Abstract
Ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif-mediated transcriptional repression is emerging as one of the principal mechanisms of plant gene regulation. The EAR motif, defined by the consensus sequence patterns of either LxLxL or DLN xxP, is the most predominant form of transcriptional repression motif so far identified in plants. Additionally, this active repression motif is highly conserved in transcriptional regulators known to function as negative regulators in a broad range of developmental and physiological processes across evolutionarily diverse plant species. Recent discoveries of co-repressors interacting with EAR motifs, such as TO PLESS (TPL) and AtSA P18, have begun to unravel the mechanisms of EAR motif-mediated repression. The demonstration of genetic interaction between mutants of TPL and AtHDA 19, co-complex formation between TPL-related 1 (TPR1) and AtHDA 19, as well as direct physical interaction between AtSA P18 and AtHDA 19 support a model where EAR repressors, via recruitment of chromatin remodeling factors, facilitate epigenetic regulation of gene expression. Here, we discuss the biological significance of EAR -mediated gene regulation in the broader context of plant biology and present literature evidence in support of a model for EAR motif-mediated repression via the recruitment and action of chromatin modifiers. Additionally, we discuss the possible influences of phosphorylation and ubiquitination on the function and turnover of EAR repressors.
Key words: EAR motif, repressome, transcriptional repression, TPL, SAP18, HDA19, phosphorylation, co-repressors, post-translational modification, histone deacetylation, chromatin modification, epigenetic regulation
Epigenetic Reprogramming of Gene Expression
All cells of multicellular organisms have essentially the same genome but may be structurally and functionally different due to differences in gene expression. Many of the differential patterns of gene expression in plants, which are established during development in response to intrinsic or external signals, arise due to changes in chromatin structure and DNA methylation status that do not involve alterations in the DNA sequence itself. Such alterations in gene expression that are either heritable transgenerationally or are stable for the remainder of the plant's life are said to be “epigenetic.”1 DNA methylation is one of the ubiquitous mechanisms of heritable epigenetic modification. A second epigenetic modification involves chromatin conformation. In all eukaryotes, the genetic information encoded by DNA is compacted into chromatin, the basic unit of which, the nucleosome, is formed by the wrapping of DNA around a histone core complex composed of an octamer consisting of two copies each of the core histones H2A, H2B, H3 and H4.2 The amino- and carboxy-terminal tails of core histones offer approximately 240 sites for posttranslational modifications, including acetylation/deacetylation and methylation of Lys and/or Arg residues, phosphorylation of Ser and/or Thr and ubiquitination and sumoylation of Lys.3–5 Many of these covalent epigenetic marks on DNA and histones can activate or repress transcription by generating open or closed chromatin conformations, respectively.1
Eukaryotic organisms have developed epigenetic mechanisms to achieve a stable yet flexible means of regulating genes and coordinating genetic pathways. An epigenetic pathway is generally initiated in response to intrinsic or external signals and is established via a complex and coordinated networking between TRs, co-regulators and chromatin modifying factors.6 TRs, which can function as either activators and/or repressors, play a central role in epigenetic reprogramming of gene expression as they are involved in perceiving and integrating external signals to establish the desired epigenetic state at the appropriate chromosomal loci to achieve the correct phenotypic response.6 In recent years, transcriptional repressors have emerged as important elements essential for establishing intricate spatio-temporal patterns of gene expression during plant development and plant responses to stress and hormonal signals. Transcriptional repressors are generally classified as active or passive repressors.7 Approximately 6% of the Arabidopsis proteome is represented by TRs of which an estimated 30% function as active transcriptional repressors.8 Unlike passive repressors, which lack an intrinsic repression domain, active repressors generally contain a distinct, small and portable repression motif(s) that inhibits activation of transcription either by modifying chromatin structure, thereby preventing binding of transcriptional activators to their target cis-elements or by interacting with and inhibiting the functions of components of the basal transcription machinery.7,9
To date, at least four different active transcriptional repression motifs have been identified in plants (table 1), including the EAR motif,10 TLLLFR motif,11 R/KLFGV motif12 and LxLxPP motif.13 In this Point-of-View article, we focus on recent findings uncovering the role and biological significance of the EAR motif in the broader context of plant gene regulation and present an overview of the literature evidence supporting involvement of epigenetic pathways in facilitating EAR motif-mediated transcriptional repression in plants.
