Abstract
Objective
O6-methylguanine-DNA methyltransferase (MG MT) is a DNA repair enzyme. MGMT promoter hypermethylation and epigenetic silencing often occur as early events in carcinogenesis. However, prognostic significance of MGMT alterations in colorectal cancer remains uncertain.
Methods
Utilizing a database of 855 colon and rectal cancers in two prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study), we detected MGMT promoter hypermethylation in 325 tumors (38%) by MethyLight and loss of MGMT expression in 37% (247/672) of tumors by immunohistochemistry. We assessed the CpG island methylator phenotype (CIMP) using eight methylation markers [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1], and LINE-1 (L1) hypomethylation, TP53 (p53), and microsatellite instability (MSI).
Results
MGMT hypermethylation was not associated with colorectal cancer–specific mortality in univariate or multivariate Cox regression analysis [adjusted hazard ratio (HR) = 1.03; 95% confidence interval (CI), 0.79–1.36] that adjusted for clinical and tumor features, including CIMP, MSI, and BRAF mutation. Similarly, MGMT loss was not associated with patient survival. MGMT loss was associated with G>A mutations in KRAS (p = 0.019) and PIK3CA (p = 0.0031).
Conclusions
Despite a well-established role of MGMT aberrations in carcinogenesis, neither MGMT promoter methylation nor MGMT loss serves as a prognostic biomarker in colorectal cancer.
Keywords: Colon cancer, MGMT, Hypermethylation, Epigenetics, Clinical outcome
Introduction
The O6-methylguanine-DNA methyltransferase (MGMT) gene encodes DNA repair protein and is frequently inactivated in colorectal cancer [1, 2]. Polymorphisms in MGMT have been associated with colorectal cancer risk [3, 4], and MGMT promoter methylation in normal colonic mucosa might be a predisposing factor for cancer as a field effect and an early event in colorectal carcinogenesis [5, 6]. MGMT promoter methylation and loss of expression have been associated with G>A mutations in a variety of genes such as KRAS, PIK3CA, TP53, and APC [7–11]. Jass [12] proposed the molecular classification based on CIMP, MSI, BRAF, KRAS, and MGMT promoter methylation, indicating that MGMT methylation is one of the key molecular alterations in colorectal cancer. In addition, MGMT has potential as a therapeutic target in human cancer [13, 14]. Collectively, it is of interest to examine a prognostic role of MGMT alteration as a tumor biomarker. In brain tumors and B-cell lymphoma, MGMT methylation or loss of MGMT has been associated with poor prognosis [15–17]. However, prognostic significance of MGMT alteration in colorectal cancer remains inconclusive due to limited statistical power of all previous studies (Table 1; all n < 200) [18–22].
Table 1.
Studies on prognostic significance of MGMT methylation (top) or loss of MGMT (bottom) in colorectal cancer
| Ref. | Authors (year) | Sample size | No. of events |
Disease stage | Chemotherapy |
MGMT methylation (vs. no methylation) or loss of MGMT (vs. expression) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OS | CS | Method | Methylated (MGMT-lost) cases (%) | p value by log-rank test | OS univariate HR (95% CI) | p value by univariate model | OS multi-variate HR (95% CI) | p value by multi-variate model | Note | |||||
| MGMT methylation | ||||||||||||||
| [18] | Nagasaka et al. (2003) | 90 | – | – | Dukes A-D | Fluoro-pyrimidines | MSP | 29 | – | – | – | 1.30a (0.27–6.47) | NS | |
| [19] | Krtolica et al. (2007) | 47 | – | – | Dukes A-D | – | MSP | 43 | NS | – | NS | – | NS | |
| [20] | Chen et al. (2009) | 117 | – | – | Dukes A-D | Adjuvant chemotherapy | MSP | 61 | – | – | – | 1.33 (0.61–2.92) | p = 0.47 | |
| [21] | Kim et al. (2010) | 131 | – | – | I–IV | Adjuvant chemotherapy | Pyro-sequencing | 21 | – | – | NS | NS | Rectal cancer only | |
| Current study | 855 | 415 | 234 | I–IV | – | q-MSP | 38 | p = 0.83 | 1.03 (0.85–1.26) | p = 0.75 | 1.08 (0.88–1.32) | p = 0.49 | ||
| Loss of MGMT | ||||||||||||||
| [22] | Kohonen-Corish et al. (2005) | 178 | 141 | – | Dukes C | Adjuvant chemotherapy | IHC | 22 | p = 0.55 | – | – | – | – | |
| Current study | 672 | 313 | 170 | I–IV | – | IHC | 37 | p = 0.54 | 0.94 (0.75–1.18) | p = 0.59 | 1.11 (0.87–1.41) | p = 0.39 | ||
CI confidence interval, CIMP CpG island methylator phenotype, CS colorectal cancer–specific survival, HR, hazard ratio, IHC immunohistochemistry, MSP methylation-specific PCR, NS not significant, OS overall survival, q-MSP quantitative methylation–specific PCR
Patients with unmethylated MGMT promoters at a 1.3-fold increased risk of death
In this study using the database of a large number (n = 855) of stage I–IV colorectal cancers, we examined the prognostic effect of MGMT promoter methylation and loss of expression. Since we concurrently assessed other molecular variables including LINE-1 hypomethylation, MSI, CIMP, and mutation in KRAS, BRAF, and PIK3CA, we could evaluate the prognostic effect of MGMT alteration after controlling for those potential confounders.
