Abstract
The majority of mammalian gene promoters are encompassed within regions of the genome called CpG islands that have an elevated level of non-methylated CpG dinucleotides. Despite over 20 years of study, the precise mechanisms by which CpG islands contribute to regulatory element function remain poorly understood. Recently it has been demonstrated that specific histone modifying enzymes are recruited directly to CpG islands through recognition of non-methylated CpG dinucleotide sequence. These enzymes then impose unique chromatin architecture on CpG islands that distinguish them from the surrounding genome. In the context of this work we discuss how CpG island elements may contribute to the function of gene regulatory elements through the utilization of chromatin and epigenetic processes.
Key words: CpG island, chromatin, histone, methylation, promoter, ZF-CxxC domain, transcription, histone lysine demethylase
CpG Islands: Enigmatic Feature of the Mammalian Genome
In mammalian genomes, approximately 70–80% of CpG dinucleotides are post-replicatively marked by the addition of a methyl group to the 5 position of the cytosine ring.1 This modification acts as a stable and heritable epigenetic mark that is generally associated with repressed chromatin states and inhibition of transcriptional initiation.2 Despite the prevalence of CpG methylation, specific regions of mammalian genomes are refractory to this modification.3,4 These regions, called CpG islands, correspond to contiguous non-methylated segments of the genome that have a higher than average level of CpG dinucleotides and GC content.5 A large fraction of CpG islands encompass transcription start sites and approximately 70% of mammalian genes are associated with CpG islands. Interestingly, unlike classical TATA box containing promoters, CpG island promoters generally utilize dispersed transcription start sites, suggesting that the CpG island may act as a generalized element for transcriptional initiation.6 Although CpG island promoters were originally considered a feature of housekeeping genes, it is now evident that many tissue-specific genes also utilize CpG island promoters.7,8
CpG islands often encompass mammalian gene promoters but their role in gene regulatory function is poorly understood. Certain transcription factors like Sp1, CREB and CTCF contain CpG in their binding site and DNA recognition is often blocked by CpG methylation.9–13 This led to the hypothesis that non-methylated CpG islands act as preferential nucleation sites for these types of transcription factors. However, given that these DNA binding factors recognize CpG dinucleotides within the context of extended DNA motifs they are only targeted to a limited subset of CpG islands.14–16 This suggests that additional, more broadly impinging mechanisms likely contribute to CpG island function.
ZF-CxxC Domain Proteins Bind Specifically to CpG Islands
In searching for additional DNA binding factors that might specifically recognize non-methylated CpG dinucleotides, Skalnik and colleagues identified a protein they called CpG Binding Protein (CGBP).17 CGBP encodes a ZF-CxxC DNA binding domain that specifically recognizes CpG dinucleotides and binding is blocked when the sequence is methylated. Therefore, CGBP could potentially recognize non-methylated CpG islands. CGBP was later renamed CxxC Finger Protein 1 (CFP1) based on the identification of an extended family of CpG binding proteins.18 Despite their in vitro DNA binding activity, the relationship between ZF-CxxC proteins and CpG islands genome-wide has largely been overlooked.17,19
Based on the intriguing possibility that ZF-CxxC domain containing proteins may specifically recognize CpG islands, we recently analyzed the genome-wide localization of the ZF-CxxC domain containing KDM2A protein.20,21 Chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) revealed that KDM2A binds greater than 90% of CpG islands genome-wide and demonstrated that the ZF-CxxC domain is a CpG island targeting module. In a related study, Bird and colleagues demonstrated via a similar ChIP-seq based approach that the CFP1 protein also specifically nucleates at CpG islands.21 Together this work demonstrates for the first time that CpG island elements are directly interpreted through recognition of non-methylated DNA. Interestingly, KDM2A and CFP1 are associated with histone modifying activities, suggesting that CpG islands may use chromatin based processes to elicit regulatory element function.