The EAR Motif
The EAR motif was the first active repression motif reported in plants. It was initially identified almost a decade ago in a subset of class II ERFs and TFIIIA-type zinc finger proteins as a small motif with a consensus of L/FDLNL/F(x)P.10 Interestingly, when this motif was tethered to transcriptional activators, they functioned as dominant repressors.14 Subsequent identification and confirmation of the repression capability of the EAR motif in several other TRs involved in diverse biological functions, including SUPERMAN,15 AUX/IAA proteins,16 HSI2 and related proteins,17,18 AGL15,19 and NIMIN proteins,20 as well as the recent discoveries of corepressors19,21 facilitating EAR motif function have unequivocally established a role for this motif in mediating transcriptional repression.
Other Active Repression Motifs in Plants
The TLLLFR motif was identified in the carboxy-terminus of AtMYBL2, a R3-MYB protein involved in negative regulation of anthocyanin biosynthesis in Arabidopsis;11 from our analysis of the proteomes of several plant species, it appears that the occurrence of this motif is restricted to AtMYBL2.8 The R/KLFGV motif was recently identified as a novel active repression motif occurring in at least 29 Arabidopsis transcription factors, including members of the ABI3/VP1, ARF, HSF and MYB families.12 The LxLxPP motif, which occurs in a few AUX/IAA proteins and other TRs from primitive as well as higher plant species, has been proposed to function as a repression motif;13 however, experimental evidence is lacking.
The Arabidopsis EAR Repressome
To obtain further insight to the potential breadth of utilization of the EAR motif in plant gene regulation, we recently conducted a comprehensive bioinformatics analysis of the Arabidopsis proteome. We have established a list of “high-confidence” Arabidopsis EAR repressors, the EAR repressome, comprising 219 TRs, which can be grouped into two categories: transcription factors (TFs, 180 candidates belonging to 18 different families) possessing distinct DNA binding domains and other transcriptional regulators (OTRs, 39 candidates belonging to three families) that do not possess a defined DNA-binding domain but are known in the literature to regulate transcription by interacting with TFs.22 Comparison of the sequences of EAR motifs and adjoining sequences from these proteins enabled refining the signature sequence patterns of the EAR motif as containing either LxLxL or DLNxxP. Our analysis suggests that the EAR motif is the most predominant form of transcriptional repression motif so far identified in plants (Table 1).8 Proteins containing this motif play key roles in diverse biological functions by negatively regulating genes involved in various developmental and physiological processes.22 Our analyses8,22 and growing evidence in the literature collectively support a role for EAR motif-containing proteins in demarcation of expression boundaries for genes involved in plant organ development and developmental transitioning, as well as regulation of stress and hormonal responses. Consequently, EAR motif-mediated repression may be considered as one of the principle mechanisms of gene regulation in plants utilized multiple times during evolution to control gene expression.
Table 1.
Active repression motifs in plants
| Repression motif | Sequence patterns | Representation among TRs* | Corepressors | Chromatin modifiers | References |
| EAR motif | LxLxL, DLNxxP | 10 to 25% | TPL, SAP18, SIN3** | HDA19 | 8, 17, 19–21, 24, 25 |
| TLLLFR motif | TLLLFR | Found only in AtMYBL2 | unknown | unknown | 8, 11 |
| R/KLFGV motif | R/KLFGV | <2% | unknown | unknown | 8, 12 |
| LxLxPP motif | LxLxPP | <3% | unknown | unknown | 8, 13 |
As determined in 12 plant species.8
Interaction between EAR motif and SIN3 is supported by genetic studies but direct physical interaction is yet to be determined.