Materials and methods
Study population
We utilized the database of two prospective cohort studies, the Nurses’ Health Study (n = 121,701 women followed since 1976) [23] and the Health Professionals Follow-up Study (n = 51,529 men followed since 1986) [23]. Participants have been sent biennial questionnaires to update information on potential risk factors and to identify newly diagnosed cancers in themselves and their first-degree relatives. We collected paraffin-embedded tumor tissue blocks of incident colorectal cancers from hospitals where participants with colorectal cancer underwent tumor resection. Hematoxylin and eosin (H&E)–stained tissue sections from all colorectal cancer cases were confirmed by a pathologist (S.O.) unaware of other data. The tumor grade was categorized as low versus high (>50 vs. ≤50% gland formation). Positive family history of colorectal cancer was defined as the presence of colorectal cancer in any first-degree relative. We excluded cases that were preoperatively treated. Based on the availability of adequate follow-up and tumor tissue data, 855 stage I–IV colorectal cancer cases diagnosed up to 2002 were included. Patients were observed until death or June 2009, whichever came first. This study was approved by the Human Subjects Committees at Harvard School of Public Health and Brigham and Women’s Hospital.
Pyrosequencing of KRAS, BRAF, and PIK3CA, and microsatellite instability (MSI) analysis
Genomic DNA was extracted from paraffin-embedded tissue. PCR and pyrosequencing targeted for KRAS (codons 12 and 13) [24], BRAF (codon 600) [25], and PIK3CA (exons 9 and 20) [11] were performed. Microsatellite instability (MSI) analysis was performed using 10 micro-satellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487) [26]. MSI-high was defined as the presence of instability in ≥30% of the markers and MSI-low/microsatellite stable (MSS) as instability in 0–29% of markers [26].
Methylation analyses for CpG islands and LINE-1
Sodium bisulfite treatment and subsequent real-time PCR were performed to quantify promoter methylation in MGMT and eight other CpG islands (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) [27, 28]; the latter eight markers have been shown to be specific for CIMP [29]. CIMP-high was defined as the presence of ≥6/8 methylated markers, CIMP-low as the presence of 1/8–5/8 methylated markers, and CIMP-0 as the absence (0/8) of methylated markers [30]. We defined and validated the cut point for MGMT promoter methylation positivity (percentage of methylated reference, or PMR> 4) as previously described [31]. LINE-1 methylation levels were quantified by PCR-pyrosequencing [32, 33].
Immunohistochemical analysis
Immunohistochemical methods for MGMT and TP53 (p53) were previously described [26], and expression patterns were interpreted by a pathologist (S.O.) unaware of other data. In agreement studies, a random selection of more than 100 cases for each marker was interpreted by a second pathologist unaware of other data (MGMT by K.S.; TP53 by K.N.). The concordance between the two observers (both p < 0.0001) was 0.86 for MGMT (κ = 0.70) and 0.87 for TP53 (κ = 0.75), indicating substantial agreement. The concordance between MGMT methylation and loss of MGMT was 81% (κ = 0.59).
Statistical analysis
We used SAS program (Version 9.1, SAS Institute, Cary, NC) for all statistical analysis. All p values were two-sided, and significance level was set at p = 0.05. The chi-square test (or Fisher’s exact test) was performed for categorical variables. For survival analysis, the Kaplan–Meier method and log-rank test were used. For analyses of colorectal cancer–specific mortality, deaths as a result of causes other than colorectal cancer were censored. To control for confounding, we used multivariate stage-matched (stratified) Cox proportional hazard models to compute hazard ratio (HR) of death according to MGMT status. To avoid residual confounding and overfitting, disease stage (I, IIA, IIB, IIIA, IIIB, IIIC, IV, unknown) was used as a stratifying variable, utilizing the “strata” option in the SAS “proc phreg” command. The multivariate model initially included age at diagnosis (continuous), sex, year of diagnosis (continuous), BMI (<30 vs. ≥30 kg/m2), family history of colorectal cancer (present vs. absent), tumor location (proximal colon vs. distal colon vs. rectum), tumor grade (low vs. high), MSI (high vs. low/MSS), CIMP (high vs. low vs. CIMP-0), LINE-1 methylation (continuous), and BRAF. A backward elimination with a threshold of p = 0.20 was used to select variables in the final model. For cases with missing information in any of the categorical variables [tumor location (0.9%), tumor grade (0.5%), MSI (3.0%), BRAF (4.8%)], we included those cases in a majority category of a given covariate to avoid overfitting. We confirmed that excluding cases with missing information in any of the covariates did not substantially alter results (data not shown). An interaction was assessed by including the cross product of the MGMT methylation (or MGMT loss) variable and another variable of interest (without data-missing cases) in a multivariate model, and the Wald test was performed. A p value for statistical significance was adjusted for multiple hypothesis testing to p = 0.0029 (=0.05/17) by Bonferroni correction.