Translating the CpG Island Signal into Unique Chromatin Architecture
KDM2A is the founding member of a recently identified family of JmjC domain containing histone lysine demethylase enzymes. KDM2A specifically catalyzes removal of methylation from histone H3 lysine 36 with a preference for the di-methyl modification state (H3K36me2).22 Mass-spectrometry analysis of histone modifications in a variety of cell types indicates that H3K36me2 is an abundant modification adorning 30–50% of histone H3.23–25 Based on the observation that KDM2A nucleates at CpG islands, we hypothesized that this enzyme could function to remove H3K36me2 from CpG island promoters. Indeed, chromatin immunoprecipitation based analysis of H3K36me2 levels revealed that while inter- and intragenic regions show high and relatively constant levels of H3K36me2, KDM2A-bound CpG islands are depleted of this mark.20 Importantly, knockdown of KDM2A by RNAi resulted in increased H3K36me2 at CpG islands indicating that KDM2A nucleation is responsible for this depletion. Like KDM2A, CFP1 modifies chromatin, but instead of removing histone methylation it associates with a histone H3 K4 methyltransferase complex (SET1 complex) to catalyze addition of the tri-methyl modification state (H3K4me3).18 Histone H3K4me3 is not broadly distributed like H3K36me2, but is instead found associated with the 5′ ends of genes.26,27 When CFP1 was depleted in cells, a reduction of H3K4me3 was observed at CFP1 bound CpG island promoters.21 Together, these observations reveal a simple yet elegant mechanism by which CpG island DNA is specifically interpreted by chromatin modifying enzymes encoding ZF-CxxC domains to create unique chromatin architecture characterized by depletion of H3K36me2 and nucleation of H3K4me3. Importantly, this architecture effectively differentiates CpG island elements from surrounding chromatin and suggests that chromatin environment may in part define CpG island function.
Transcriptionally Permissive Chromatin Architecture at CpG Islands
ZF-CxxC domain protein binding at CpG island promoters does not directly correlate with gene expression, fitting with the fact that ZF-CxxC proteins are recruited by the underlying non-methylated CpG content and not by the transcriptional state of the associated gene.20,21 This is consistent with in vivo footprinting studies illustrating that active transcription has little or no effect on regions of protection at CpG islands.28 By virtue of the fact that ZF-CxxC domains recognize DNA sequence it appears that they create CpG island chromatin architecture that is independent and therefore upstream of traditional transcription factor mediated gene activation or silencing. This prompts the question, what role does ZF-CxxC domain mediated CpG island chromatin architecture play in CpG island function?
To consider the implications of CpG island chromatin for gene function, it is useful to examine each of the unique CpG island chromatin features individually. Histone lysine methylation marks can act as binding sites for specific effector proteins that possess plant homeodomain (PHD) fingers or chromatin organization modifier (chromo) domains. A number of studies suggest that H3K4me3 acts as a binding site for the PHD finger of proteins that support transcription initiation, such as TFIID,29 nucleosome remodeling factor (NURF)30 and the ING4 containing histone acetyltransferase (HAT) complex.31 In contrast, experiments in yeast suggest that H3K36me2 is inhibitory to transcriptional initiation by acting as a binding site for the chromodomain of the EAF3 component of the RPD3S histone deacetylase (HDAC) complex.32–35 In addition to ZF-CxxC domain mediated chromatin features, early biochemical experiments examining CpG island chromatin revealed a distinct depletion of Histone H1 at these elements.36 Histone H1 is considered repressive to transcription,37–40 possibly due to stabilization of a higher order chromatin structure.41 Together these features appear to create a CpG island chromatin environment that is an ideal substrate for transcriptional initiation.
How Do CpG Islands Contribute to Regulatory Element Function?