Epigenetic Mechanisms of EAR Motif-Mediated Transcriptional Repression
Transcriptional repression by chromatin modification is one of the principal mechanisms employed by eukaryotic active repressors.23 In yeast and mammalian cells, histone deacetylation plays an important role in active transcriptional repression9,24 and in these systems transcriptional co-repressors such as SIN3 and SAP18 are postulated to establish a physical link between HDACs and DNAbound active repressors.9 Putative orthologues of SIN3 and SAP18 have been identified in Arabidopsis.25,26 Our analysis has identified putative orthologs of SAP18 and SIN3 in evolutionarily diverse plant species, including primitive species such as Physcomitrella patens (a moss), Selaginella moellendorffii (a lycophyte) and Chlamydomonas reinhardtii (a unicellular green alga; Kagale S and Rozwadowski K, unpublished). Furthermore, the existence of several HDACs and their importance in histone deacetylation and transcriptional repression during plant growth and development has been well established,27 suggesting that active repression mechanisms in plants employ HDAC complexes, analogous to yeast and animal systems.
Evidence in the literature supports a role for AtSAP18, AtSIN3 and AtHDA19 (an HDAC) in transcriptional repression by EAR motif-containing proteins in Arabidopsis.19,25,26 For instance, the EAR motif containing class II ERFs, such as ERF3 and ERF4, which are known to function as active repressors in vitro and in vivo,10,28–30 have been shown to physically interact with AtSAP18, which in turn interacts and forms a repression complex with AtHDA19.26 AtERF7, another EAR motif-containing class II ERF protein, is also known to recruit AtHDA19 via a physical interaction with AtSIN3.25 The in planta coexpression of AtERF3, AtSAP18 and AtHDA19 or AtERF7, AtSIN3 and AtHDA19 in transient repression assays results in greater transcriptional repression of reporter genes as compared to when these proteins are expressed alone,25,26 suggesting a role for AtSAP18, AtSIN3 and AtHDA19 in ERF-mediated transcriptional repression possibly via histone deacetylation. Although the EAR motifs in AtERF3, 4 and 7 have been shown to be responsible for the repression capability of these proteins, the direct involvement of the EAR motifs in mediating their interaction with AtSAP18 or AtSIN3 has not been determined. A more recent study has revealed that the EAR motif in AGL15, a MADS-domain transcription factor, mediates the physical interaction of AGL15 with AtSAP18,19 supporting a role for the EAR motif in regulating gene expression via recruitment of an HDAC complex. Unlike ERF repressors that contain a DLNxxP type of EAR motif, AGL15 contains a LxLxL (LQLGL) type of EAR motif.22 Replacement of the Leu residues within this motif with Ala disrupts the interaction between AGL15 and AtSAP18,19 demonstrating that the interaction between these two proteins is EAR motif dependent. These observations clearly establish a role for the EAR motif of AGL15 in recruitment of AtSAP18/HDAC complex to the promoters of AGL15 target genes. Thus, current information suggests a model where EAR motifs mediate gene repression through recruitment of an HDAC complex. Notably, as found for AGL15, the Leu residues within the LxLxL type of EAR motifs in SUPERMAN and AUX/IAA proteins in Arabidopsis have also been shown to be important for their repression activity.16,31 Considering these findings, it is possible that other members of the 165 Arabidopsis EAR repressome candidates that contain a LxLxL type EAR motif(s)22 could potentially interact with AtSAP18 and recruit an HDAC complex to perform transcriptional repression functions. It would be of interest to determine if the DLNxxP types of EAR motifs in ERFs and other candidates of the EAR repressome aid in recruiting AtSAP18 and the HDAC complex in a similar manner.
