Abstract
Included among the quantitative high throughput screens (qHTS) conducted in support of the U.S. Tox21 program are those being evaluated for the detection of genotoxic compounds. One such screen is based on the induction of increased cytotoxicity in 7 isogenic chicken DT40 cell lines deficient in DNA repair pathways compared to the parental DNA repair-proficient cell line. To characterize the utility of this approach for detecting genotoxic compounds and identifying the type(s) of DNA damage induced, we evaluated nine of 42 compounds identified as positive for differential cytotoxicity in qHTS (actinomycin D, adriamycin, alachlor, benzotrichloride, diglycidyl resorcinol ether, lovastatin, melphalan, trans-1,4-dichloro-2-butene, tris(2,3-epoxypropyl)isocyanurate) and one non-cytotoxic genotoxic compound (2-aminothiamine) for (1) clastogenicity in mutant and wild-type cells; (2) the comparative induction of γH2AX positive foci by melphalan; (3) the extent to which a 72-hr exposure duration increased assay sensitivity or specificity; (4) the use of 10 additional DT40 DNA repair-deficient cell lines to better analyze the type(s) of DNA damage induced; and (5) the involvement of reactive oxygen species in the induction of DNA damage. All compounds but lovastatin and 2-aminothiamine were more clastogenic in at least one DNA repair-deficient cell line than the wild-type cells. The differential responses across the various DNA repair-deficient cell lines provided information on the type(s) of DNA damage induced. The results demonstrate the utility of this DT40 screen for detecting genotoxic compounds, for characterizing the nature of the DNA damage, and potentially for analyzing mechanisms of mutagenesis.
Keywords: DT40 DNA repair-deficient cell lines, quantitative high throughput screens, cytotoxicity, genotoxicity, chromosomal aberrations, γH2AX positive foci
Introduction
The number of chemical compounds introduced into commercial use has increased dramatically over the last few decades and comprehensive toxicological profiles on many of these compounds are lacking [NRC, 2007]. In response to this lack of toxicological information, the original members of the U.S. Tox21 community (the U.S. National Toxicology Program [NTP], the National Institutes of Health Chemical Genomics Center [NCGC], the U.S. Environmental Protection Agency [EPA]), have been evaluating quantitative high throughput screens (qHTS) [Inglese et al., 2006] that could be used to help prioritize compounds for more extensive toxicological testing, to elucidate mechanisms of action, and ultimately for predicting the potential for adverse health effects in humans [Collins et al., 2008; Kavlock et al., 2009].
Among the toxicological endpoints of interest to the U.S. Tox21 partners is genotoxicity. Induction of DNA damage in eukaryote cells by physical and chemical agents is generally quantified by a number of methods, including those that evaluate (1) the frequency of chromosomal aberrations (CAs), micronuclei, or gene mutations [Cimino, 2006]; (2) the extent of DNA migration measured using the comet assay [Tice et al., 2000], or (3) the level of γH2AX positive foci detected in nuclear DNA [Mah et al., 2010]. None of these assays are currently suitable for qHTS. However, assays that measure the repair response of cells to DNA damage might be suitable for qHTS. DNA damage is repaired by DNA repair pathways that are specific for different classes of damage [Branzei et al., 2009]. For example, DNA double strand breaks (DSBs) are repaired by homologous recombination (HR) and non-homologous end joining (NHEJ) DNA repair pathways. Base damage is repaired by nucleotide excision repair (NER) and base excision repair (BER). Repair of DNA interstrand cross-links is accomplished via the Fanconi anemia (FA) and translesion DNA synthesis (TLS) pathways. In addition, check point mechanisms facilitate DNA repair by triggering cell cycle arrest, allowing for damage to be repaired prior to the entry of cells into S-phase or mitosis.
One approach that is being investigated for qHTS is the use of DT40 cell lines deficient in their ability to repair DNA damage. DT40 cells originate from a chicken B-lymphocyte line, isolated in 1985 from an avian leucosis virus-induced bursal lymphoma [Baba et al., 1985]. These cells possess several characteristics that make them particularly well-suited to studies involving DNA repair pathways. First, DT40 cells with a cell cycle time of ~8 hrs proliferate more rapidly than most mammalian cell lines [Hori et al., 2008; Zhao et al., 2007]. Second, during logarithmic growth, ~70% of DT40 cells are in S phase in contrast to the ~50% for mammalian cells [Takao et al., 1999]. Third, DT40 cells lack the G1/S checkpoint that is normally present in mammalian cells [Takao et al., 1999]. As a result, DT40 cells are exquisitely sensitive to the induction of DNA damage by genotoxic chemicals and, compared to mammalian cells, DNA repair activity contributes a greater extent to their survival. Another advantage of DT40 cells is the efficiency with which targeted gene disruption can be accomplished [Buerstedde and Takeda, 1991]. Through the use of targeted gene disruption techniques, a non-functioning mutant for every DNA repair pathway has been created, with each clone having the same genetic background as the parental wild-type DT40 cells.
Recently, the NCGC conducted an evaluation of a novel qHTS approach for identifying genotoxicity potential based on the detection of increased cytotoxicity in at least one of seven isogenic DT40 DNA repair-deficient cell lines (covering six DNA repair pathways) compared to that exhibited by the repair-proficient parental cell line under the same exposure conditions [Ji et al., 2009; R. Tice, manuscript in preparation]. The premise behind this strategy is that a decrease in DNA repair competency will increase the sensitivity of cells to the cytotoxic effects of DNA damage. The DNA repair-proficient parental cell line serves as the control in this screen, providing high sensitivity and specificity [Evans et al., 2010]. Furthermore, the use of a panel of DNA repair-deficient cell lines allows for characterization of the nature of the DNA lesions caused by genotoxic chemicals [Mizutani et al., 2004; Nojima et al., 2005; Wu et al., 2006]. In the initial DT40 qHTS study, the NCGC screened a library of 1408 compounds provided by the NTP (the complete list of compounds in this library is available at http://www.ncbi.nlm.nih.gov/sites/entrez?db=pcsubstance&term=NTPHTS) for increased cytotoxicity in a set of seven DT40 DNA repair-deficient cell lines compared to the parental DNA repair-proficient cell line. These DNA repair-deficient cell lines included mutants for ATM, FancC, Polβ (2 clones), Rad54/Ku70, Rev3, and Ubc13 (Table 1). Polβ is active in BER, FancC is active in DNA interstrand cross-link repair, Rad54/Ku70 and Ubc13 are active in DSB repair, Rev3 and Ubc13 are active in TLS repair, and ATM is active in the DNA damage checkpoint (Table I and references therein). In the initial qHTS study, cells from each DT40 cell line (mutant and wild-type) were exposed in a 1536-well plate format to the 1408 compounds over a 14-point concentration range from 0.59 nM to 92 μM (single wells per concentration) for 24 hrs in the absence of metabolic activation. At the end of the exposure period, the extent of cytotoxicity was determined by measuring the levels of intracellular adenosine triphosphate (ATP) in each well. Possible genotoxicity was based on the presence of a significant increase in cytotoxicity in one or more of the DNA repair-deficient cell lines compared to that observed in the parental DNA repair-proficient cell line. Cytotoxicity was quantified as the concentration of a compound that induced a 50% decrease in ATP levels (i.e., the IC50). A significant increase in cytotoxicity in one or more of the DNA repair-deficient cell lines versus the parental cell line was defined as a difference of at least 6-fold in IC50 values (mutant clone < DNA repair-proficient clone). Based on this criterion, 42 compounds were identified as exhibiting possible genotoxic activity in the absence of metabolic activation. Confirmation qHTS studies were conducted to verify the initial qHTS finding for these 42 compounds using 24-point titrations at concentrations ranging from 11 pM to 92 μM, with each concentration tested in triplicate. In total, five independent confirmatory studies were conducted, three times using a 24-hr exposure duration and two times using a 48-hr exposure duration. The 48-hr experiments were conducted to evaluate the effect of increased exposure duration on the results observed at 24 hrs. In the more comprehensive qHTS confirmation studies, a statistically significant decrease in IC50 values was used within each experiment to identify compounds inducing differential cytotoxicity in at least one DNA repair-deficient cell line. Based on this criterion, all 42 compounds were classified as potential direct-acting genotoxicants.
Table I.