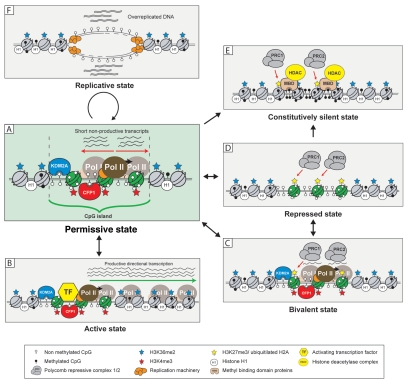
Up to 70% of mammalian genes are associated with CpG islands, almost all of which are free from DNA methylation. Their association with promoter and regulatory elements suggest they likely contribute to gene regulatory processes, but the presence of a CpG island alone does not define transcriptional output. Therefore, what do CpG islands and CpG island chromatin architecture contribute to gene regulation? Although the defined answers to these questions remain unclear, an emerging body of evidence support the possibility that CpG islands functionally impinge upon transcriptional potential through chromatin architecture and act as platforms for additional levels of transcriptional and epigenetic control. Below we discuss in more detail the nature of CpG island chromatin and how the function of additional signaling and chromatin modification pathways alter the default CpG island chromatin state leading to a series of more activated or repressed configurations (Fig. 1).
Figure 1.
The various chromatin states of CpG islands. CpG islands have the capacity to adopt multiple chromatin states each with varying propensities for gene activation or repression (see main text for a description of these). Central to the capability of CpG islands to adopt and transition between these states is the default permissive state (A), partly imposed by the ZF-CxxC proteins KDM2A and CFP1. Nucleosomes within CpG islands are depicted in green and nucleosomes outside of CpG islands are grey. Straight arrows represent pathways of transition between individual CpG island states. Circular arrow represents a cell cycle involving initiation of DNA replication from CpG island ORIs.
The permissive state.
The default CpG island chromatin environment appears to have features that are permissive to transcriptional initiation, in part through the function of ZF-CxxC proteins.20,21 We hypothesize that this system functions to highlight promoter regions within the vast expanse of the mammalian genome and focus nucleation of the transcriptional machinery at the 5′ ends of genes. In support of this idea RNA pol II is enriched not only at the promoters of actively transcribed genes but many genes that are apparently silent.27 Additionally, short bidirectional transcripts emanate from the 5′ regions of genes, including some with no detectable full-length mRNA.42–46 Interestingly, there is a strong correlation between RNA pol II enrichment, bidirectional transcripts and CpG island promoters (Fig. 1A).27,42–44,46 One explanation for this relationship is that chromatin structure at CpG islands creates an accessible environment that favors binding of RNA pol II and other components of the basal transcription machinery over surrounding non-regulatory chromatin. In the absence of additional regulatory factors that define activated directional transcriptional output, the CpG island may only permit futile and non-productive initiation/termination cycles of transcription that result in short bidirectional transcripts. In order to transition from this default state to a transcriptionally active state, additional regulatory signals outside of chromatin architecture alone are presumably required.
The active state.
CpG islands appear to create a permissive chromatin environment but this does not define productive transcriptional output. This implies a specific signal must initiate the transition from this permissive state to the active state characterized by productive and directional transcription (Fig. 1B). This process likely relies on the function of sequence-specific DNA binding transcription regulators. For example, the immediate early genes, most of which have CpG island promoters and are usually silent or expressed at very low levels, can be rapidly activated in response to external stimuli such as lipopolysaccharide (LPS).47 LPS stimulates the Toll-like receptor 4 pathway, resulting in activation of transcription factors such as NFκB and AP-1 that bind immediate early gene CpG island promoters and permit gene activation.48,49 If the CpG island chromatin environment is permissive to transcription, one would envisage that activation of CpG island genes would be kinetically more favorable than genes that lack this environment. In support of this contention, induction of CpG island containing immediate early genes occurs rapidly and without the requirement of the chromatin remodeling machinery. In contrast, induction of non-CpG islands containing immediate early genes relies on the function of the ATP-dependent chromatin remodeling factor BRG-1 and suggests that chromatin environment at these genes is a barrier to activation.27 These observations indicate that CpG island chromatin is permissive to transcription but that transcription factors ultimately define productive transcriptional output.