The recent discoveries of physical interactions between TPL and the EAR motifs in several TRs in Arabidopsis, such as IAA12/BDL21 and NINJA32 have uncovered another novel component of EAR motif-mediated gene regulation. IAA12/BDL contains a LxLxL type of EAR motif in the N-terminal region and belongs to the AUX/IAA family of transcriptional repressors, which are known to act as negative regulators of auxin signaling. 16 Mutation of the Leu residues within the EAR motif of IAA12/BDL to Ala abrogates its interaction with TPL, suggesting that the Leu residues within the EAR motif are necessary and sufficient for facilitating the interaction between AUX/IAAs and TPL.21 Through genetic analysis of a tpl-1/bdl-1 double mutant and in planta transcriptional repression assays, TPL was shown to influence the repression ability of IAA12, thus revealing the biological significance of EAR-dependent recruitment of TPL in AUX/IAA-regulated transcriptional repression.21 Similar to AUX/IAA proteins, NINJA, which contains a LxLxL type of EAR motif in the amino-terminal region and functions as a negative regulator of jasmonic acid responses, also physically interacts with TPL in an EAR motifdependent manner.32 AFP proteins, which function as negative regulators of abscisic acid responses, also contain a LxLxL type of EAR motif in the amino-terminal region and have recently been shown to interact with TPL;32 however, whether this interaction is EAR motif-dependent remains to be determined. Although, a genetic interaction between TPL and a DLNxxP type of EAR motif has not been reported, we have recently detected physical interaction between TPL and five DLNxxP type of EAR motif-containing proteins in Arabidopsis (Kagale S and Rozwadowski K, unpublished) and are currently analyzing if these interactions are EAR motif dependent. As the EAR repressome in Arabidopsis has already been defined,22 it would be interesting to determine which of the members of this collection of TRs utilize TPL to facilitate transcriptional repression and whether there is overlap with those interacting with AtSAP18 or distinct categories of EAR repressome members recruit each of these co-repressors. A yeast two-hybrid screen performed using full-length TPL as bait has revealed that TPL interacts with several members of the Arabidopsis EAR repressome involved in diverse biological functions (Kagale S and Rozwadowski K, unpublished). These findings collectively support TPL having an important role in facilitating repression functions of at least a subset of EAR repressors in Arabidopsis. Evolutionary conservation of this gene regulation system was recently shown in a study of maize where REL2, a transcriptional co-repressor and maize orthologue of Arabidopsis TPL, was found to interact with RA1, a LxLxL type of EAR motifcontaining C2H2-zinc finger transcription factor involved in regulation of the fate of auxiliary meristems during development of the inflorescence, tassel and ear, in an EAR motif-dependent manner.33
Through elegant genetic interaction studies, Long et al.34 have shown that TPL works in conjunction with AtHDA19 to repress expression of root promoting genes in the apical half of the embryo and enable proper shoot pole formation. T-DNA insertion alleles of AtHDA19 were found to enhance the penetrance of tpl-1 and display similar apical embryonic defects, suggesting that TPL and AtHDA19 act on the same targets.34 The possible role for HDACs in TPL function is further alluded to by the structural and potentially functional similarities between TPL and the Groucho/Tup1 family of transcriptional co-repressors, which are known to facilitate recruitment of HDACs to the regulatory regions of genes targeted by several distinct types of active repressors found in yeast, flies, worms and humans.35–37 Furthermore, AtHDA19 was recently shown to co-immunoprecipitate with AtTPR1, the most closely related of the four paralogues of TPL in Arabidopsis (93% amino acid sequence identity with TPL), supporting that AtTPR1 associates with AtHDA19 in vivo.38 Notably, the CTLH domain of AtTPR1 and TPL, responsible for physical interaction with EAR motifs,21,32 share 98% identity, supporting the likelihood that AtTPR1 also interacts with EAR proteins. Analysis of physical interaction between TPL and AtHDA19 by yeast two-hybrid assay revealed that these proteins do not directly interact (Kagale S and Rozwadowski K, unpublished), suggesting that an unknown adapter protein may be involved in facilitating their association. The genetic and in vivo association of TPL and AtTPR1, respectively, with AtHDA19 and the ability of TPL to interact with several candidates of the EAR repressome suggest a novel epigenetic link between the EAR motif, TPL, AtTPR1 and chromatin modification via histone deacetylation.