Panel of Isogenic DNA Repair-Deficient DT40 Cell Lines
| Name1 | Function | Reference |
|---|---|---|
| ATM2 | Checkpoint kinase, arrests cell cycle when chromosomal breaks are present. | Takao et al. (1999) |
| Brca2 | HR dependent DSB repair. | Hatanaka et al. (2005) |
| CtIP(S332A/−/−) | Eliminating covalently bound polypeptides from DSBs. | Nakamura et al. (2010) |
| CtIP(+/−/−) | Control clone for CtIP(S332A/−/−) | Nakamura et al. (2010) |
| FancC2 | Eliminating ICLs. | Kitao et al. (2006), Gurtan and D'Andrea (2006) |
| FancD2 | Eliminating ICLs. | Yamamoto et al. (2005) |
| Fen1 | BER, processing 5'flap in long patch and lagging strand DNA replication. | Matsuzaki et al. (2002) |
| Ku70 | Initial step for NHEJ dependent DSB repair. | Takata et al. (1998) |
| Nbs1/p70 | HR and initiation step for NHEJ. | Nakahara et al. (2009) |
| Pol-β2,3 | BER, repairs of basal damages caused by oxidative stress and environmental factors. | Horton et al. (2008) |
| Rad54 | HR dependent DSB repair. | Bezzubova et al. (1997) |
| Rad54/Ku702 | HR and NHEJ DSB repair | Takata et al. (1998) |
| Rev32 | TLS polymerase, involved in HR and also works epistatically with FancC in cross-linking tolerance. | Sonoda et al. (2003) |
| Rev3/FancC | TLS and ICLs repair. | Nojima et al. (2005) |
| Ubc132 | UBC13 is related to the initial step of HR and TLS. | Zhao et al. (2007) |
| XPA | Initial step of NER. | Okada et al. (2002) |
Abbreviations: BER = base excision repair, BRCA2 = breast cancer Type 2, CtIP = tumor suppressor protein, DSB = double strand break, FANCC = Fanconi anemia complementation group C, FANCD2 = Fanconi anemia complementation group D2, FEN1 = flap-endonuclease-1, HR = homologous DNA recombination, ICL = interstrand cross-linking, Ku70 = protein encoded by the XRCC6 gene that is required for non-homologous end joining, Nbs1/p70 = Nijmegen breakage syndrome, NER = nucleotide excision repair, NHEJ = non-homologous end joining, Pol-β = DNA polymerase-β, Pol-ζ = DNA polymerase-ζ, Rad54 = DNA-dependent ATPase, Rev3 = catalytic subunit of DNA polymerase ζ, TLS = translesion DNA synthesis, Ubc13 = Ubiquitin (Ub)-conjugating enzyme 13, XPA = xeroderma pigmentosum, complementation group A
DNA repair-deficient mutant clones used in qHTS and chromosome analysis.
Two isogenic clones were used.
To further characterize the ability of this qHTS approach to identify direct-acting genotoxic compounds, we evaluated a subset of nine compounds from among the 42 potential genotoxicants for their ability to induce CAs across the panel of DNA repair-deficient and repair-proficient isogenic cell lines. Compounds were selected that exhibited different patterns of activity among the seven DT40 DNA repair-deficient clones. The nine compounds selected were actinomycin D, adriamycin, alachlor, benzotrichloride, diglycidyl resorcinol ether, lovastatin, melphalan, trans-1,4-dichloro-2-butene, and tris(2,3-epoxypropyl) isocyanurate. We also tested 2-aminothiamine, which has been reported to be mutagenic in vitro in mouse lymphoma cells in the absence of metabolic activation [Cameron et al., 1985] but was not detected as differentially cytotoxic in any of the DNA repair-deficient cell lines under the experimental conditions used. We measured the number of CAs in metaphase cells obtained from cultures of each cell line exposed to each compound for 24 hrs. For each of the nine positive compounds, the concentrations tested were based on the IC50 value obtained in the confirmation qHTS studies; 2-aminothiamine was tested at concentrations up to 92 μM, the maximum concentration tested in the qHTS cytotoxicity assays. For melphalan, we evaluated also for the induction of DNA double strand breaks, as measured by the presence of increased numbers of γH2AX-positive foci in a DNA repair-deficient cell line mutant for FancC compared to the parental cell line. We then expanded our characterization of this approach for detecting genotoxic compounds by (1) evaluating the extent to which a longer exposure duration of 72 hrs would increase the sensitivity or specificity of the assay, (2) expanding the analysis of differential cytotoxicity to additional DT40 DNA repair-deficient clones, and (3) evaluating the involvement of reactive oxygen species (ROS) in the induction of DNA damage. We demonstrate here that isogenic DNA repair-deficient DT40 clones allow for reliable detection of genotoxic compounds as well as for characterization of the nature of the DNA lesions induced.
Materials and Methods
Cell lines and culture conditions
The DT40 cell lines were obtained from the Laboratory of Radiation Genetics, Graduate School of Medicine in Kyoto University, Japan. The wild-type cell line and the DNA repair-deficient cell lines differ only by the presence/absence of an endogenous DNA repair gene; other than this difference, their genetic backgrounds are identical (i.e., they are isogenic). Ultimately, 16 DNA repair-deficient clones were used in this study (Table I). The disrupted genes in the DNA repair-deficient cell lines include the ataxia telangiectasia-mutated (ATM) gene for the DNA damage checkpoint; breast cancer Type 2 (Brca2), Rad54, Ubiquitin (Ub)-conjugating enzyme 13 (Ubc13), and Nijmegen breakage syndrome (Nbs1p70) genes for HR; Ku70 gene for NHEJ; Fanconi anemia complementation group C (FancC) and group D2 (FancD2) genes for the repair of interstrand cross-links (ICLs); Ubc13 and Rev3 genes for TLS; the DNA polymerase β (Polβ) and Flap-endonuclease-1 (Fen1) genes for the BER pathway; xeroderma pigmentosum complementation group A (XPA) for the NER pathway; and CtIP and Nbs1 genes for removal of polypeptides at DNA double strand break ends. In addition, isogenic DNA repair-deficient cell lines with disruption of two DNA repair genes (Rad54/Ku70 genes for HR and NHEJ, Rev3/FancC genes for TLS and ICLs repair) were used. The seven isogenic mutant cell lines (ATM, FancC, two clones of Polβ, Rad54/Ku70, Rev3, and Ubc13) used for the initial and confirmation qHTS studies and CA analysis at the NCGC are identified in Table 1 by footnote. The ten other DNA repair-deficient cell lines were used in additional differential cytotoxicity studies conducted at the Laboratory of Radiation Genetics, Graduate School of Medicine in Kyoto University, Japan. All cell lines were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 1% chicken serum (Sigma-Aldrich), 0.1% 55 mM mercaptoethanol (Invitrogen), and 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen). The cell lines were maintained at 39°C under a humidified atmosphere and 5% CO2
Compounds
Unless otherwise noted, the following compounds were purchased from Sigma-Aldrich: actinomycin D (Chemical Abstracts Services Registry Number [CASRN] = 50-76-0; purity = 98%), adriamycin (CASRN = 25316-40-9; purity = 98%), alachlor (CASRN = 15972-60-8; purity = 99.2%), benzotrichloride (CASRN = 98-07-7; purity = 99.6%), lovastatin (CASRN = 75330-75-5; purity = 98% from Sigma or >98% from ENZO Life Science, Plymouth Meeting, PA, USA), melphalan (CASRN = 148-82-3; purity = 95%), trans-1,4-dichloro-2-butene (CASRN = 110-57-6; purity = 98%), tris(2,3-epoxypropyl) isocyanurate (CASRN = 2451-62-9; purity = 99.5 %), and 2-aminothiamine (CASRN = 96-50-4; purity >90%). Diglycidyl resorcinol ether (CASRN = 101-90-6; purity = 94%) was obtained from Alpha Aesar (Ward Hill, MA, USA). All compounds were dissolved in dimethyl sulfoxide (DMSO; Fisher Scientific, Pittsburgh, PA), at either 10 or 20 mM prior to use.
qHTS differential cytotoxicity studies conducted at the NCGC
In the initial qHTS study, DT40 cells were dispensed at 2,000 cells/5 μL/well in 1536-well white/solid bottom plates (Greiner Bio-One North America, NC, USA) using a Flying Reagent Dispenser (FRD; Aurora Discovery, CA, USA), followed by addition of 23 nL of 1408 compounds at 14 concentration ranging from 0.59 nM to 92 μM into the assay plates by using a pin tool (Kalypsys, San Diego, CA, USA). Tetra octyl ammonium bromide at 92 μM was used as the positive control in the screen. The positive control was arrayed as follows: Column 1, concentration-response titration of tetra octyl ammonium bromide from 1.4 nM to 46 μM; Column 2, 92 μM tetra octyl ammonium bromide; Column 3, DMSO only; and Column 4, 23 μM tetra octyl ammonium bromide. The concentration-response titration for tetra octyl ammonium bromide was used to evaluate plate-to-plate consistency, based on the calculated IC50. The compound stock solutions were at 10 mM dissolved in DMSO; this translates into a 46 μM maximum concentration using the NCGC standard dilution process that creates final concentrations ranging from 0.59 nM to 46 μM (with a corresponding DMSO concentration of 0.45%) in 1536-well plates. To achieve a maximum final compound concentration of 92 μM (with a corresponding DMSO concentration of 0.9%), 23 nL was transferred twice from the highest concentration mother plate into each well of the assay plate. These concentrations are typically far lower than those used in traditional in vitro genotoxicity assays, but are more relevant to the concentrations that could reasonably be achieved in the plasma of an exposed human. After the assay plates were incubated for 24 hrs at 39°C, 5 μL per well of CellTiter-® (Promega, Madison, WI, USA) reagent was added into each well of the assay plate. The plates were incubated for 30 min at room temperature and the luminescence intensity of the plates was measured using a ViewLux plate reader (Perkin Elmer, Shelton, CT, USA). In this assay, the luminescent signal is proportional to the amount of ATP and thus to the number of metabolically competent cells [Crouch et al., 1993]. To confirm the results of the initial qHTS study, 42 compounds re-purchased from commercial vendors were re-tested at 24-point titrations with concentration ranging from 11 pM to 92 μM in the cytotoxicity assay, using the same protocol as described above except the 24 titrations were all within one 1536-well plate. The stock solution of each compound used for these experiments was 20 mM. The confirmation qHTS studies were performed five times in total with two different incubation times, three times for 24 hrs and two times for 48 hrs.