In addition, we constructed multivariate logistic regression analysis model to assess the independent effect of MGMT loss on G>A mutations in KRAS or PIK3CA (as a binary outcome variable). The model initially included age at diagnosis (continuous), sex, year of diagnosis (continuous), BMI (<30 vs. ≥30 kg/m2), family history of colorectal cancer (present vs. absent), tumor location (proximal vs. distal), MSI (high vs. low/MSS), CIMP (high vs. low/0), LINE-1 methylation (continuous), and MGMT loss, and a backward elimination with a threshold of p = 0.20 was done to select variables in the final model.
Results
MGMT methylation and loss of MGMT in colorectal cancer
MGMT promoter methylation was detected in 325 (38%) of 885 tumors, and loss of MGMT was detected in 247 (37%) of 672 tumors. There was a significant association between MGMT promoter methylation and CIMP status (p < 0.0001). Loss of MGMT was significantly associated with PIK3CA mutation (p = 0.0031) and inversely associated with TP53 expression (p = 0.0004; Table 2).
Table 2.
Clinical and molecular characteristics in colorectal cancer with MGMT promoter methylation/loss of MGMT
| Clinical or molecular feature | All cases |
MGMT promoter methylation |
MGMT status |
||||
|---|---|---|---|---|---|---|---|
| (−) | (+) | p value | Expressed | Lost | p value | ||
| Total n | 855 | 530 | 325 | 425 | 247 | ||
| Sex | 0.23 | 0.71 | |||||
| Male (HPFS) | 367 (43%) | 236 (45%) | 131 (40%) | 173 (41%) | 97 (39%) | ||
| Female (NHS) | 488 (57%) | 294 (55%) | 194 (60%) | 252 (59%) | 150 (61%) | ||
| Mean age ± SD | 66.2 ± 8.4 | 66.2 ± 8.6 | 66.3 ± 8.1 | 0.72 | 66.3 ± 8.4 | 65.9 ± 7.9 | 0.60 |
| Body mass index (BMI) | 0.38 | 0.093 | |||||
| <30 kg/m2 | 714 (83%) | 438 (83%) | 276 (85%) | 347 (82%) | 214 (87%) | ||
| ≥30 kg/m2 | 141 (17%) | 92 (17%) | 49 (15%) | 78 (18%) | 33 (13%) | ||
| Family history of colorectal cancer in any 1st degree relative | 0.47 | 0.28 | |||||
| Absent | 646 (76%) | 396 (75%) | 250 (77%) | 319 (75%) | 176 (71%) | ||
| Present | 209 (24%) | 134 (25%) | 75 (23%) | 106 (25%) | 71 (29%) | ||
| Year of diagnosis | 0.68 | 0.031 | |||||
| Prior to 1995 | 400 (47%) | 245 (46%) | 155(48%) | 182 (43%) | 127 (51%) | ||
| 1995–2004 | 455 (53%) | 285 (54%) | 170 (52%) | 243 (57%) | 120 (49%) | ||
| Tumor location | 0.90 | 0.014 | |||||
| Proximal colon (cecum to transverse) | 381 (45%) | 239 (46%) | 142 (44%) | 197 (47%) | 108 (44%) | ||
| Distal colon (splenic flexure to sigmoid) | 273 (32%) | 168 (32%) | 105 (33%) | 125 (30%) | 96 (39%) | ||
| Rectum | 192 (23%) | 117 (22%) | 75 (23%) | 100 (24%) | 40 (16%) | ||
| Disease stage | 0.86 | 0.70 | |||||
| I | 199 (23%) | 124 (23%) | 75 (23%) | 94 (22%) | 57 (23%) | ||
| II | 259 (30%) | 156 (29%) | 103 (32%) | 133 (31%) | 77 (31%) | ||
| III | 228 (27%) | 146 (28%) | 82 (25%) | 110 (26%) | 69 (28%) | ||
| IV | 102 (12%) | 65 (12%) | 37 (11%) | 57 (13%) | 24 (9.7%) | ||
| Unknown | 67 (7.8%) | 39 (7.4%) | 28 (8.6%) | 31 (7.3%) | 20 (8.1%) | ||
| Tumor grade | 0.63 | 0.23 | |||||
| Low | 771 (91%) | 480 (91%) | 291 (90%) | 389 (92%) | 220 (89%) | ||