Evolutionary Conservation of EAR-Mediated Gene Regulation
We have previously shown that the EAR motif is highly conserved across evolutionarily diverse plant species22 and is detected in 10–25% of TRs belonging to multiple gene families across various plant proteomes.8 Furthermore, comparative bioinformatics analyses have revealed that TPL- and AtSAP18-related proteins identified from primitive as well as higher plant species are highly similar in sequence and structural properties (Kagale S and Rozwadowski K, in preparation), supporting the possibility that the co-repressor functions amongst these proteins are conserved. The evolutionary conservation of the EAR motif and associated co-repressors such as TPL and AtSAP18, combined with the success of EAR motif derived dominant repressor technology in different plant species,14,39 collectively support EAR-mediated repression of gene expression being a general regulatory mechanism in plants. Indeed, the recent study in maize demonstrating the role of an EAR motif-dependent repressor complex between RA1 and REL2 (a TPL orthologue) in the control of meristem fate33 has provided genetic and molecular evidence for EAR motif-TPL-mediated repression mechanisms being evolutionarily conserved.
Transcriptional and Post-Translational Modulation of EAR-Mediated Gene Regulation
The Arabidopsis genes encoding EAR repressors have been shown to be differentially regulated by various developmental, hormonal and environmental signals.22 Thus, it is possible that EAR-mediated gene regulation is modulated at least in part by differential spatio-temporal expression of EAR repressors. Conversely, the TPL gene family and AtSAP18 in Arabidopsis, as represented in publicly available transcriptome datasets,40 appear to be broadly expressed in most plant tissues and are either not affected or marginally affected by stress or hormonal signals, suggesting a broad and general role for these co-repressors in a range of biological processes. The lack of substantial differential transcriptional response amongst TPL gene family members suggests that the function of these genes may be regulated at a post-transcriptional, translational or post-translational level.
Post-translational modifications such as phosphorylation and ubiquitination can potentially have either a positive or negative influence on EAR-mediated gene regulation, as they may affect conformation, protein-protein interactions, subcellular localization and turnover of the EAR repressors or associated co-repressors. Interestingly, Ser and Thr residues adjacent or integral to the EAR motif have been detected as being phosphorylated in at least five proteins belonging to the Arabidopsis EAR repressome, including IAA9, ERF10, BEH4, DEAR4 and a C2H2 family protein.22 Additionally, the Ser and Thr residues in IAA9, BEH4 and DEAR4 that were determined to be phosphorylated are conserved in corresponding orthologs across evolutionarily diverse plant species.8 Furthermore, analysis of the Arabidopsis phosphoproteome41,42 revealed that at least three residues in TPL, including Tyr133, Ser214 and Thr286, are identified by mass spectrometry as being phosphorylated. Our analysis indicates that Tyr133 and Thr286 of TPL are highly conserved in TPL proteins across diverse plant species (Kagale S and Rozwadowski K, in preparation). Overall, these results suggest that phosphorylation may have a role in regulating the functions of EAR repressors as well as TPL, and may possibly provide another level of regulation controlling EAR-mediated repression. We have recently identified a protein kinase that interacts with the EAR motif and two protein kinases as well as a phosphatase that interact with TPL (Kagale S and Rozwadowski K, unpublished). The potential roles of these kinases and the phosphatase in regulating the functions of the EAR motif or TPL are currently being explored. Phosphorylation-mediated regulation of co-repressor function has also been demonstrated for the animal Groucho/TLE family proteins,43–45 which are structurally similar to TPL.