qHTS data analysis
Data were analyzed as described previously [Xia et al., 2008]. Briefly, data were normalized and corrected using the DMSO controls (0%) and positive control compound (tetra octyl ammonium bromide; −100%), and concentration response curves were fit to the Hill equation yielding concentrations of half-maximal inhibition (IC50) and maximal cytotoxicity (efficacy) values. The concentration response curves were sorted into four major classes (1–4) using previously published criteria [Xia et al., 2008]. Curve classes were further subdivided to provide more detailed classification. Briefly, compounds with class 1.1, 1.2, 2.1 or 2.2 (efficacy >50%) curves were considered active; class 4 compounds showed no concentration response and were defined as inactive; and compounds with other curve classes were defined as inconclusive. Compounds with an IC50 value at least six times lower in at least one DNA repair-deficient cell line than that obtained in the parental DNA repair-proficient cell line were classified as active and selected for confirmation studies. The 6-fold IC50 criteria was based on individual compound IC50 values from selected cell lines (Rad54/Ku70, 2 polβ clones) run in duplicate during the initial qHTS screen; in these experiments, it was determined that this difference is significantly greater (p<0.05) than what can be expected from experimental variation. In the 42-compound confirmation studies where each compound was tested in triplicate, compounds were considered differentially cytotoxic if they showed significantly higher potency, based on their respective IC50 values, in the DNA repair-deficient cell line than in the parental DNA repair-proficient cell line by student's t-test at p<0.05.
Chromosomal aberration analysis
For CA analysis, we used the same seven DNA repair-deficient clones (ATM, FancC, Polβ [2 clones], Rad54/Ku70,Rev3, and Ubc13 mutants) used for the differential cytotoxicity qHTS assays. We modified a previously described protocol to obtain and prepare metaphase cells from each DT40 cell line [Sonoda et al., 1998]. Briefly, DT40 cells were incubated with RPMI1640 medium with 10% heat inactivated FBS, 1% chicken serum, and 0.1% 50 mM mercaptoethanol at 39°C for 24 hrs. To arrest cells in metaphase, 0.1% Colcemid (GIBCO-BRL, Grand Island, NY, USA) was added 3 hrs before harvest. Cells were transferred to 15 mL centrifuge tubes (Greiner Bio-One North America, Monroe, NC, USA), pelleted by centrifugation, resuspended in 1 mL of 75 mM KCl for 15 min at room temperature, pelleted again, and then fixed in 5 mL of a freshly prepared 3:1 mixture of methanol/acetic acid (i.e., Carnoy's solution). The cell suspension was dropped onto cleaned glass slides and air dried for 30 min. The slides were stained with 5% Harleco Giemsa stain solution (EMD Chemicals, Gibbstown, NJ, USA) for 10 min, and dried after being rinsed carefully with water.
The chicken karyotype consists of 80 chromosomes, including 11 major autosomal macrochromosomes, the ZW sex chromosomes, and 67 microchromosomes [Sonoda et al., 1998]. For CA analysis, 50 Giemsa-stained metaphase cells per concentration tested were scored at 1000x magnification with scoring limited to the 11 major macrochromosomes and the Z chromosome [Sonoda et al., 1998]. CAs were classified as chromosome or chromatid gaps, breaks, and exchanges (triradial, quadriradial, ring, dicentric or other) according to the International System for Human Cytogenetic Nomenclature (ISCN) system [ISCN, 1985]. A gap is classified as a clear non-staining region on a chromatid or chromosome that is equal to or less than the width of a chromatid, and a break is a discontinuity of the chromatid or chromosome that shows a clear misalignment of the distal fragment. Iso-chromatid breaks and chromosomal breaks were not distinguished from each other. The percentage of mitotic cells in 1000 cells per culture was determined and used to calculate the mitotic index (MI). A one-tailed Fisher's exact test was used to analyze the difference in the number of CAs between each DT40 DNA repair-deficient cell line and the parental DNA repair-proficient cell line using GraphPad Prism 4 for Windows (GraphPad Inc., San Diego, CA, USA).
Detection of γH2AX sites in nuclear DNA
To evaluate for the differential induction of DNA double strand breaks by melphalan in the DT40 DNA repair-deficient cell line mutant for FancC vs. the parental DNA repair-proficient cell line, we determined the number of γH2AX positive foci in nuclear DNA. Experimental conditions for immunocytochemical analysis were as described previously [Takata et al., 2001]. Briefly, cells from the DT40 cell line mutant for FancC and the parental DNA repair-proficient cell line (1.0 ×105 cells) treated with melphalan at 0.5 μM and 1.5 μM for 1 hr were collected on a slide glass using a Cytospin3 (Shandon, Pittsburgh, PA, USA). The concentrations of melphalan were the same as those used for the CA analysis. Cells were cross-linked with 4% para-formaldehyde (Nacalai tesque, Kyoto, Japan) for 10 min at room temperature. To permeabolize the cells, 0.1%NP40/PBS (nonidet p-40, Nacalai tesque) was loaded onto the slides. After blocking with 3% BSA/PBS (bovine serum albumin F-V, pH 5.2; Nacalai tesque), the cells were treated with specific primary anti-γH2AX monoclonal mouse antibodies (1:1000, Millipore, Billerica, MA, USA) for 45 mins under humidified conditions at 37°C, followed by secondary Alexa 488-conjugated anti-mouse IgG antibodies (1:1000, Molecular Probes, Eugene, OR, USA) for 45 mins. The nucleus of at least 50 morphologically intact cells was examined per dose group at 1000× magnification for the number of γH2AX-positive foci. This experiment was conducted twice and the data combined across the two experiments. For descriptive purposes, the mean and median values for the number of γH2AX-positive foci per cell were calculated (data not shown). A one-tailed Fisher's exact test was used to analyze the difference in the number of cells with >4 γH2AX-positive foci between the DNA repair-deficient cell line mutant for FancC and the parental DNA repair-proficient cell line using GraphPad Prism 4 for Windows (GraphPad Inc.). This conservative criterion is based on previous studies conducted with this assay (data not published).
Follow-up cytotoxicity and ROS studies conducted at Kyoto University
Cytotoxicity was also evaluated after exposure of various isogenic DT40 cell lines to these compounds for 72 hrs. In these experiments, 10,000 cells/1000 μL/well were dispensed into 24-well tissue culture plates and treated with 1–5 mL of each compound dissolved in DMSO, followed by incubation for 72 hrs at 39(C. The concentration range used for each compound was based on the maximum concentration used for the CA analyses conducted in NCGC. Initially, five compound concentrations (1/100-, 1/10-, 1-, 10-, and 100- fold of the maximum concentration tested for CA induction) of each compound were tested to identify the optimal concentration range to be used in the 72-hr cytotoxicity test. Selection of the final concentration range was based on achieving at least 10% cytotoxicity in the parental DNA repair-proficient cell line. Using from three to six concentrations per compound, differential cytotoxicity experiments were conducted using triplicate wells per concentration to estimate the concentration that induced an IC10 (i.e., the concentration that induced a decrease in viability of 10%). Differential cytotoxicity in these experiments was based on an IC10 rather than an IC50 because of the relatively few concentrations tested over a narrow concentration range (i.e., 3 to 6 vs 14 or 24 at the NCGC). Cells in 100 μL volumes were transferred to solid 96-well plates, and 100 μL of CellTiter-Glo® reagent was added to each well. The plates were incubated at room temperature for 10 min, and the luminescence intensity of each well was determined using a multiplex luminometer (Fluoroskan Ascent FL; Thermo Fisher Scientific Inc., MA, USA). A compound was considered to exhibit increased cytotoxicity in one or more DNA repair-deficient cell lines if the calculated IC10 value compared to that calculated for the parental DNA repair-proficient cell line was at least 1.4-fold different. To evaluate the involvement of ROS in the induction of DNA damage by some of the 10 compounds, N-acetyl-L-cysteine (NAC; Sigma-Aldrich), a reactive oxygen species scavenger, was added 2 hrs prior to treatment.