| High | 79 (9.3%) | 47 (8.9%) | 32 (9.9%) | 33 (7.8%) | 26 (11%) | ||
| MSI status | 0.19 | 0.21 | |||||
| MSS/MSI-low | 705 (85%) | 442 (87%) | 263 (83%) | 361 (86%) | 201 (83%) | ||
| MSI-high | 124 (15%) | 70 (13%) | 54 (17%) | 57 (14%) | 42 (17%) | ||
| CIMP status | <0.0001 | 0.0077 | |||||
| CIMP-0 | 399 (47%) | 280 (53%) | 119 (37%) | 218 (51%) | 96 (39%) | ||
| CIMP-low | 325 (38%) | 184 (35%) | 141 (43%) | 145 (34%) | 107 (43%) | ||
| CIMP-high | 131 (15%) | 66 (12%) | 65 (20%) | 62 (15%) | 44 (18%) | ||
| KRAS mutation | 0.75 | 0.019 | |||||
| (−) | 529 (63%) | 332 (64%) | 197 (62%) | 281 (67%) | 136 (56%) | ||
| Non G>A mutation | 185 (22%) | 111 (22%) | 74 (23%) | 86 (20%) | 66 (27%) | ||
| G>A | 121 (14%) | 73 (14%) | 48 (15%) | 53 (13%) | 41 (17%) | ||
| BRAF mutation | 0.92 | 0.11 | |||||
| (−) | 703 (87%) | 434 (86%) | 269 (86%) | 347 (84%) | 209 (89%) | ||
| (+) | 111 (13%) | 68 (14%) | 43 (14%) | 64 (16%) | 26 (11%) | ||
| PIK3CA mutation | 0.079 | 0.0031 | |||||
| (−) | 627 (84%) | 397 (86%) | 230 (81%) | 335 (88%) | 167 (79%) | ||
| Non G>A mutation | 53 (7.1%) | 26 (5.7%) | 27 (9.5%) | 23 (6.1%) | 16 (7.6%) | ||
| G>A | 64 (8.6%) | 36 (7.8%) | 28 (9.8%) | 21 (5.5%) | 28 (13%) | ||
| Mean LINE-1 methylation (%) ± SD | 61.4 ± 9.5 | 61.3 ± 9.8 | 61.6 ± 9.0 | 0.70 | 60.9 ± 9.4 | 61.2 ± 8.8 | 0.63 |
| TP53 expression | 0.60 | 0.0004 | |||||
| (−) | 482 (57%) | 295 (56%) | 187 (58%) | 219 (52%) | 160 (66%) | ||
| (+) | 362 (43%) | 228 (44%) | 134 (42%) | 206 (48%) | 84 (34%) | ||
CIMP CpG island methylator phenotype, CRC colorectal cancer, HPFS Health Professionals Follow-up Study, MSI microsatellite instability, MSS microsatellite stable, NHS Nurses’ Health Study, SD standard deviation
MGMT methylation/loss and G>A mutations in KRAS and PIK3CA
Because functional loss of MGMT may contribute to G>A mutations [7–11], we examined the relations between MGMT methylation (or loss) and G>A mutations in KRAS and PIK3CA (Table 2). MGMT loss was significantly associated with G>A mutations in KRAS (p = 0.019) and PIK3CA (p = 0.0031) (by a priori hypothesis testing), while MGMT methylation was not. In multivariate logistic regression analysis to assess independent effect of MGMT loss on G>A mutations, MGMT loss remained significantly associated with G>A mutations in KRAS (adjusted OR, 1.58; 95% CI, 1.07–2.34; p = 0.021) and PIK3CA (adjusted OR, 2.55; 95% CI, 1.40–4.68; p = 0.0024).
MGMT methylation, loss of MGMT, and survival of patients with colorectal cancer
Among the 855 patients, there were 415 deaths including 234 colorectal cancer–specific deaths. The median follow-up time for censored patients was 13.0 years. For either colorectal cancer–specific or overall mortality, MGMT methylation was not significantly associated with patient outcome in log-rank test, or univariate or multivariate stage–matched Cox regression analysis (Table 3, Fig. 1). Likewise, loss of MGMT was not significantly associated with colorectal cancer–specific or overall mortality in univariate or multivariate stage–matched analysis (Table 3, Fig. 1).
Table 3.
