in K562 cells after hemin induction. (b) Lin28 expression declined along with induction of erythroid differentiation. (c) The let-7-family expression was upregulated in the later stages of erythroid differentiation, as measured by qRT-PCR. (d) The erythroid-specific markers CD235a and γ-globin showed a lower level of expression in Lin28-K562 cells and a higher level of expression both in let-7a-K562 and miR-181a-K562 cells as compared with the controls when induced toward erythroid differentiation, as measured by qRT-PCR. (e) Results from qRT-PCR show that miR-181a and b lacked significant changes when K562 cells were driven toward erythroid differentiation. (f) Lin28 was the direct target of miR-181a for driving K562 cells toward erythroid differentiation as evidenced by the finding that overexpression of Lin28 abolished the effect of miR-181a on the erythroid differentiation process. The error bars represent S.D. t-tests were used for statistical analysis; * indicates P<0.05 and ** indicates P<0.01
In K562 cells that stably overexpressed miR-181a, CD235a and γ-globin were significantly elevated in response to hemin induction (Figure 6d). Lin28 is also the direct target of miR-181a in the process of erythroid differentiation, as evidenced by attenuation of the positive effect of miR-181a on cell differentiation by Lin28 overexpression (Figure 6f). Surprisingly, miR-181, especially miR-181b, maintained relatively steady expression levels across the course of hemin treatment (Figure 6e), which implies that miR-181 might not be a natural ‘molecular switch' to turn off the inhibitory effect of Lin28 during erythroid differentiation. Nevertheless, the non-specific impact of miR-181 may point to a potential application for miRNAs in cancer therapeutic applications through promotion of cell differentiation.
Discussion
Hematopoiesis is a life-long, highly regulated, multistage process in which self-renewing hematopoietic stem/HPCs give rise to all blood cell lineages. In this study, we showed that Lin28 expression was decreased whereas let-7 expression inversely increased during the MK differentiation process. A mutated pre-let-7a transcript can bypass the post-transcriptional control of Lin28, resulting in promotion of MK differentiation as well as an increase at the expression levels of other let-7s by downregulating Lin28. Moreover, the interactivity of Lin28 and let-7 has a similar impact on erythroid differentiation in K562 cells. Together with previous studies,8, 9, 11, 12 our findings demonstrate that Lin28 and let-7 have important roles in hematopoietic differentiation.
MiRNAs have been demonstrated to have important roles in many biological events such as development, differentiation, apoptosis, and tumorigenesis. Single miRNAs have been shown to regulate hundreds to thousands of cognate genes at the translational level; likewise, a single gene can be targeted by multiple miRNAs. For example, the 3′-UTR of Lin28 mRNA includes as many as 3270 nucleotides that provide plenty of potential binding sites for miRNA regulation. TargetScan predicts that there are 15 miRNA-binding sites within the conserved 3′-UTR sequence of Lin28 in all vertebrates, and that 195 miRNAs potentially target human Lin28. This prediction suggests that individual miRNAs may regulate Lin28 to achieve specific cell lineages in different manners. Lin-4 was the first characterized miRNA, and it has also been shown to bind to the complementary sequence within the 3′-UTR of Lin28 mRNA and regulate its stage-specific activity. Deletion of the lin-4 complementary sequence produces a dominant gain-of-function phenotype of Lin28 and causes delayed development in animals.28 Additionally, elevated expression of miR-125 during human neuronal differentiation of embryonic carcinoma cells was found to be attributed to the downregulation of Lin28 by binding to two conserved miR-125-responsive elements within the 3′-UTR of Lin28, which caused a reduction in the translational efficiency and mRNA abundance.29 In this study, we first report that miR-181 can target Lin28. During MK differentiation, miR-181 alters ahead of Lin28 and let-7; increased expression of miR-181 may promote cell differentiation by downregulating Lin28 and sequentially elevating let-7. MiR-181 is preferentially expressed in the B-lymphoid cells of the mouse bone marrow, and ectopic expression in hematopoietic stem/HPCs was shown to lead to an increased fraction of B-lineage cells in tissue culture differentiation assays and in adult mice.30 Expression of miR-181 generally decreases, thus causing its target gene, T-cell leukemia/lymphoma-1 (TCL1), to be expressed in B-cell chronic lymphocytic leukemia (B-CLL) samples. Overexpression of the oncogene TCL1 is reportedly a critical causal event in the pathogenesis of the aggressive form of B-CLL.31, 32 The question whether Lin28 involves the effects of miR-181 in this mouse model requires further investigation, although these results support the importance of miR-181 in cell differentiation.