Results and Discussion
To further characterize the utility of the DT40 qHTS differential cytotoxicity assay for detecting direct-acting genotoxic compounds, we evaluated the ability of nine compounds classified as positive in the qHTS studies for their ability to differentially induce CAs across a panel of seven isogenic DNA repair-deficient cell lines and the DNA repair-proficient parental cell line. Except for lovastatin, each of the nine compounds is known to be a direct-acting genotoxicant from standard in vitro genotoxicity assays (Table II). However, selection of these compounds for further characterization was not based on historical genetic toxicology information but rather on the pattern of differential cytotoxicity responses obtained across the seven DNA repair-deficient cell lines. In addition to the nine positive compounds, we also tested 2-aminothiamine; this compound was negative for differential cytotoxicity over the concentration range tested in qHTS but has been reported to be a direct-acting mutagen in vitro [Cameron et al., 1985]. Our expectation was that the DNA repair-deficient cell lines that exhibited a significant increase in cytotoxicity compared to the parental DNA repair-proficient cell line would also exhibit more clastogenic damage. In addition, we conducted other studies on a case-by-case basis to evaluate whether a compound induced DNA DSB and/or whether ROS was involved in the induction of differential cytotoxicity. Finally, for the nine compounds, we evaluated the effect of increasing the exposure duration from 24 or 48 hrs to 72 hrs on the nature and extent of differential cytotoxicity detected in different DNA repair-deficient cell lines, including ten additional DT40 DNA repair-deficient cell lines with knock-outs for other DNA repair genes. The purpose of these latter studies was to determine if increasing the exposure duration increased assay sensitivity and to provide more information about the nature of the DNA lesions induced by the different compounds. The results obtained using different exposure durations are provided in Figure 1.
Table II.
Known Rodent Carcinogenicity and Genotoxicity of Compounds Selected for Additional Investigation1
| Compound2 | CASRN | Purity | Rodent Carcinogenicity | In Vitro Mammalian Cell Genotoxicity3 | Ames Test Mutagenicity3 | Known DNA repair pathway (Reference) |
|---|---|---|---|---|---|---|
| Actinomycin D | 50-76-0 | 98% | Yes | positive | negative | Nbs1 (Porcedda et al., 2006) |
| Adriamycin | 25316-40-9 | 98% | Yes | positive | positive | NER, HR (Spencer et al., 2008) |
| Alachlor | 15972-60-8 | 99.2% | Yes | positive | - | Excision repair (Surrallés et al., 1995) |
| Benzotrichloride | 98-07-7 | 98% | Yes | positive | negative4 | none |
| Diglycidyl resorcinol ether | 101-90-6 | 94% | Yes | positive | positive | none |
| Lovastatin | 75330-75-5 | ≥98% | Yes | negative | negative | none |
| Melphalan | 148-82-3 | 95% | Yes | positive | positive | Fanconi anemia pathway (Nojima et al., 2005) |
| Trans-1,4-Dichloro-2-butene | 110-57-6 | 98% | Yes | positive | positive | none |
| Tris(2,3-epoxypropyl)isocyanurate | 2451-62-9 | 99.5 % | Yes | positive | positive | none |
| 2-Aminothiamine | 96-50-4 | >90% | Yes | positive | negative4 | none |
Abbreviations: CASRN = Chemical Abstracts Registry Service Number, ICL = interstrand cross-linking, Nbs1/p70 = Nijmegen breakage syndrome, NHEJ = non-homologous end joining.
Diglycidyl resorcinol ether was purchased from Alpha Aesar; the other compounds were purchased from Sigma-Aldrich, except that lovastatin was purchased from Sigma-Aldrich and then again from ENZO Life Science.
In vitro mammalian cell genotoxicity (e.g., tests for chromosomal aberrations, sister chromatid exchanges, or micronuclei in cultured Chinese hamster cells or mitogen-stimulated human lymphocytes, mutagenicity in mouse lymphoma cells) and Ames mutagenicity test results only in the absence of metabolic activation; test results were obtained from EPA ACToR and United States National Library of Medicine; TOXNET.
Positive in the presence of metabolic activation.
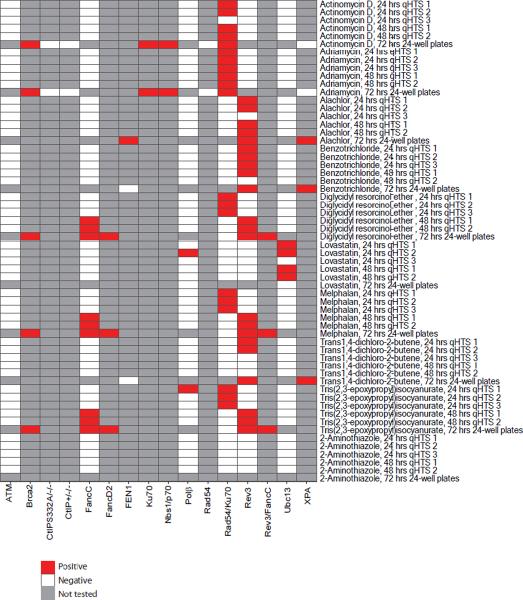
Figure 1.
Heat-map representing the presence of increased cytotoxicity in DT40 DNA repair-deficient cell lines compared to the parental DNA repair-proficient cell line at different culture times. In the follow-up study after the primary screening, all of the compounds listed in this heat-map were tested for inducing cytotoxicity after treatment for 24 hrs in three independent experiments (qHTS 1, qHTS 2, and qHTS 3), for 48 hrs in two independent experiments (qHTS 1 and qHTS 2), and for 72 hrs in one experiment (72-hr 24-well plates).
A compound that showed statistically significant differential toxicity against a mutant line is considered positive and colored red; a compound that did not show significant differential toxicity is considered negative and colored white; and compounds not tested are colored grey.
To confirm the genotoxicity of the nine positive compounds identified from qHTS and the lack of genotoxicity of the one negative compound, we determined the number of CAs in metaphase cells collected from the different DT40 DNA repair-deficient cell lines and the parental DNA repair-proficient cell line after exposure to each chemical for 24 hrs. We also determined the MI of each culture to assess the extent of cytotoxicity. The concentration of each compound selected for CA analysis was based on the IC50 determined in the confirmation qHTS studies. The results of these and additional mechanistic studies are detailed below.
Actinomycin D and Adriamycin
DSBs are the most toxic form of DNA damage, as a single unrepaired DSB could result in cell death, and inaccurate DSB repair can result in chromosomal rearrangements. Ionizing radiation (IR) directly induces DSBs, while UV photoproducts cause DSBs through interference with DNA replication. The former type of DSB is repaired preferentially by NHEJ, while DSBs caused by interference with replication are mainly repaired by HR. Thus, the comparative sensitivity of cell lines deficient in NHEJ and HR to compounds can be used to assess the mechanism underlying DSB formation.
Both actinomycin D and adriamycin are chemotherapeutic agents. Adriamycin is a topoisomerase II poison; it interferes with the dissociation of topoisomerase II from DNA thereby generating DSBs covalently associated with topoisomerase II [Tewey et al., 1984]. NHEJ has a major role and HR a minor role in repairing such DSBs. Actinomycin D is commonly used for the treatment of childhood cancers such as Wilms' tumor and Ewing's sarcoma [Estlin et al., 2003]. This compound inhibits mRNA synthesis by interfering with RNA polymerase [Bensaude et al., 1999]. The genotoxicity of actinomycin D is detectable by the micronucleus assay in mice (NTP unpublished data) and in cultured Chinese hamster lung cell lines [Hashimoto et al., 2010] as well as in the yeast DEL assay [Kirpnick et al., 2005], but not by the Ames reverse mutation assay [Suter et al., 1982]. However, unlike adriamycin, the mechanism by which actinomycin D induces genomic DNA damage remains has not been elucidated.