MGMT promoter methylation/loss of MGMT in colorectal cancer and patient mortality
| Total n | Colorectal cancer–specific mortality |
Overall mortality |
|||||
|---|---|---|---|---|---|---|---|
| Deaths/person-years | Univariate HR (95% CI) | Multivariate HR (95% CI) | Deaths/person-years | Univariate HR (95% CI) | Multivariate HR (95% CI) | ||
| Colorectal cancer | |||||||
| MGMT promoter | |||||||
| Unmethylated | 530 | 148/4,996 | 1 (referent) | 1 (referent) | 256/4,996 | 1 (referent) | 1 (referent) |
| Methylated | 325 | 86/3,002 | 0.95 (0.73–1.24) | 1.03 (0.79–1.36) | 159/3,002 | 1.03 (0.85–1.26) | 1.08 (0.88–1.32) |
| MGMT | |||||||
| Expressed | 425 | 116/3,930 | 1 (referent) | 1 (referent) | 200/3,930 | 1 (referent) | 1 (referent) |
| Lost | 247 | 58/2,450 | 0.83 (0.61–1.14) | 1.08 (0.85–1.37) | 116/2,450 | 0.94 (0.75–1.18) | 1.11 (0.87–1.41) |
| Colon cancer | |||||||
| MGMT promoter | |||||||
| Unmethylated | 407 | 115/3,807 | 1 (referent) | 1 (referent) | 199/3,832 | 1 (referent) | 1 (referent) |
| Methylated | 247 | 57/2,336 | 0.80 (0.58–1.09) | 0.85 (0.61–1.18) | 118/2,354 | 0.96 (0.77–1.21) | 1.00 (0.79–1.26) |
| MGMT | |||||||
| Expressed | 322 | 87/2,993 | 1 (referent) | 1 (referent) | 151/2,993 | 1 (referent) | 1 (referent) |
| Lost | 204 | 47/2,020 | 0.82 (0.58–1.17) | 1.03 (0.71–1.49) | 97/2,020 | 0.96 (0.74–1.24) | 1.12 (0.86–1.46) |
| Rectal cancer | |||||||
| MGMT promoter | |||||||
| Unmethylated | 117 | 31/1,129 | 1 (referent) | 1 (referent) | 53/1,129 | 1 (referent) | 1 (referent) |
| Methylated | 75 | 27/652 | 1.47 (0.87–2.46) | 1.37 (0.79–2.36) | 38/652 | 1.24 (0.82–1.89) | 1.17 (0.75–1.84) |
| MGMT | |||||||
| Expressed | 100 | 27/920 | 1 (referent) | 1 (referent) | 46/920 | 1 (referent) | 1 (referent) |
| Lost | 40 | 9/416 | 0.77 (0.36–1.64) | 0.96 (0.41–2.27) | 16/416 | 0.77 (0.44–1.36) | 1.05 (0.57–1.93) |
The multivariate, stage-matched (stratified) Cox regression model initially included the MGMT promoter methylation or loss of MGMT variable, sex, age at diagnosis, year of diagnosis, tumor location, obesity, family history of colorectal cancer, tumor grade, CIMP, MSI, BRAF, and LINE-1 methylation. A backward elimination with threshold of p = 0.20 was used to select variables in the final models. CI confidence interval, HR hazard ratio
Fig. 1.
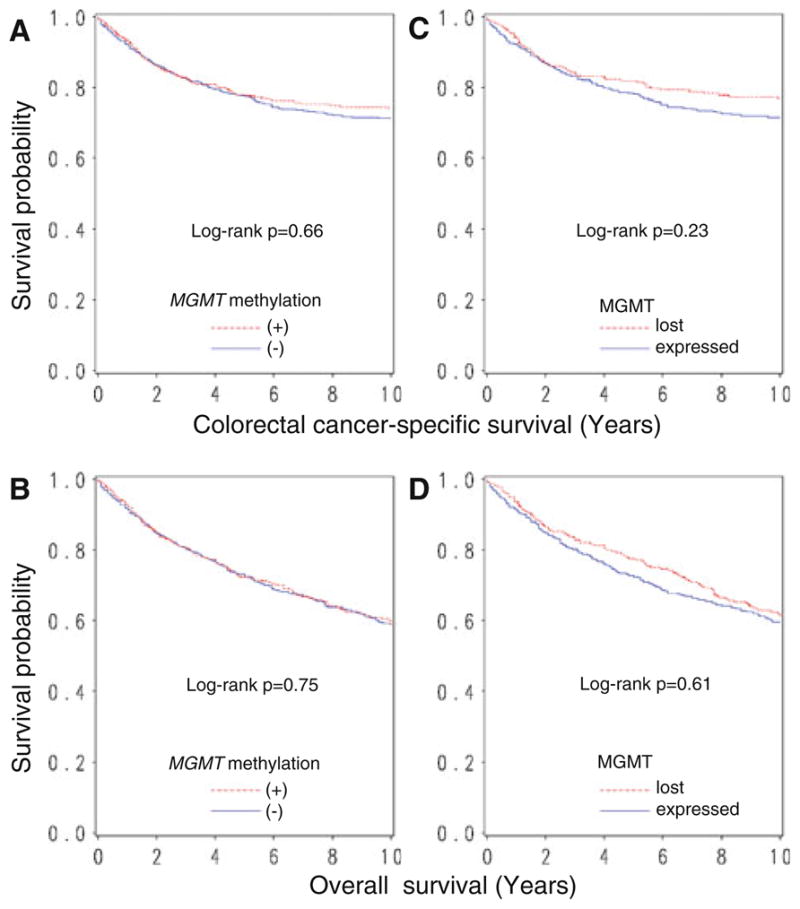
Kaplan–Meier curves for colorectal cancer–specific survival (upper panel) and overall survival (lower panel), according to MGMT promoter methylation (a, b) or loss of MGMT (c, d) in colorectal cancer
We analyzed the prognostic effect of MGMT methylation or loss of MGMT in colon cancer and rectal cancer separately, since clinical management for patients with colon cancer differ from that for patients with rectal cancer. MGMT methylation (or MGMT loss) was not significantly associated with colorectal cancer–specific or overall mortality in either patients with colon cancer or rectal cancer (Table 3).