Poly-ubiquitination and proteolytic degradation of EAR repressors has emerged as a general theme in plant hormone signaling.30,46–50 For example, the AUX/IAA proteins are known to be degraded via the proteasomal degradation pathway in an auxin dependent manner.47,49 Similarly, the JAZ proteins, which are known to interact with the EAR repressor NINJA and function as negative regulators of jasmonic acid signaling, are also degraded by the 26S proteasome in the presence of jasmonic acid.46 Evidence for compartmentalization of ERF repressors into nuclear bodies and their proteolytic degradation as a means of regulating ethylene responses has also been reported.30 Ubiquitination and proteasomal degradation thus appear to provide an elegant mechanism for relieving the negative effects of EAR repressors on gene expression, especially when their actions are no longer required.
Concluding Remarks
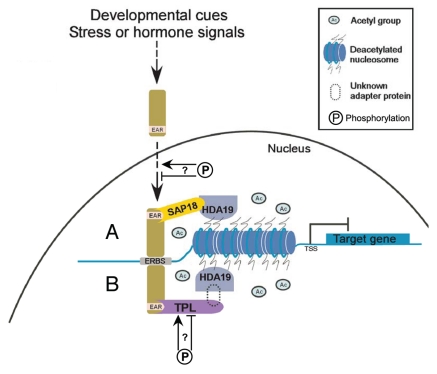
Several lines of evidence in the literature support repression of transcription by EAR motif-containing proteins being governed by epigenetic mechanisms resulting from chromatin modifications, mainly through the recruitment and actions of co-repressors, such as AtSAP18 and TPL, as well as an HDAC, AtHDA19. In light of these findings, we present a model for epigenetic regulation of gene expression (Fig. 1) in which EAR repressors play a central role in coordinating responses to environmental and developmental stimuli by facilitating HDAC-mediated chromatin modification of target loci through recruitment of co-repressors such as AtSAP18 and TPL or related proteins.
Figure 1.
Model for EAR motif-mediated transcriptional repression in plants. EAR repressors, which are known to respond to various developmental, as well as stress and hormonal signals, at the transcriptional level play a central role in integrating these signals at precise locations on the chromosome by virtue of their ability to bind DNA directly or their ability to physically interact with other DNA binding proteins. EAR repressors suppress the expression of target genes probably through chromatin modification of regulatory regions by histone deacetylation via physically interacting with co-repressors such as (A) SAP18, known to directly interact with HDA19 potentially forming a repression complex, or (B) TPL, which is known to function in conjunction with HDA19. An unknown adapter protein is speculated to facilitate the association between TPL and HDA19 since evidence for their direct interaction is lacking but a genetic interaction between TPL and HDA19 has been established and in vivo co-complex formation between the highly related TPR1 and HDA19 has been demonstrated. Phosphorylation of some EAR repressors or the co-repressor TPL has been detected and may further influence repression complex formation or function by potentially affecting the activity of these proteins. TSS, transcription start site; ERBS, EAR repressor binding site.
Plant genomes encode several other co-repressors and factors associated with chromatin remodeling. Future efforts aimed at identifying and analyzing the function of additional novel co-repressors of the EAR motif and associated chromatin regulatory mechanisms are essential to advance our understanding of the repression mechanisms utilized by EAR repressors. Another challenge for the immediate future would be to detect and map the “EAR epigenome” including dynamic changes in the chromatin structure associated with the regulatory regions of single or multiple target loci of EAR repressor(s).
Acknowledgements
Funding for this research provided by Agriculture and Agri-Food Canada to K.R.