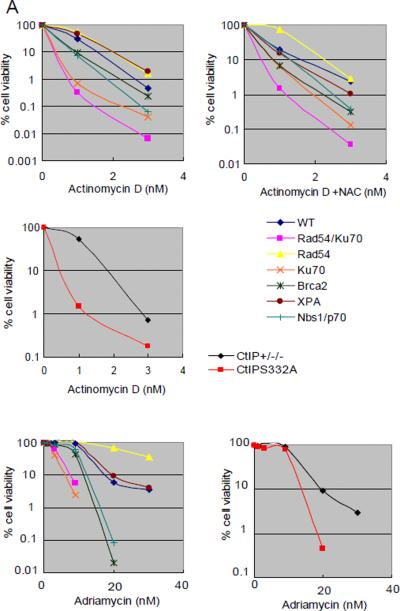
Similar to what was observed with another topoisomerase II poison, VP-16 [Adachi et al., 2003], NHEJ deficient Ku70 and Rad54/Ku70 mutant cells are hypersensitive to adriamycin (Fig 2A). However, adriamycin increased the number of CAs in Rad54/Ku70 mutant cells only (Fig 3B). Brca2 but not Rad54 mutant cells are also hypersensitive to adriamycin (Fig 2A), indicating that HR initially works at the DNA-topo cleavable complex site, which is subsequently replaced by the NHEJ pathway. These observations agree with the notion that this compound directly induces DSBs but not through interference with replication. We analyzed Nbs1−/−/− cells expressing a truncated form of Nbs1 (Nbs1−/−/−/Nbs1p70 cells) and CtIPS332A/−/− cells, because both Nbs1 and CtIP are involved in the elimination of chemical modifications which are oligonucleotides covalently bound to polypeptides from DSBs such as topoisomerase II, thereby facilitating subsequent DSB repair. CtIP+/−/− cells are used as control cells when CtIPS332A/−/− cells are analyzed, because CtIPS332A/−/− cells are generated from CtIP+/−/− cells and posses the same background genotype as the parental cells. Nbs1−/−/−/Nbs1p70 cells as well as CtIPS332A/−/− cells indeed showed increased sensitivity to adriamycin, in comparison to wild-type and CtIP+/−/− control cells, respectively (Fig 2A). These observations are consistent with a previous report [Nakamura et al., 2010], and indicate that adriamycin induces DSBs that require the removal of the polypeptides as well as NHEJ repair.
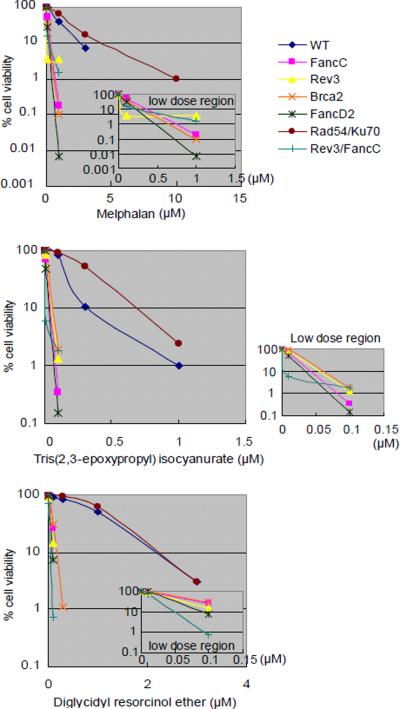
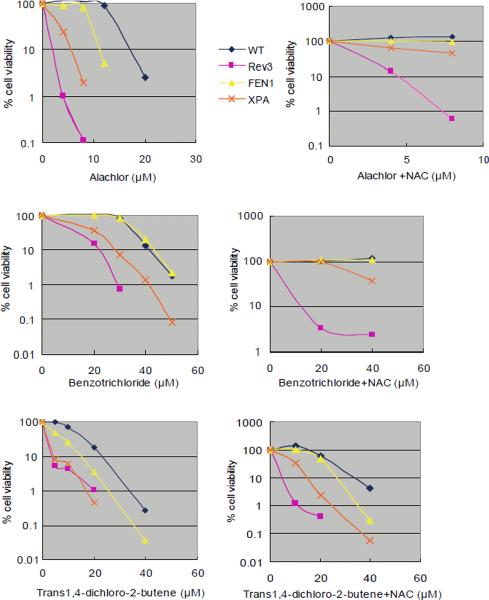
Figure 2.
Sensitivity of DT40 isogenic DNA repair-deficient cell lines and the parental DNA repair-proficient cell line to chemical compounds.
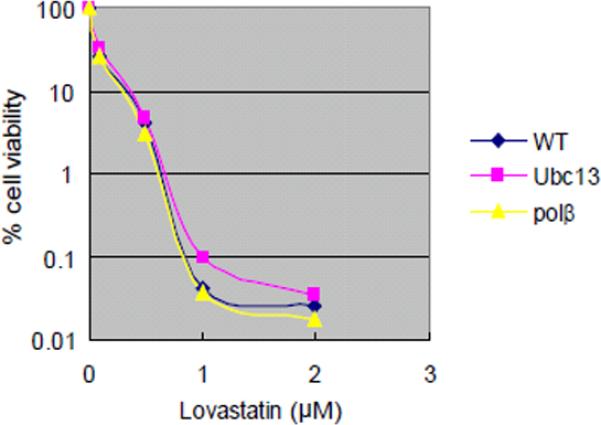
Analysis of cell number by measuring levels of cellular ATP after a 72-hr exposure to actinomycin D and adriamycin (A); melphalan, diglycidyl resorcinol ether, tris(2,3-epoxypropyl)isocyanurate (B); alachlor, benzotrichloride, and trans-1,4-dichloro-2-butene (C); and lovastatin (D); (A–C) actinomycin D, alachlor, benzotrichloride, and trans-1,4-dichloro-2-butene were treated with or without antioxidant (1 mM N-acetyl-L-cysteine [NAC]). CtIPS332A cells are compared with CtIP+/−/− control cells (A).
The concentrations of each compound are displayed on the x-axis in a linear scale, whereas the corresponding extent of cytotoxicity (presented as % cell viability) is displayed on the y-axis in a logarithmic scale. On this log scale, the standard deviations for the data are within the symbol for each data point and are excluded for ease of viewing.
Figure 3.
Frequencies of CAs in DT40 isogenic DNA repair-deficient cell lines and the parental DNA repair-proficient cell line.
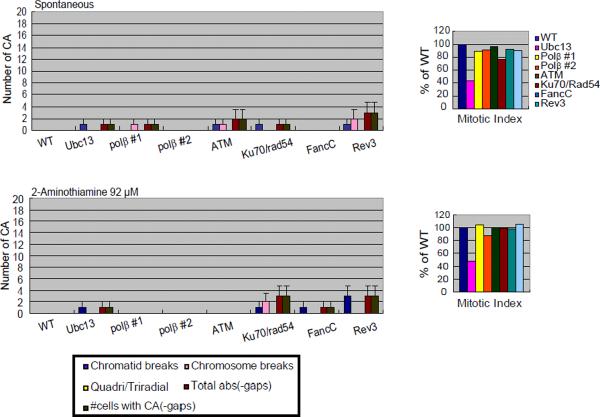
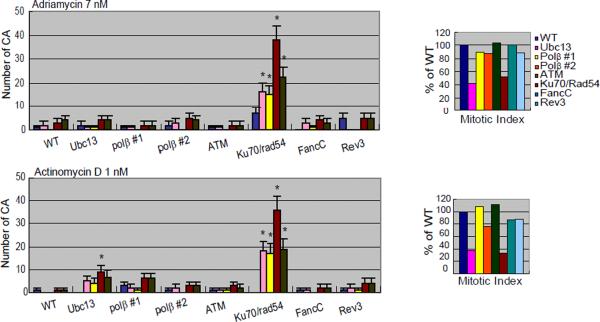
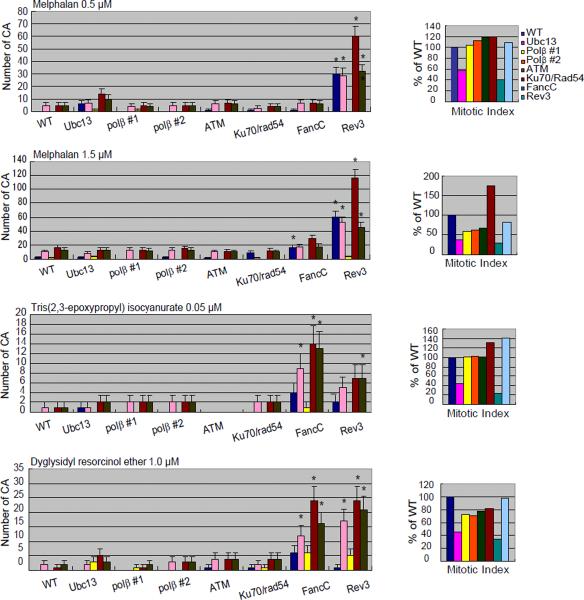
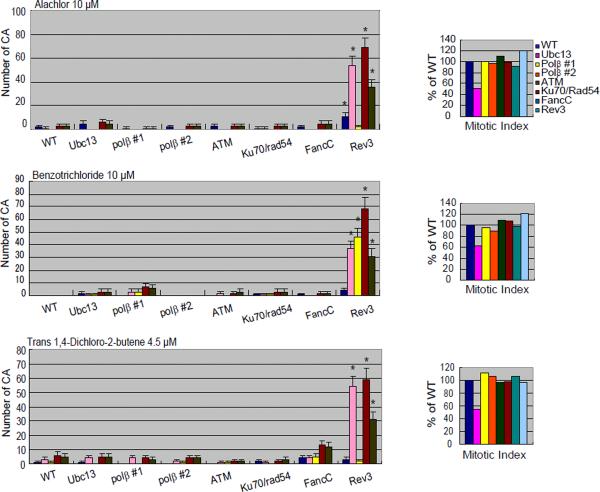
Analysis of numbers of CAs without any compounds (spontaneous; A) or after a 24-hr exposure to 2-aminothiazole (A); actinomycin D and adriamycin (B); melphalan, diglycidyl resorcinol ether, tris(2,3-epoxypropyl)isocyanurate (C); alachlor, benzotrichloride, and trans- 1,4-dichloro-2-butene (D).