There was no significant modifying effect on the prognostic influence of MGMT methylation (or MGMT loss) by Table 2 Clinical and molecular characteristics in colorectal cancer with MGMT promoter methylation/loss of MGMT any of the other variables including sex, age, year of diagnosis, BMI, family history of colorectal cancer, tumor location, stage, tumor grade, CIMP, MSI, BRAF, KRAS, PIK3CA, LINE-1 methylation, and TP53 (all Pinteraction > 0.02).
Discussion
We conducted this study to examine whether MGMT promoter methylation or loss of expression in colorectal cancer has any prognostic role. This question has remained inconclusive due to limited statistical power of all previous studies on this topic [18–22]. Given the crucial roles of MGMT aberrations in colorectal carcinogenesis or a potential use of MGMT as a therapeutic target in human cancer, the assessment of MGMT alteration (i.e., MGMT promoter methylation or loss of MGMT) and clinical outcome using a large number of cancers is needed. Utilizing the database of 855 clinically and molecularly annotated colorectal cancers in the two large prospective cohort studies, we found that MGMT alteration was not associated with patient prognosis in colorectal cancer. In addition, we assessed the prognostic effect of MGMT promoter methylation (or MGMT loss), controlling for other molecular features, including CIMP, MSI, and BRAF mutation, all of which have been documented to be critical in colorectal carcinogenesis.
Studying molecular variants and somatic changes is important in cancer research [34–42]. Recent studies have shown that MGMT promoter polymorphism (rs16906252) is associated with MGMT methylation in colorectal cancer [43], in normal colorectal mucosa [44], and in peripheral blood cells from normal individuals [45]. Epigenetic silencing of MGMT by promoter methylation in normal colonic mucosa may be a predisposing factor for cancer as a field effect and an early event in colorectal carcinogenesis [5, 6]. In addition, MGMT methylation might be a valuable biomarker in plasma for early detection of colorectal cancer [46].
Studies on colorectal cancer have shown that MGMT methylation is associated with MGMT loss [5, 6, 47]. In agreement with these studies [5, 6, 47], our current study showed a good concordance (81%, κ = 0.59, p < 0.0001) between MGMT methylation and MGMT loss. MGMT methylation and loss of MGMT were not perfectly correlated perhaps due to a few reasons. First, loss of MGMT expression may be caused not only by promoter methylation but also by other mechanisms such as a gene deletion or mutation. Second, in rare cases, promoter methylation may be present in only one MGMT allele, and the MGMT protein may be expressed from the second allele. Third, there may be other molecules (such as miRNA) that may downregulate MGMT.
MGMT promoter methylation or loss of expression in colorectal cancer has been associated with G>A mutations in KRAS [7, 9, 10, 44, 48], TP53 [8, 9], and PIK3CA [11]. Our current study is the first one to perform multivariate analysis and show that MGMT loss is associated with G>A mutations in KRAS and PIK3CA, independent of potential confounders. Thus, our current study supports the concept that loss of MGMT contributes to G>A mutations of KRAS and PIK3CA.
Previous studies [18–22] have shown no prognostic significance of MGMT methylation (or loss of MGMT) in colorectal cancers (Table 1). These previous studies [18–22] on prognostic significance of MGMT methylation (or MGMT loss) are limited by low statistical power (n < 200). In contrast to the prior studies [18–22], our study examined both MGMT promoter methylation and loss of MGMT expression in a much larger cohort of colorectal cancers. In the current study, MGMT methylation was found in 38% of colorectal cancer. Previous studies [18–22] have shown a large variation in the frequency of MGMT promoter methylation (21–61%; Table 1). This variation might be caused by differences in study samples and/or methylation detection methods (MSP vs. quantitative MethyLight vs. Pyrosequencing) and might also be in part due to a chance variation in the small studies.
With regard to the predictive role of MGMT aberrations, Braun et al. [34] examined a predictive role of loss of MGMT expression (among other markers) in 1,125 patients with metastatic colorectal cancer who underwent different chemotherapy treatment arms (fluorouracil vs. fluorouracil/irinotecan vs. fluorouracil/oxaliplatin) and found no predictive role of MGMT aberrations.
