Abstract
Motivation: Finding protein-protein interaction (PPI) information from literature is challenging but an important issue. However, keyword search in PubMed® is often time consuming because it requires a series of actions that refine keywords and browse search results until it reaches a goal. Due to the rapid growth of biomedical literature, it has become more difficult for biologists and curators to locate PPI information quickly. Therefore, a tool for prioritizing PPI informative articles can be a useful assistant for finding this PPI-relevant information.
Results: PIE (Protein Interaction information Extraction) the search is a web service implementing a competition-winning approach utilizing word and syntactic analyses by machine learning techniques. For easy user access, PIE the search provides a PubMed-like search environment, but the output is the list of articles prioritized by PPI confidence scores. By obtaining PPI-related articles at high rank, researchers can more easily find the up-to-date PPI information, which cannot be found in manually curated PPI databases.
Availability: http://www.ncbi.nlm.nih.gov/IRET/PIE/
Contact: sun.kim@nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Researchers keep track of protein-protein interaction (PPI) information by searching literature online or using PPI database services. When using PPI databases, well-summarized information can be obtained. But, newly discovered evidence may be missed due to the rapid growth of the biomedical literature and time-consuming manual curation process (Blaschke et al., 2005). For online literature search, people commonly find relevant information in PubMed by exploring a combination of keywords, e.g. protein names, journal names or author names. This step can be time consuming; however, the interactive query-retrieval process by manual effort can reach better and more focused PPI information. Automatic article recommendation for PPI information can be, therefore, positioned between PPI database services and manual literature search because it provides more effective retrieval by suggesting PPI informative articles (PPI articles) from simple user queries.
PPI extraction tasks require several prerequisite steps such as gene mention, gene normalization, PPI article filtering or PPI experimental method extraction. Although article filtering is an essential step among those tasks, it has been often neglected by previous protein-interaction extraction systems. Improving PPI article classification enables better literature navigation for biologists (Krallinger et al., 2009), effective assistance for curators in manually updating repositories (Dowell et al., 2009) and better literature-mining system development for text mining researchers (Leitner et al., 2010).
PIE the search is an online web service to assist in finding PPI articles from PubMed. PIE the search provides the following novel features distinguished from other PPI services: First, it navigates PPI-specific articles for biologists and curators. Second, it provides a compact PubMed-search environment to help easy access for PubMed users. Third, users can easily find the up-to-date PPI information, which has not been curated in PPI databases. Since the proposed system implements the recent competition-winning approach in BioCreative (BC) III (Krallinger et al., 2010), it also guarantees the state-of-the-art performance among various methods.
2 SYSTEM AND FUNCTIONALITY
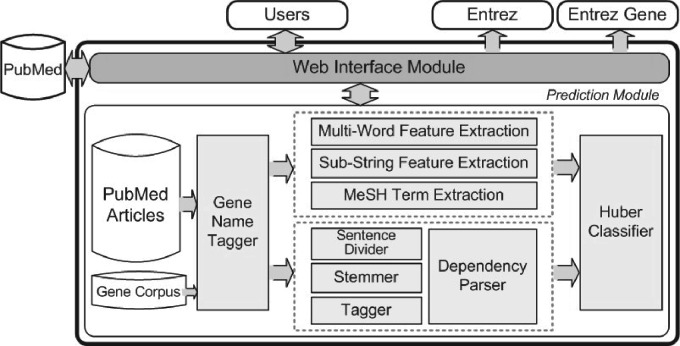
Figure 1 shows the overall architecture of the PIE the search system. The web interface module manages the whole process of PPI article prediction for users. For user queries, PubMed IDs are first retrieved through online PubMed services. PPI confidence scores are calculated for retrieved articles, and articles are re-ranked based on scores. For protein name queries, this process does not guarantee highly ranked articles that contain query-specific PPIs. However, it is still likely to have useful PPI information related to protein queries. The prediction module learns and classifies PubMed articles (Kim and Wilbur, 2011). To effectively capture PPI patterns from biomedical literature, our approach utilizes both word and syntactic features in a machine learning framework. Dependency parsing, gene mention tagging and term-based features are utilized along with a Huber classifier.
Fig. 1.
Overall architecture of PIE the search.
Since PIE the search is designed to provide only compact, but necessary features for PPI article search, its use is very straightforward, especially for PubMed users. It accepts PubMed input formats including All Fields, Author, Journal, MeSH Terms, Publication Date, Title and Title/Abstract with Boolean operations (AND, OR and NOT). However, the output is the list of articles prioritized by PPI confidence scores. Search results can be sorted by either PPI scores or dates. With the date sorting, only articles with PPI scores > 0.1 will appear. For convenient use, there are no page changes between search results and detailed article information. One typical routine in PubMed search is to follow full paper links after article information pages. Hence, full paper and PubMed links are also shown on the same page. In addition, some gene/protein names which contributed for PPI prediction are underlined and linked to Entrez and Entrez Gene. Another feature of PIE the search is a Keyword Cloud. Key noun phrases used for PPI article prediction are pooled and listed based on frequencies. This function helps users view abstracts in a nutshell.