The numbers of CAs are displayed on the y-axis in a linear scale. CAs were scored in 50 metaphase cells. The mitotic index (MI) in each DNA repair-deficient cell line is presented as the percentage of the MI obtained for the parental DNA repair-proficient cell line (1000 cells were scored per sample). Cells were cultured with each compound for 24 hrs, with Colcemid present for the last 3 hrs. Quadri/Triradial = the numbers of complex rearrangements; total abs (-gaps) = the total number of aberrations excluding gaps; # cells with CA (-gap) shows the number of cells that have at least one CA. Error bars show mean ± standard deviation. DNA repair-deficient cell lines with significant increase in the number of CA compared to the parental DNA repair-proficient cell line at p <0.05 (one-tailed Fisher's exact test) are indicated by an asterisk.
Actinomycin D significantly reduced the viability of Rad54/Ku70 mutant cells in both the 24 hrs and the 48 hrs qHTS experiments (Fig 1). Consistent with these data, increased numbers of CA were observed selectively in Rad54/Ku70 mutant cells (Fig 3B). We wished to investigate whether Rad54-dependent HR or Ku70-dependent NHEJ plays a major role in repairing actinomycin D-induced DSBs. Thus, we analyzed the sensitivity of Ku70 and Brca2 or Rad54-deficient HR mutant cells to actinomycin D using a 72-hr exposure duration. The Ku70 mutant cells showed increased sensitivity, whereas the Rad54 mutant cells did not exhibit significantly increased sensitivity to actinomycin D (Fig 2A). Brca2 mutant cells showed mild sensitivity to actinomycin D. Thus, actinomycin D-induced DSBs are repaired primarily by NHEJ and partially by HR, as are adriamycin-induced DSBs. To gain insight into the nature of actinomycin D-induced DSBs, we also analyzed Nbs1−/−/−/Nbs1p70 mutant cells as well as CtIPS332A/−/− cells. The Nbs1p70 and CtIPS332A/−/− clones showed significantly higher sensitivity to actinomycin D, when compared to wild-type and CtIP+/−/− cells, respectively (Fig 2A). Hence, we conclude that like adriamycin, actinomycin D generates DSBs carrying a chemical modification that needs to be removed prior to HR- and NHEJ-dependent DSB repair. This chemical modification may not be attributable to ROS, as pretreatment with NAC prior to addition of actinomycin D did not rescue the cells from increased cytotoxicity. Taken together, our data indicate that actinomycin D directly induces DSBs that likely carry chemical modification.
Tris(2,3-epoxypropyl) isocyanurate, Diglycidyl Resorcinol Ether, and Melphalan
Melphalan, an alkylating nitrogen mustard extensively used as an anti-neoplastic agent, damages DNA by forming mono-adducts as well as cross-links [Lawley et al., 1996]. Cross-link damage is classified into three types: interstrand cross-links, intrastrand cross-links, and protein-DNA cross-links. Interstrand cross-links (ICLs), the most harmful type of damage, are repaired collaboratively by the FA and TLS pathways [Nojima et al., 2005]. In contrast, cellular tolerance to the other types of cross-links depends on TLS but not on the FA pathway. Thus, the sensitivity of the FA mutant cells provides diagnostic information on whether a given genotoxic compound generates ICLs. Indeed, cellular sensitivity to melphalan was significantly increased in three FA mutant cells - Fanc-C, Fanc-D1 (Brca2), and Fanc-D2 - after incubation with melphalan for 72 hrs (Fig 2B). We found that these FA mutants also exhibited increased sensitivity to tris(2,3-epoxypropyl)isocyanurate and diglycidyl resorcinol ether, indicating that these compounds also induce ICLs (Fig 2B).
To verify that the cell sensitivity reflects reduced capability of DNA repair, we measured the number of CAs in different DNA repair-deficient clones exposed to diglycidyl resorcinol ether, tris(2,3-epoxypropyl)isocyanurate, and melphalan. Tris (2,3-epoxypropyl) isocyanurate and diglycidyl resorcinol ether induced higher numbers of CAs in the Fanc-C mutant cells in comparison with wild-type cells (Fig 3C). This observation is consistent with induction of ICLs by these two compounds, leading to chromosomal breaks. Thus, a panel of DT40 mutants including mutants for FA can identify mutagenic chemical compounds that generate ICLs.
After evaluating the original qHTS data obtained using a 24- and 48-hr exposure duration, we hypothesized that the sensitivity of the assay would be increased by increasing the exposure duration to 72 hrs. For example, these three chemical compounds reduced cellular growth of the Rad54/Ku70 mutant cells after a 24- but not a 48-hr incubation, while they reduced cellular growth of the Rev3 and FancC mutant cells after a 48- but not a 24-hr incubation (Fig 1). To analyze the effect of chemicals on cellular viability more comprehensively, we extended the incubation time to 72 hrs. In contrast to the results obtained at 24-hr, the 48- and 72-hr exposures showed similar results (Fig 1). The CA data (obtained at 24 hrs) are consistent with the differential cytotoxicity data from the 48- but not the 24-hr exposure duration, as increased numbers of CA were observed in the Rev3 and FancC mutant cells, but not in the Rad54/Ku70 mutant cells (Fig 3C). Thus, increases in cytotoxicity observed at 24-hr exposure in some DNA repair-deficient cell lines might not accurately represent genotoxicity.
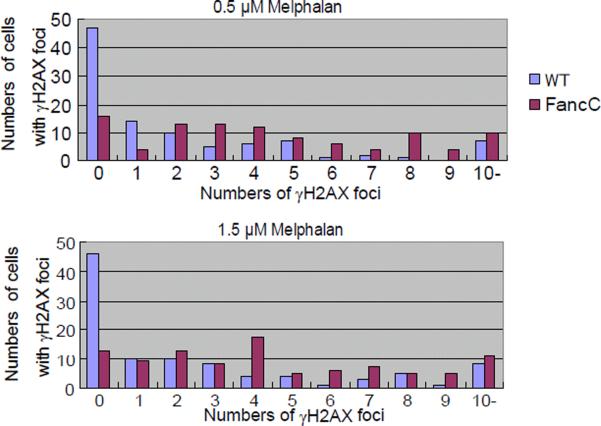
During the evaluation of the qHTS data, we also realized that an analysis of CA in metaphase cells at 24 hrs might yield false-negative data. For example, although melphalan reduced the viability of the FancC mutant cells more than that of parental DNA repair-proficient cells and the MI for these cells was only one-third the MI of the parental wild-type cells, treatment with 0.5 μM melphalan induced very similar numbers of CAs in these cell lines (Fig 3C). This discordance might be attributable to increased levels of cell cycle arrest in FancC mutant cells carrying ICLs (i.e., the ability of the damaged cells to progress to mitosis was greatly reduced). This is consistent with the finding that FancC mutant cells treated with melphalan displayed an increased number of γH2AX-positive foci, which represent sites of DNA damage, compared with wild-type cells. The mean/median values for the number of number of γH2AX-positive foci induced by melphalan, after combing the data from the two independent experiments, in the parental DNA-repair proficient and the FancC mutant cells are 2.2/1 and 4.6/4 at 0.5 μM, respectively, and 2.5/1 and 4.8/4 at 1.5 μM, respectively (Fig 4). The increase in the number of cells with >4 γH2AX-positive foci in the FancC mutant cells exposed to melphalan (54/100 cells vs. 24/100 for the parental DNA repair-proficient cells at 0.5 μM; 57/100 cells vs. 26/100 for the parental DNA repair-proficient cells at 1.5 μM cells) is statistically significant at both concentrations of melphalan by a one-tailed Fisher's exact test at p <0.001.
Figure 4.
Extent of induction of γH2AX positive foci in DT40 FancC mutant and parental DNA repair-proficient cells by melphalan.
Cells were stained with antiphosphorylated H2AX (γH2AX) after a 1 hr exposure to 0.5 μM and 1.5 μM melphalan. The numbers of cells with 0, 1, 2, 3, 4, …..>10 γH2AX foci combined from two independent experiments are shown on the y-axis (50 cells scored per sample, 100 cells scored total). The numbers of foci are shown on the x-axis.