There are limitations in this study. For example, data on cancer treatment were limited. Nonetheless, it is unlikely that chemotherapy use substantially differed according to MGMT status in tumor, since such data were unavailable for treatment decision making. In addition, our multivariate survival analysis adjusted for disease stage as finely as I, IIA, IIB, IIIA, IIIB, IIIC, IV on which treatment decision making was mostly based. As another limitation, beyond cause of mortality, data on cancer recurrences were unavailable in these cohort studies. Nonetheless, colorectal cancer–specific survival might be a reasonable surrogate of colorectal cancer–specific outcome.
There are advantages in utilizing the database of the two prospective cohort studies, the Nurses’ Health Study and the Health Professionals Follow-up Study, to examine prognostic significance of tumor biomarkers. Anthropometric measurements, family history, cancer staging, and other clinical, pathologic, and tumoral molecular data were prospectively collected, blinded to patient outcome. Cohort participants who developed cancer were treated at hospitals throughout the United States and thus more representative colorectal cancers in the US population than patients in one to a few academic hospitals. There were no demographic difference between cases with tumor tissue analyzed and those without tumor tissue analyzed [23]. Finally, our rich tumor database enabled us to simultaneously assess pathologic and tumoral molecular correlates and control for confounding by a number of tumoral molecular alterations.
In conclusion, our findings suggest that MGMT promoter methylation or loss of expression is not a prognostic biomarker in colorectal cancer, despite its well-established role in carcinogenesis.
Acknowledgments
We deeply thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires, and hospitals and pathology departments throughout the United States for providing us with medical records and tumor tissue specimens. This work was supported by U.S. National Institute of Health (NIH) grants P01 CA87969 (to SE Hankinson), P01 CA55075 (to WC Willett), P50 CA127003 (to CSF), K07 CA122826 (to SO), and R01 CA151993 (to SO), and in part by grants from the Bennett Family Fund and from the Entertainment Industry Foundation National Colorectal Cancer Research Alliance. Y.B. was supported by a fellowship grant from the Uehara Memorial Foundation. K.N. and M.S. were supported by fellowship grants from the Japan Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- BMI
Body mass index
- CI
Confidence interval
- CIMP
CpG island methylator phenotype
- HR
Hazard ratio
- MGMT
O6-methylguanine-DNA methyltransferase
- MSI
Microsatellite instability
- MSS
Microsatellite stable
- OR
Odds ratio
Footnotes
Conflicts of interest No conflicts of interest exist.
Contributor Information
Kaori Shima, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Teppei Morikawa, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Yoshifumi Baba, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Katsuhiko Nosho, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Maiko Suzuki, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Mai Yamauchi, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Marika Hayashi, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA.
Edward Giovannucci, Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA. Departments of Epidemiology and Nutrition, Harvard School of Public Health, Boston, MA, USA.
Charles S. Fuchs, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA. Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
Shuji Ogino, Email: shuji_ogino@dfci.harvard.edu, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA, USA. Department of Pathology, Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA, USA. Center for Molecular Oncologic Pathology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital, Harvard Medical School, 44 Binney St., Room JF-215C, Boston, MA 02115, USA.
References
- 1.Asaka S, Arai Y, Nishimura Y, et al. Microsatellite instability-low colorectal cancer acquires a KRAS mutation during the progression from Dukes’ A to Dukes’ B. Carcinogenesis. 2009;30:494–499. doi: 10.1093/carcin/bgp017. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalo V, Lozano JJ, Munoz J, et al. Aberrant gene promoter methylation associated with sporadic multiple colorectal cancer. PLoS One. 2010;5:e8777. doi: 10.1371/journal.pone.0008777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tranah GJ, Bugni J, Giovannucci E, et al. O6-methylguanine-DNA methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of colorectal cancer in the nurses’ Health Study and Physicians’ health study (United States) Cancer Causes Control. 2006;17:721–731. doi: 10.1007/s10552-006-0005-y. [DOI] [PubMed] [Google Scholar]
- 4.Zhong Y, Huang Y, Huang Y, et al. Effects of O6-methylguanine-DNA methyltransferase (MGMT) polymorphisms on cancer: a meta-analysis. Mutagenesis. 2010;25:83–95. doi: 10.1093/mutage/gep050. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 6.Nagasaka T, Goel A, Notohara K, et al. Methylation pattern of the O6-methylguanine-DNA methyltransferase gene in colon during progressive colorectal tumorigenesis. Int J Cancer. 2008;122:2429–2436. doi: 10.1002/ijc.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 8.Esteller M, Risques RA, Toyota M, et al. Promoter hy-permethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigen-esis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- 9.Halford S, Rowan A, Sawyer E, Talbot I, Tomlinson I. O(6)-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut. 2005;54:797–802. doi: 10.1136/gut.2004.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogino S, Kawasaki T, Kirkner GJ, et al. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56:1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 13.Hegi ME, Sciuscio D, Murat A, Levivier M, Stupp R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin Cancer Res. 2009;15:5026–5031. doi: 10.1158/1078-0432.CCR-08-1169. [DOI] [PubMed] [Google Scholar]