The bivalent state.
In addition to permissive and active CpG islands, embryonic stem (ES) cells have an intriguing subset of CpG island genes that are characterized by nucleation of the polycomb mediated silencing machinery (Fig. 1C). These CpG islands retain the permissive H3K36me2 depleted/H3K4me3 enriched chromatin environment, but are also modified by histone H3 lysine 27 trimethylation (H3K27me3) through the function of the polycomb repressive complex 2 (PRC2).26,50 This class of promoters are referred to as bivalent and are often associated with genes that must be actively silenced in ES cells but will often be activated later during development and lineage specific commitment. Interestingly, PRC2 nucleation and H3K27me3 profiles usually correspond to the underlying boundaries of the CpG island element suggesting that information within the CpG island itself may contribute to this profile.26,51 The function of the polycomb system at CpG islands appears to constrain the permissive capacity of the CpG island chromatin environment and ensure the associated gene is not expressed. The mechanisms underlying this repression are not entirely clear, but there is experimental evidence that the polycomb repressive complex 1 (PRC1) restrains RNA pol II elongation at bivalent promoters.52 Interestingly, a recent study demonstrated that short RNAs transcribed from bivalent CpG islands form hairpin structures that are directly recognized by polycomb components and aid in targeting PRC2 to these genes.44 Therefore, PRC2 may have hijacked non-productive transcripts emanating from CpG islands as a gene-specific polycomb targeting mechanism. Clearly more work is required to understand how polycomb complexes recognize specific CpG islands to create bivalency.
The repressed state.
In some tissues, expression of a given CpG island gene may very rarely or never be required. Therefore, maintenance of permissive or bivalent chromatin architecture may not be entirely beneficial. In mouse brain there is a population of CpG islands (<10%) that lacks DNA methylation yet does not display significant ZF-CxxC protein nucleation.21 In general, it appears these genes are subject to polycomb-mediated repression and are modified by H3K27me3, but unlike bivalent genes in ES cells they lack H3K4me3 (Fig. 1D). This suggests in some instances ZF-CxxC protein binding and permissive chromatin architecture are excluded from CpG islands, perhaps through an as of yet undiscovered polycomb dependent silencing mechanism. Therefore, in some instances it appears that CpG islands transition from the default permissive state to a repressed state lacking ZF-CxxC dependent chromatin architecture as a means of actively repressing gene expression.
The constitutively silent state.
A small number of CpG islands, such as those silenced during X chromosome inactivation or genomic imprinting, acquire DNA methylation during cellular differentiation. DNA methylation blocks recruitment of ZF-CxxC domain proteins and this abrogates the permissive chromatin environment usually defined by these factors.20,21 At the same time methylated CpG acts as binding sites for methyl CpG-binding domain proteins (MBDs) that recruit HDAC complexes and are potent repressors of transcription (Fig. 1E).2,53,54 This results in a very stable and epigenetically heritable form of silencing and makes reactivation of the affected gene extremely difficult. It still remains mechanistically unclear how this subset of CpG islands become susceptible to DNA methylation, but evidence from studies in cancer tissues indicate that a polycomb-silenced intermediate state may ultimately give rise to the constitutively repressed and DNA methylated state.55,56 It remains poorly defined whether this is also true of CpG islands that acquire DNA methylation normally during development. Therefore, it will be important to understand if polycomb silencing is related to acquisition of DNA methylation.
The replicative state.