The number of cells with >4 γH2AX foci is significantly increased in FancC mutant cells compared with the parental DNA repair-proficient cells (54/100 cells vs. 24/100 for the parental DNA repair-proficient cells at 0.5 μM; 57/100 cells vs. 26/100 for the parental DNA repair-proficient cells at 1.5 μM; one-tailed Fisher's exact test, p<0.001).
Alachlor, Benzotrichloride, and Trans-1,4-Dichloro-2-butene
Alachlor, benzotrichloride, and trans-1,4-dichloro-2-butene selectively reduced the viability of cells deficient in translesion DNA synthesis polymerase ζ (Rev3) (Fig 1). Rev3 mutant cells exhibited increased cytotoxicity after treatment with trans-1,4-dichloro-2-butene for 24 and 72 hrs. Consistently, all three compounds induced greater numbers of CAs in Rev3 mutant cells when compared with wild-type cells (Fig 3D). These observations indicate that cells deficient in Rev3 are very useful for detecting genotoxic compounds. Moreover, translesion DNA synthesis polymerase ζ accounts for a majority of spontaneously arising mutations in yeast [Lawrence et al., 2001]. The critical role of this polymerase in cellular tolerance to these three chemical compounds indicates that they induce DNA lesions that stall the replicative DNA polymerase and, as a result, can give rise to mutations through error-prone DNA synthesis by DNA polymerase ζ during bypass of the DNA lesions.
To gain insight into the nature of the DNA lesions induced by these three compounds, we analyzed the cellular sensitivity of XPA and Fen-1 mutants. XPA functions in the NER pathway and eliminates bulky DNA damage such as UV photoproducts. Fen-1, as well as DNA polymerase β, contributes to the BER pathway [Asagoshi et al., 2010], which eliminates a variety of chemical modifications such as alkylation and oxidation. A single damaged base is removed and repaired by BER, and damaged bases along with surrounding nucleotides are together corrected in NER. When cultured for 72 hrs after the start of treatment, XPA as well as Rev3 mutant cells are more sensitive to these three compounds while Fen1 mutant cells are more sensitive to alachlor and trans-1,4-dichloro-2-butene only (Fig 2C). These data indicate that DNA damage induced by benzotrichloride is repaired by the NER pathway, while DNA damage induced by alachlor and trans-1,4-dichloro-2-butene is repaired by the NER and BER pathways. NAC pretreatment did not rescue the increased loss in viability caused by these compounds (Fig 2C); therefore, we conclude that the DNA damage is not caused by ROS.
Lovastatin
Lovastatin is clinically used as an anti-hyperlipidemia agent and has not been reported to be genotoxic [EPA ACToR, 2011]. Lovastatin induced differential cytotoxicity in the Ubc13 mutant cell line at 24 and 48 hrs, but did not induce a significant increase in the numbers of CAs in any DT40 cell line evaluated (data not shown). Furthermore, in contrast to the differential cytotoxicity results obtained at 24 and 48 hrs, lovastatin did not induce increased cytotoxicity in the Ubc13 mutant cell line nor in the Polβ mutant cell lines after a 72-hr exposure (Fig 1, Fig 2D). These results suggest that short-term exposure data obtained with some DT40 DNA repair-deficient cell lines may not reflect genotoxicity.
2-aminothiamine
2-aminothiamine did not reduce the viability of any cell line when tested at concentrations up to 92 μM, the maximum concentration tested in this study (Fig 1). This compound is reported as positive for mutagenicity in the Salmonella assay with metabolic activation only and mouse lymphoma assay with or without metabolic activation [Cameron et al., 1985]. One likely explanation for the lack of increased cytotoxicity relates to the difference in the concentration range tested; the maximum concentration tested in the DT40 cells was 92 μM, while the lowest mutagenic concentration in the mouse lymphoma studies in the absence of metabolic activation was more than 3 mM. Thus, testing at higher concentrations might have resulted in a positive response in the DT40 screen.
Conclusion
We observed a significantly increased number of CAs in one or more isogenic DT40 DNA repair-deficient cell lines compared to the DNA repair-proficient parental cell line after treatment with adriamycin, actinomycin D, alachlor, benzotrichloride, diglycidyl resorcinol ether, melphalan, trans-1,4-dichloro-2-butene, and tris(2,3-epoxypropyl)isocyanurate, but not with lovastatin or 2-aminothiamine. In this study, we evaluated the number of induced CA as a method for demonstrating the relevance of a qHTS approach for genotoxicity based on the detection of increased cytotoxicity in isogenic mutant DT40 clones, each with a specific DNA repair pathway deficiency, compared to the DNA repair-proficient parental cell line. The number of compounds identified as positive in the initial screen (i.e., those with an IC50 6-fold lower than that obtained in the parental cell line) is limited by the inherent variation within each assay. Some compounds with less than a 6-fold IC50 difference are also likely genotoxic. Furthermore, there are several other limitations associated with the use of a qHTS strategy to identify genotoxic compounds. The main one is the inability to incorporate xenobiotic metabolism (e.g., S9 mix) into a 1536-well plate format as this screening platform requires the use of homogenous assays without any wash steps in order to ensure data quality. In the confirmation qHTS studies, the reproducibility of 42 differentially cytotoxic compounds tested in the qHTS confirmation assays was good in terms of compound activity, which was 97% for the 24-hr assays and 95% for the 48-hr assays. The average reproducibility of the calculated IC50 values was within 1.7 fold for the 24-hr assays and within 1.5 fold for the 48-hr assays.
Isogenic DT40 cell lines deficient in various DNA repair pathways provide a means for the rapid identification of genotoxic compounds and for evaluating their mechanism of action. The ability to screen extensive compound libraries for genotoxic activity and also to produce detailed information on the type(s) of DNA damage induced has the potential to replace traditional in vitro genotoxicity assays. However, there is first the need to better characterize the rate of false positive and false negative calls that would be associated with this approach, and to identify which DNA repair-deficient clones are the most useful by virtue of having the greatest sensitivity and specificity.
The spectrum of sensitivity to each compound among the various DNA repair-deficient mutants is useful for understanding the type of DNA damage induced. For example, actinomycin D induced increased numbers of CAs in the Rad54/Ku70 mutant cell line, suggesting that it induces DSBs. Melphalan, an alkylating agent, induced increased numbers of CAs in the FancC mutant cell line, suggesting that it induces ICLs. Alachlor, benzotrichloride, and trans-1,4-dichloro-2-butene induced increased numbers of CAs in Rev3-deficient cells, suggesting that they induce DNA strand lesions that cause replication stalling. The relationship between DT40 mutant cell lines deficient in different DNA repair genes/pathways and compounds that selectively induced increased cytotoxicity in one or more of these mutant cell lines provides a useful data source for insight into the nature of chemically-induced DNA damage.
For four of the compounds (adriamycin, actinomycin D, alachlor, and benzotrichloride), there was a consistent pattern of increased cytoxicity at 24, 48, and 72 hrs hrs and increased clastogenicity at 24 hrs in the same DNA repair-deficient cell line. For trans-1,4-dichloro-2-butene, although not differentially cytotoxic at 48 hrs, the same DNA repair-deficient cell line expressed both differential cytotoxicity at 24 and 72 hrs and increased clastogenicity at 24 hrs. For diglycidyl resorcinol ether, melphalan, and tris(2,3-epoxypropyl)isocyanurate, the increase in clastogenic activity was in DNA repair-deficient cell lines that exhibited increased cytotoxicity at 48 and 72 hrs but not at 24 hrs. In contrast, lovastatin induced differential cytotoxicity in the Ubc13 mutant cell line at both 24 and 48 hrs but not 72 hrs, and the lack of increased clastogenicity in any DNA repair-deficient cell line in consistent with the lack of differential cytotoxicity at the 72-hr sample time. Thus, the results of the 72-hr differential cytotoxicity assay were the most consistent with the results obtained for CA or γH2AX analysis. This indicates that increasing the exposure duration of DT40 cell lines to 72 hrs would be expected to provide the greatest sensitivity and specificity. However, due to the small volumes/well used in 1536-well plates (~5 μL), the maximum exposure duration possible in qHTS is ~48 hrs suggesting that qHTS studies use this exposure duration, while follow-up studies focused on verifying the qHTS results and elucidating the nature of the induced DNA damage use lower throughput methods more amenable to longer exposure durations. In the future, this approach may supplant the more time consuming analysis of CA or γH2AX positive foci to identify direct-acting genotoxic compounds while being capable of providing critical information about the nature of the induced DNA damage.