- 14.Watson AJ, Sabharwal A, Thorncroft M, et al. Tumor O(6)-methylguanine-DNA methyltransferase inactivation by oral lomeguatrib. Clin Cancer Res. 2010;16:743–749. doi: 10.1158/1078-0432.CCR-09-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 16.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligoden-droglial tumors: a report from EORTC brain tumor group study 26951. J Clin Oncol. 2009;27:5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteller M, Gaidano G, Goodman SN, et al. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst. 2002;94:26–32. doi: 10.1093/jnci/94.1.26. [DOI] [PubMed] [Google Scholar]
- 18.Nagasaka T, Sharp GB, Notohara K, et al. Hypermethylation of O6-methylguanine-DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res. 2003;9:5306–5312. [PubMed] [Google Scholar]
- 19.Krtolica K, Krajnovic M, Usaj-Knezevic S, et al. Comethylation of p16 and MGMT genes in colorectal carcinoma: correlation with clinicopathological features and prognostic value. World J Gastroenterol. 2007;13:1187–1194. doi: 10.3748/wjg.v13.i8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SP, Chiu SC, Wu CC, et al. The association of methylation in the promoter of APC and MGMT and the prognosis of Taiwanese CRC patients. Genet Test Mol Biomarkers. 2009;13:67–71. doi: 10.1089/gtmb.2008.0045. [DOI] [PubMed] [Google Scholar]
- 21.Kim JC, Choi JS, Roh SA, et al. Promoter methylation of specific genes is associated with the phenotype and progression of colorectal adenocarcinomas. Ann Surg Oncol. 2010;17:1767–1776. doi: 10.1245/s10434-009-0901-y. [DOI] [PubMed] [Google Scholar]
- 22.Kohonen-Corish MR, Daniel JJ, Chan C, et al. Low microsatellite instability is associated with poor prognosis in stage C colon cancer. J Clin Oncol. 2005;23:2318–2324. doi: 10.1200/JCO.2005.00.109. [DOI] [PubMed] [Google Scholar]
- 23.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Brahmandam M, Cantor M, et al. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 27.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 35.Weijenberg MP, Luchtenborg M, de Goeij AF, et al. Dietary fat and risk of colon and rectal cancer with aberrant MLH1 expression, APC or KRAS genes. Cancer Causes Control. 2007;18:865–879. doi: 10.1007/s10552-007-9032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samowitz WS, Curtin K, Wolff RK, et al. Microsatellite instability and survival in rectal cancer. Cancer Causes Control. 2009;20:1763–1768. doi: 10.1007/s10552-009-9410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berndt SI, Huang WY, Yeager M, et al. Genetic variants in frizzled-related protein (FRZB) and the risk of colorectal neoplasia. Cancer Causes Control. 2009;20:487–490. doi: 10.1007/s10552-008-9274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsilidis KK, Helzlsouer KJ, Smith MW, et al. Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control. 2009;20:1739–1751. doi: 10.1007/s10552-009-9427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglesias D, Nejda N, Azcoita MM, et al. Effect of COX2–765G>C and c.3618A>G polymorphisms on the risk and survival of sporadic colorectal cancer. Cancer Causes Control. 2009;20:1421–1429. doi: 10.1007/s10552-009-9368-1. [DOI] [PubMed] [Google Scholar]
- 40.Figueiredo JC, Levine AJ, Lee WH, et al. Genes involved with folate uptake and distribution and their association with colorectal cancer risk. Cancer Causes Control. 2009;21:597–608. doi: 10.1007/s10552-009-9489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Guelpen B, Dahlin AM, Hultdin J, et al. One-carbon metabolism and CpG island methylator phenotype status in incident colorectal cancer: a nested case-referent study. Cancer Causes Control. 2010;21:557–566. doi: 10.1007/s10552-009-9484-y. [DOI] [PubMed] [Google Scholar]
- 42.Feik E, Baierl A, Hieger B, et al. Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps and colorectal cancer risk. Cancer Causes Control. 2009;21:91–97. doi: 10.1007/s10552-009-9438-4. [DOI] [PubMed] [Google Scholar]
- 43.Ogino S, Hazra A, Tranah GJ, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28:1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 44.Hawkins NJ, Lee JH, Wong JJ, et al. MGMT methylation is associated primarily with the germline C>T SNP (rs16906252) in colorectal cancer and normal colonic mucosa. Mod Pathol. 2009;22:1588–1599. doi: 10.1038/modpathol.2009.130. [DOI] [PubMed] [Google Scholar]
- 45.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res (Phila Pa) 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- 46.Lee BB, Lee EJ, Jung EH, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 47.Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 48.de Vogel S, Weijenberg MP, Herman JG, et al. MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: indications for distinct pathways and sequence of events. Ann Oncol. 2009;20:1216–1222. doi: 10.1093/annonc/mdn782. [DOI] [PubMed] [Google Scholar]