3 RESULTS AND DISCUSSION
The prediction module in PIE the search is trained by all available BC datasets except for the BC3 test set (Kim and Wilbur, 2011). The performance of the PIE system was evaluated on the BC3 test set in terms of F1, MCC1 and AUC iP/R1 measures (Krallinger et al., 2010). This test set contains 910 PPI and 5090 non-PPI articles, which is unbalanced reflecting a real-world situation. The proposed system provides 0.6258 F1, 0.5610 MCC and 0.6834 AUC iP/R, whereas the medians of BC3 participant results are 0.5353 F1, 0.4563 MCC and 0.5367 AUC iP/R. The PIE system significantly outperformed the other approaches on all measures.
Since PIE the search is a web-based ranking system, the performance at top-ranked articles is more important than overall classification performance. At rank 10 (P@10), 100 (P@100) and 200 (P@200), our system achieves 100, 94 and 91.50% precision, respectively. Our system also produces over 95% precision at 10 recall. Even though the ratio between PPI and non-PPI articles in the PubMed database is more skewed than the BC3 test set, the ranking performance of PIE the search shows its usefulness as a PPI article search engine.
To understand how accurately our system ranks PPI articles and how users respond to this service, we further performed manual evaluation. A total of 10 biologists were asked to judge the top 10 search results from PubMed and PIE the search using their own queries. Each user performed searches for five different queries, and precision was calculated based on their assessments. As a result, PubMed achieved 25.40% precision on average. Meanwhile, PIE the search achieved 81.60 and 75.89 precision on average for results sorted by PPI scores and dates options, respectively. For the question of how satisfactory is this service as a PPI article search tool, the biologists responded with a rating of 4.4 on average on a 1 (bad) to 5 (good) scale. The Supplementary Material describes the manual evaluation in detail.
While most PPI extraction services provide PPI-centered information, PIE the search pursues a more general strategy for biologists and curators, i.e. a topic-specific search service by ranking PPI articles in PubMed. Even though it places more responsibility on users to choose useful query terms, it increases the chance to get the correct PPI articles. If one wants to find core PPI information from gene or protein names, other extraction services may be a good choice. However, PIE the search provides more up-to-date PPI information by directly searching PubMed with guaranteed high classification performance.
Taking a user-friendly perspective, the interface adopts a very easy and compact search scenario. Moreover, the PIE system provides a batch access through CGI programs, which can help other bio-text mining researchers develop similar prediction systems or perform performance comparisons. A tutorial of how to use PIE the search can be found at the homepage.
4 CONCLUSION
PIE the search is a web service designed for searching PPI articles from PubMed, which employs word and syntactic features in a machine learning framework. Compared to previous PPI article classification approaches, this method actively utilizes syntactic information. The Priority Model (Tanabe and Wilbur, 2006) and Huber classifiers are also a distinctive choice for effectively handling PubMed data. PIE the search is already practical as a ranking system since it provides high classification performance at top-ranked articles. The web service is freely accessible and the local PubMed database in PIE is being updated monthly.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all the participants for their contribution to the manual evaluation. The authors also would like to thank Natalie Xie for valuable comments on web implementation of the system.
Funding: Intramural Research Program of the National Institutes of Health, National Library of Medicine (to S.K. and W.J.W.); Basic Science Research Program through the National Research Foundation of Korea (NRF) (NRF-2011-0002437) (to D.K.).
Conflict of Interest: none declared.
Footnotes
1MCC and AUC iP/R measures are further explained in the Supplementary Material.
REFERENCES
- Blaschke C., et al. Evaluation of BioCreAtIvE assessment of task 2. BMC Bioinfomratics. 2005;6(Suppl. 1):S16. doi: 10.1186/1471-2105-6-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell K.G., et al. Integrating text mining into the MGI biocuration workflow. Database. 2009;2009:bap019. doi: 10.1093/database/bap019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Wilbur W.J. Classifying protein-protein interaction articles using word and syntactic features. BMC Bioinfomratics. 2011;12(Suppl. 8):S9. doi: 10.1186/1471-2105-12-S8-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M., et al. Creating reference datasets for systems biology applications using text mining. Ann. N. Y. Acad. Sci. 2009;1158:14–28. doi: 10.1111/j.1749-6632.2008.03750.x. [DOI] [PubMed] [Google Scholar]
- Krallinger M., et al. Proceedings of the BioCreative III. Bethesda, MD, USA: 2010. Results of the BioCreative III (interaction) article classification task; pp. 17–23. [Google Scholar]
- Leitner F., et al. The FEBS Letters/BioCreative II.5 experiment: making biological information accessible. Nat. Biotechnol. 2010;28:897–899. doi: 10.1038/nbt0910-897. [DOI] [PubMed] [Google Scholar]
- Tanabe L., Wilbur W. J. Proceedings of the HLT-NAACL BioNLP Workshop on Linking Natural Language and Biology. New York, USA: 2006. A priority model for named entities; pp. 33–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.