Abbreviations
- ERF
ethylene-responsive element binding factor
- EAR motif
ERF-associated amphiphilic repression motif
- TR
transcriptional regulator
- HDAC
histone deacetylase
- AUX/IAA
auxin/indole-3-acetic acid
- HSI2
high level expression of sugar inducible gene 2
- AGL15
agamous-like 15
- NIMIN
NIM1-interacting
- ABI3/VP1
abscisic acid insensitive 3/viviparous 1
- ARF
auxin response factor
- HSF
heat shock factor
- SIN3
SWI-independent 3
- SAP18
SIN3-associated polypeptide of 18 kDa
- TPL
TOPLESS
- TPR1
toplessrelated 1
- BDL
bodenlos
- NINJA
novel interactor of JAZ
- AFP
ABI5 binding protein
- BEH4
BES1 homolog 4
- DEAR4
dehydration-responsive element binding protein 1/C-repeat binding factor
- JAZ
jasmonate ZIM-domain
- HDA19
histone deacetylase 19
- RA1
ramosa1
- REL2
ramosa enhancer locus2
References
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Chinnusamy V, Gong Z, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 6.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 8.Kagale S, Rozwadowski K. Small yet effective: The Ethylene responsive element binding factor-associated Amphiphilic Repression (EAR) motif. Plant Signal Behav. 2010;5:691–694. doi: 10.4161/psb.5.6.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 10.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:195919–195968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda M, Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 2009;50:970–975. doi: 10.1093/pcp/pcp048. [DOI] [PubMed] [Google Scholar]
- 13.Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, et al. The evolution of nuclear auxin signalling. BMC Evol Biol. 2009;9:126. doi: 10.1186/1471-2148-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 15.Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 2002;514:351–354. doi: 10.1016/s0014-5793(02)02435-3. [DOI] [PubMed] [Google Scholar]
- 16.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukagoshi H, Morikami A, Nakamura K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA. 2007;104:2543–2547. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2005;138:675–685. doi: 10.1104/pp.104.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill K, Wang H, Perry SE. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008;53:172–185. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- 20.Weigel RR, Pfitzner UM, Gatz C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell. 2005;17:1279–1291. doi: 10.1105/tpc.104.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 22.Kagale S, Links MG, Rozwadowski K. Genomewide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiel G, Lietz M, Hohl M. How mammalian transcriptional repressors work. Eur J Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 24.Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 25.Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song CP, Galbraith DW. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol Biol. 2006;60:241–257. doi: 10.1007/s11103-005-3880-9. [DOI] [PubMed] [Google Scholar]
- 27.Hollender C, Liu Z. Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139:949–959. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- 31.Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun. 2004;321:172–178. doi: 10.1016/j.bbrc.2004.06.115. [DOI] [PubMed] [Google Scholar]
- 32.Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, et al. The control of axillary meristem fate in the maize ramosa pathway. Development. 2010;137:2849–2856. doi: 10.1242/dev.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler CJ, Ponce A, Courey AJ. Groucho-mediated repression may result from a histone deacetylasedependent increase in nucleosome density. PLoS One. 2010;5:e10166. doi: 10.1371/journal.pone.0010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shikata M, Ohme-Takagi M. The utility of transcription factors for manipulation of floral traits. Plant Biotechnol. 2008;25:31–36. [Google Scholar]
- 40.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-northerns, Expression Angling and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 41.Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, et al. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An Update. Nucleic Acids Res. 2010;38:828–834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, et al. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36:1015–1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nuthall HN, Husain J, McLarren KW, Stifani S. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol Cell Biol. 2002;22:389–399. doi: 10.1128/MCB.22.2.389-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuthall HN, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol Cell Biol. 2004;24:8395–8407. doi: 10.1128/MCB.24.19.8395-8407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buscarlet M, Hermann R, Lo R, Tang Y, Joachim K, Stifani S. Cofactor-activated phosphorylation is required for inhibition of cortical neuron differentiation by Groucho/TLE1. PLoS ONE. 2009;4:e8107. doi: 10.1371/journal.pone.0008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol. 2008;11:486–494. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 48.Huq E. Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci. 2006;11:4–7. doi: 10.1016/j.tplants.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Maraschin Fdos S, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59:100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- 50.Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010;61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]