Recently, evidence provided by miRNA array profiling data demonstrated elevation of miR-181a and miR-181b during MK differentiation of K562 cells; however, the function and mechanism of miR-181 on MK hematopoiesis remained unknown.33 In this study, using K562 cells and CD34+ HPCs, we verified that upregulated miR-181a is capable of specifically promoting MK hematopoiesis, especially involving the transition of MK progenitors to megakaryocytes, through disruption of the feedback circuit between Lin28 and let-7. However, miR-181 was obviously not responsible for inducing erythroid differentiation, which suggests that it is not a specific factor in erythroid hematopoiesis like Lin28 and let-7.
The oncogenic characteristics of Lin28 and its homolog, Lin28b, have been recognized because of the poor prognosis in different human cancers associated with their elevated expression.19, 34 Lin28 can also stimulate cellular transformation,19 facilitate cancer cell metastasis,34 and improve chemotherapeutic drug resistance by interfering with oct4.35, 36 It has been noticed that expression of an oncogene, c-Myc, is associated with frequent absence of let-7 in most of the prevalent human cancers,37, 38 and restoration of let-7 is projected to prevent tumorigenesis.14 Taken together, Lin28 seems to have a central role as a pluripotent factor in maintaining a low differentiation stage, which is essential for tumorigenesis. In this study, Lin28 has demonstrated a significant impact on both MK and erythroid differentiation. As presented above, inhibition of Lin28 using let-7, miR-181, or other miRNAs can assist cells in moving toward high differentiation stages after induction. Unlike let-7, miR-181 may not be a specific marker that is naturally involved in erythroid differentiation; however, enforced elevation of miR-181a expression in K562 cells still effectively downregulated Lin28 and drove cells toward erythroid differentiation. This result demonstrates the feasibility of targeting Lin28 using specific or non-specific agents such as miRNAs for future therapeutic use.
In summary, our results demonstrate that miR-181 functions as a ‘molecular switch' to turn on the Lin28 and let-7 circuit in the maintenance of cell differentiation. This study will improve our understanding of the mechanisms of miRNAs' diverse involvement in cellular processes at different levels and emphasize future miRNA-oriented targeting therapy.
Materials and Methods
Cell culture
The K562, PA-1, HCT116, MDA-MB-231, MG63, U2OS, and 293TN cell lines were incubated at 37 °C in a humidified atmosphere of 5% CO2 using various media supplemented with 10% (v/v) fetal bovine serum (HyClone, Logan, UT, USA). Details are provided in Supplementary Table S1. Frozen human umbilical cord blood CD34+ progenitor cells and the associated reagents, including StemSpan SFEM defined medium and the cytokine cocktail CC220, were purchased from StemCell Technologies (Vancouver, BC, Canada). CC220 contains a combination of recombinant human cytokines (thrombopoietin, stem cell factor, IL-6, and IL-9) formulated to support the expansion of megakaryocyte progenitors and the production of megakaryocytes and platelets in cultures of human hematopoietic cells. TPA and hemin (Sigma-Aldrich, St. Louis, MO, USA) were used for induction of MK and erythroid differentiation by using concentrations of 15 μM and 25 μM, respectively.
Lentivector construction, packaging, and cell infection
Target fragments containing the sequences for pre-miR-181a, pre-let-7a, or Lin28 ORF were ligated with the self-inactivating transfer vector plasmid pWPXL. The primers used for construction, reverse transcription (RT), and PCR are summarized in Supplementary Table S2. Two micrograms of each construct were co-transfected into 293TN cells using 10 μg of a packaging plasmid mixture (System Bioscience, Mountain View, CA, USA) according to the manufacturer's instructions. The lentivirus-containing supernatants were collected, filtered, and concentrated by PEG precipitation (System Bioscience) at 48 h after transfection. To establish cells that stably overexpressed the target genes, K562 cells were infected with the respective lentivirus and then sorted at least 72 h after infection using a BD Biosciences (Bedford, MA, USA) FACS Aria II cell sorter. A total of 3 × 105 CD34+ hematopoietic progenitor cells (StemCell Technologies) were infected with a high-titer, miR-181a-expressing lentivirus overnight. Cells were then rinsed with phosphate-buffered saline (PBS) and treated with the CC220 cytokine cocktail to induce MK differentiation.