The majority of work on CpG islands has focused on understanding their relationship with gene expression. Interestingly, another documented feature of CpG islands is that they often correspond to origins of DNA replication (ORIs).57,58 The permissive nature of CpG island chromatin may in analogy to acting a nucleation site for the transcriptional machinery, also preferentially act as assembly points for the DNA replication machinery. Intriguingly, it has been reported during S phase that CpG island ORIs undergo several rounds of short abortive replication, resulting in over-replication of these regions compared to other parts of the genome (Fig. 1F).59 This phenomenon appears analogous to the short bidirectional transcripts initiating from CpG island promoters and could result from non-productive initiation of ORIs in the absence of factors needed for processive replication. More work is clearly required to understand what features of CpG islands make these preferential targets for initiating DNA replication.
Conclusion: The Many States of CpG Islands
The traditional view of gene promoters is one of defined transcription start sites that are either active or silent. However, genome-wide maps of RNA pol II binding and high resolution analysis of RNA transcripts reveal that CpG island promoters do not conform to this ideal. Rather than defining the active or repressed transcriptional state, CpG island elements utilize chromatin-based processes to create environments that contribute to the transcriptional potential of the associated gene (Fig. 1). From a default permissive state, CpG islands appear to act as a platform on which additional signaling and chromatin modification systems alter and modify this default state to help define the functional output of the associated gene (Fig. 1). Despite recent advances in our understanding of CpG islands, there are still significant gaps in our knowledge of these abundant yet poorly understood elements. For example, the mechanisms by which CpG islands are kept free of DNA methylation in an otherwise heavily methylated genome remain a mystery. A recent study suggests that TET1, an enzyme that hydroxylyates methylated DNA,60,61 may be involved in the process of regulatory element demethylation.62 It is interesting to note that Tet1 also possesses a ZF-CxxC domain61 making it tempting to speculate that this domain targets DNA demethylase activity to CpG islands. With respect to CpG island chromatin architecture, more mechanistic investigation is required to directly define how chromatin impacts upon transcriptional regulation. Through defining these fundamental mechanisms, an understanding of how DNA methylation profiles and CpG islands provide a framework for gene regulatory processes will be achieved.
Acknowledgements
We would like to thank Professor Francisco Antequera and Dr. Sarah Cooper for critical reading of this manuscript. Work in R.J.K.'s lab is supported by the Wellcome Trust.
Abbreviations
- ZF-CxxC
zinc finger, CxxC
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- LPS
lipopolysaccharide
- ES cell
embryonic stem cell
- PRC
polycomb repressive complex
- MBD
methyl CpG binding domain
- ORI
origin of replication
- PHD
plant homeodomain
References
- 1.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 4.Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 7.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 10.Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 11.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGA CGT CA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 14.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 15.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 19.Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 23.Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone H3 variants. J Proteome Res. 2008;7:4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 25.Robin P, Fritsch L, Philipot O, Svinarchuk F, Ait-Si-Ali S. Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv391h and G9a. Genome Biol. 2007;8:270. doi: 10.1186/gb-2007-8-12-r270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuadrado M, Sacristan M, Antequera F. Species-specific organization of CpG island promoters at mammalian homologous genes. EMBO Rep. 2001;2:586–592. doi: 10.1093/embo-reports/kve131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 31.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 33.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, et al. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 37.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 38.Levine A, Yeivin A, Ben-Asher E, Aloni Y, Razin A. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J Biol Chem. 1993;268:21754–21759. [PubMed] [Google Scholar]
- 39.Shimamura A, Sapp M, Rodriguez-Campos A, Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989;9:5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolffe AP. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 1989;8:527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 49.Yao J, Mackman N, Edgington TS, Fan ST. Lipopolysaccharide induction of the tumor necrosis factor-alpha promoter in human monocytic cells. Regulation by Egr-1, c-Jun and NFkappaB transcription factors. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 50.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 51.Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci USA. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 53.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 54.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 55.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 56.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 57.Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez M, Antequera F. Overreplication of short DNA regions during S phase in human cells. Genes Dev. 2008;22:375–385. doi: 10.1101/gad.445608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5 mC to 5 hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]