The results obtained on extensive compound libraries also have the potential to provide clues for clinical research. For example, inhibitors of Rev3 or FancC could sensitize tumor cells to killing by melphalan, and inhibitors of the NHEJ pathway could increase the effectiveness of adriamycin and actinomycin D therapy. Also, TLS inhibition or HR inhibition could confer cancer cells with DNA repair deficiencies that would increase the sensitivity of the cells to DNA damaging agents. Therefore, compounds identified through these screens as TLS or HR inhibitors, or inducers of hyper-HR may have the potential to become future cancer therapies. Thus, this novel qHTS approach using DT40 cells might one day enable improvements in combination chemotherapy applications.
Acknowledgments
This work was supported in part by the Intramural Research Programs (Interagency agreement #Y2-ES-7020-01) of the National Toxicology Program, National Institute of Environmental Health Sciences.
References
- Adachi N, Suzuki H, Iiizumi S, Koyama H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J Biol Chem. 2003;278(38):35897–902. doi: 10.1074/jbc.M306500200. [DOI] [PubMed] [Google Scholar]
- Asagoshi K, Tano K, Chastain PD, Adachi N, Sonoda E, Kikuchi K, Koyama H, Nagata K, Kaufman DG, Takeda S, Wilson SH, Watanabe M, Swenberg JA, Nakamura J. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res. 2010;8(2):204–215. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba TW, Giroir BP, Humphries EH. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985;144(1):139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bensaude O, Bonnet F, Cassé C, Dubois MF, Nguyen VT, Palancade B. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD) Biocheem Cell Biol. 1999;77(4):249–255. [PubMed] [Google Scholar]
- Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89(2):185–93. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- Buerstedde J, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67(1):179–88. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2009;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Cameron T, Hughes T, Kirby P, Palmer K, Fung V, Dunkel V. Mutagenic activity of 5 thiazole compounds in the Salmonella/microsome and mouse lymphoma TK+/− assays. Mutat Res. 1985;155(1–2):17–25. doi: 10.1016/0165-1218(85)90020-5. [DOI] [PubMed] [Google Scholar]
- Cimino MC. Comparative overview of current international strategies and guidelines for genetic toxicology testing for regulatory purposes. Environ Mol Mutagen. 2006;47:362–390. doi: 10.1002/em.20216. [DOI] [PubMed] [Google Scholar]
- Collins F, Gray G, Bucher J. Toxicology. Transforming environmental health protection. Science. 2008;319(5865):906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch S, Kozlowski R, Slater K, Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods. 1993;160(1):81–8. doi: 10.1016/0022-1759(93)90011-u. [DOI] [PubMed] [Google Scholar]
- EPA. ACToR (Aggregated Computational Toxicology Resource) 2011 doi: 10.1016/j.taap.2007.12.037. Available at http://actor.epa.gov/actor/faces/ACToRHome.jsp. [DOI] [PubMed]
- Estlin EJ, Veal GJ. Clinical and cellular pharmacology in relation to solid tumours of childhood. Cancer Treat Rev. 2003;29(4):253–273. doi: 10.1016/s0305-7372(02)00109-3. [DOI] [PubMed] [Google Scholar]
- Evans TJ, Yamamoto KN, Hirota K, Takeda S. Mutant cells defective in DNA repair pathways provide a sensitive high-throughput assay for genotoxicity. DNA Repair (Amst) 2010;9(12):1292–1298. doi: 10.1016/j.dnarep.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Gurtan A, D'Andrea A. Dedicated to the core: understanding the Fanconi anemia complex. DNA Repair (Amst) 2006;5(9–10):1119–25. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Nakajima Y, Matsumura S, Chatani F. An in vitro micronucleus assay with size-classified micronucleus counting to discriminate aneugens from clastogens. Toxicol In Vitro. 2010;24(1):208–216. doi: 10.1016/j.tiv.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Hatanaka A, Yamazoe M, Sale J, Takata M, Yamamoto K, Kitao H, Sonoda E, Kikuchi K, Yonetani Y, Takeda S. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25(3):1124–34. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer C, Welburn J, Dong Y, McEwen B, Shang W, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135(6):1039–52. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J, Watson M, Stefanick D, Shaughnessy D, Taylor J, Wilson S. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18(1):48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISCN (An International System for Human Cytogenetic Nomenclature) Report of the Standing Committee on Human Cytogenetic Nomenclature. Birth Defects Orig Artic Ser. 1985;21(1):1–117. [PubMed] [Google Scholar]
- Ji K, Kogame T, Choi K, Wang X, Lee J, Taniguchi Y, Takeda S. A novel approach using DNA-repair-deficient chicken DT40 cell lines for screening and characterizing the genotoxicity of environmental contaminants. Environ Health Perspect. 2009;117(11):1737–44. doi: 10.1289/ehp.0900842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R, Austin C, Tice R. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009;29(4):485–7. doi: 10.1111/j.1539-6924.2008.01168.x. discussion 492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpnick Z, Homiski M, Rubitski E, Repnevskaya M, Howlett N, Aubrecht J, Schiestl R. Yeast DEL assay detects clastogens. Mutat Res. 2005;582(1–2):116–34. doi: 10.1016/j.mrgentox.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kitao H, Yamamoto K, Matsushita N, Ohzeki M, Ishiai M, Takata M. Functional interplay between BRCA2/FancD1 and FancC in DNA repair. J Biol Chem. 2006;281(30):21312–20. doi: 10.1074/jbc.M603290200. [DOI] [PubMed] [Google Scholar]
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat Res. 1996b;355(1–2):13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- Lawrence C, Maher V. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos Trans R Soc Lond B Biol Sci. 2001;356(1405):41–6. doi: 10.1098/rstb.2000.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30(14):3273–7. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A, Okada T, Shibutani S, Sonoda E, Hochegger H, Nishigori C, Miyachi Y, Takeda S, Yamazoe M. Extensive chromosomal breaks are induced by tamoxifen and estrogen in DNA repair-deficient cells. Cancer Res. 2004;64(9):3144–3147. doi: 10.1158/0008-5472.can-03-3489. [DOI] [PubMed] [Google Scholar]
- Nakahara M, Sonoda E, Nojima K, Sale J, Takenaka K, Kikuchi K, Taniguchi Y, Nakamura K, Sumitomo Y, Bree R, et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009;5(1):e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui K, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6(1):e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli B, Bishop D, Hirano S, Ohzeki M, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65(24):11704–11. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- NRC . A Vision for Toxicity Testing in the 21st Century. The National Academies Press; Washington, DC: 2007. [Google Scholar]
- Okada T, Sonoda E, Yamashita Y, Koyoshi S, Tateishi S, Yamaizumi M, Takata M, Ogawa O, Takeda S. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J Biol Chem. 2002;277(50):48690–5. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- Porcedda P, Turinetto V, Lantelme E, Fontanella E, Chrzanowska K, Ragona R, De Marchi M, Delia D, Giachino C. Impaired elimination of DNA double-strand break-containing lymphocytes in ataxia telangiectasia and Nijmegen breakage syndrome. DNA Repair (Amst) 2006;5(8):904–13. doi: 10.1016/j.dnarep.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao G, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik N, van Gent D, Takata M, et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22(12):3188–97. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki M, Buerstedde J, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17(2):598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D, Bilardi R, Koch T, Post G, Nafie J, Kimura K, Cutts S, Phillips D. DNA repair in response to anthracycline-DNA adducts: a role for both homologous recombination and nucleotide excision repair. Mutat Res. 2008;638(1–2):110–21. doi: 10.1016/j.mrfmmm.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Surrallés J, Xamena N, Creus A, Marcos R. The suitability of the micronucleus assay in human lymphocytes as a new biomarker of excision repair. Mutat Res. 1995;342(1–2):43–59. doi: 10.1016/0165-1218(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Suter W, Jaeger I. Comparative evaluation of different pairs of DNA repair-deficient and DNA repair-proficient bacterial tester strains for rapid detection of chemical mutagens and carcinogens. Mutat Res. 1982;97(1):1–18. doi: 10.1016/0165-1161(82)90015-2. [DOI] [PubMed] [Google Scholar]
- Takao N, Kato H, Mori R, Morrison C, Sonada E, Sun X, Shimizu H, Yoshioka K, Takeda S, Yamamoto K. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene. 1999;18(50):7002–9. doi: 10.1038/sj.onc.1203172. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki M, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17(18):5497–508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki M, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson L, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21(8):2858–66. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki Y. The single cell gel/comet assay: Guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226(4673):466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- Wu X, Takenaka K, Sonoda E, Hochegger H, Kawanishi S, Kawamoto T, Takeda S, Yamazoe M. Critical roles for polymerase zeta in cellular tolerance to nitric oxide-induced DNA damage. Cancer Res. 2006;66(2):748–754. doi: 10.1158/0008-5472.CAN-05-2884. [DOI] [PubMed] [Google Scholar]
- Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116(3):284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, Buerstedde J, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25(1):34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Sonoda E, Barber L, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25(5):663–75. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]