Quantitative real-time PCR
Total RNA was isolated by using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to our established protocols.39 Two micrograms of total RNA were used for cDNA synthesis using a high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Foster City, CA, USA) in accordance to the conditions described previously.39 Random primers were used for mRNA RT, and specific primers were pre-designed for the miRNA RT reactions. qRT-PCR was performed using a Bio-Rad IQ-5 real-time PCR System (Bio-Rad, Hercules, CA, USA) using a SYBR master mix (Roche, Indianapolis, IN, USA). The reaction mixture containing 10 μl of 2 × SYBR master mix, 1 μl of primers, 1 μl of cDNA, and 7 μl of nuclease-free water underwent 30 PCR cycles consisting of denaturation for 10 s at 94 °C and annealing and prolongation for 30 s each at 58 °C. Gene expression was analyzed by the 2−ΔΔCT method.40 GAPDH and U6 were used as the endogenous controls for relative quantitation analyses for mRNA and miRNA, respectively.
Luciferase reporter assay
Luciferase reporter plasmids were constructed by inserting the target 3′-UTRs of Lin28 downstream from the luciferase gene in the pMIR-REPORT vector (Ambion, Austin, TX, USA). HCT116 cells at a density of 1.2 × 104 per well in 24-well plates were co-transfected with pMIR-REPORT luciferase reporters and miRNA precursors (Ambion) using the Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours after transfection, cells were harvested and lysed with passive lysis buffer (Promega, Madison, WI, USA). Luciferase activity was measured by using a dual luciferase reporter assay (Promega).
Differentiation determination
The specific markers for hematopoietic lineage were examined by both qRT-PCR and flow cytometry. Previously described protocols were used for qRT-PCR. For flow cytometry, cells were immunostained with 1 μg of CD34-PE-Cy7, CD41-Pacific Blue, and CD61-AF647 antibodies (BioLegend, San Diego, CA, USA) and analyzed on a BD Biosciences FACS Canto II flow cytometer. To detect the formation of a polyploid nucleus during MK differentiation, cells were fixed with 75% ethanol, resuspended in cold PBS, and stained with 0.5 mg/ml propidium iodide for 30 min after RNaseA treatment. Finally, intracellular DNA was measured by flow cytometry using the FACS Canto II instrument.
Modulation of miR-181a and let-7
For overexpression of miR-181a, K562 cells were transfected with 5 μl of 10 μM pre-miR-181a or with control pre-miR oligonucleotides (Ambion). For inhibition of miR-181a and let-7a, K562 cells were transfected with 15 nM LNA-181a, LNA-let-7a, and a non-target LNA-control (Exiqon, Woburn, MA, USA). K562 cells were transfected with LNA or pre-miR molecules using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Briefly, 1 μl of Oligofectamine was mixed with 249 μl of Opti-MEM (Invitrogen) and incubated for 10 min at room temperature. LNA or pre-miR molecules were mixed with 240 μl of Opti-MEM and incubated for 10 min at room temperature. The Oligofectamine and the LNA or pre-miR mix were pooled and allowed to stand for 15 min at room temperature before adding the complex into a six-well plate. Cells were then incubated at 37 °C for the designated time period.
Western blotting
Equal amounts (25 μg) of samples were resolved by SDS-PAGE on 10% gels. Proteins were probed using rabbit anti-Lin28 and anti-Lin28b polyclonal antibodies (Abcam, Cambridge, MA, USA) and a mouse anti-α-tubulin monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), then incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). Proteins were visualized by using a chemiluminescence detection system, using the Super Signal substrate (Thermo Scientific, Rockford, IL, USA).
Acknowledgments
This study was supported by institutional start-up funding awarded to Dr. Yaguang Xi. We thank Dr. Ming Tan, Dr. Aparna Mitra, Ms Amy Brown, and Mrs Caitlin Wainscott for valuable discussion and editorial assistance.
Glossary
- miRNA
microRNA
- TUT4
terminal uridylyl transferase-4
- MK
megakaryocytic
- TPA
phorbol-12-myristate-13-acetate
- GFP
green fluorescent protein
- HPC
hematopoietic progenitor cell
- TCL1
T-cell leukemia/lymphoma-1
- B-CLL
B-cell chronic lymphocytic leukemia
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Guo J, Li ZC, Feng YH. Expression and activation of the reprogramming transcription factors. Biochem Biophys Res Commun. 2009;390:1081–1086. doi: 10.1016/j.bbrc.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Alitalo R. Induced differentiation of K562 leukemia cells: a model for studies of gene expression in early megakaryoblasts. Leuk Res. 1990;14:501–514. doi: 10.1016/0145-2126(90)90002-q. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano JE, Jr, Shivdasani RA. Megakaryocytes and beyond: the birth of platelets. J Thromb Haemost. 2003;1:1174–1182. doi: 10.1046/j.1538-7836.2003.00290.x. [DOI] [PubMed] [Google Scholar]
- Leary JF, Ohlsson-Wilhelm BM, Giuliano R, LaBella S, Farley B, Rowley PT. Multipotent human hematopoietic cell line K562: lineage-specific constitutive and inducible antigens. Leuk Res. 1987;11:807–815. doi: 10.1016/0145-2126(87)90065-8. [DOI] [PubMed] [Google Scholar]
- Whalen AM, Galasinski SC, Shapiro PS, Nahreini TS, Ahn NG. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger SR, Zutter MM, Sturgill-Koszycki S, Santoro SA. Induced cell surface expression of functional alpha 2 beta 1 integrin during megakaryocytic differentiation of K562 leukemic cells. Exp Cell Res. 1992;202:28–35. doi: 10.1016/0014-4827(92)90400-3. [DOI] [PubMed] [Google Scholar]
- Tetteroo PA, Massaro F, Mulder A, Schreuder-van Gelder R, von dem Borne AE. Megakaryoblastic differentiation of proerythroblastic K562 cell-line cells. Leuk Res. 1984;8:197–206. doi: 10.1016/0145-2126(84)90143-7. [DOI] [PubMed] [Google Scholar]
- Bertolini F, Battaglia M, Pedrazzoli P, Da Prada GA, Lanza A, Soligo D, et al. Megakaryocytic progenitors can be generated ex vivo and safely administered to autologous peripheral blood progenitor cell transplant recipients. Blood. 1997;89:2679–2688. [PubMed] [Google Scholar]
- Maharaj D, Steinberg JP, Gouvea JV, Gieser PW. Changes in endogenous TPO levels during mobilization chemotherapy are predictive of CD34+ megakaryocyte progenitor yield and identify patients at risk of delayed platelet engraftment post-PBPC transplant. Bone Marrow Transplant. 1999;23:539–548. doi: 10.1038/sj.bmt.1701618. [DOI] [PubMed] [Google Scholar]
- Meldgaard Knudsen L, Jensen L, Jarlbaek L, Hansen PG, Hansen SW, Drivsholm L, et al. Subsets of CD34+ hematopoietic progenitors and platelet recovery after high dose chemotherapy and peripheral blood stem cell transplantation. Haematologica. 1999;84:517–524. [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Calin GA, Pekarsky Y, Croce CM. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best Pract Res Clin Haematol. 2007;20:425–437. doi: 10.1016/j.beha.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z, et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- Saiki Y, Ishimaru S, Mimori K, Takatsuno Y, Nagahara M, Ishii H, et al. Comprehensive analysis of the clinical significance of inducing pluripotent stemness-related gene expression in colorectal cancer cells. Ann Surg Oncol. 2009;16:2638–2644. doi: 10.1245/s10434-009-0567-5. [DOI] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ongkeko WM, Chen L, Yang ZF, Lu P, Chen KK, et al. Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4–AKT–ATP-binding cassette G2 pathway. Hepatology. 2010;52:528–539. doi: 10.1002/hep.23692. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a downregulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.