Abstract
In vitro assays identified the Golgi peripheral protein GRASP65 as a Golgi stacking factor that links adjacent Golgi cisternae by forming mitotically regulated trans-oligomers. These conclusions, however, require further confirmation in the cell. In this study, we showed that the first 112 amino acids at the N-terminus (including the first PDZ domain, PDZ1) of the protein are sufficient for oligomerization. Systematic electron microscopic analysis showed that the expression of non-regulatable GRASP65 mutants in HeLa cells enhanced Golgi stacking in interphase and inhibited Golgi fragmentation during mitosis. Depletion of GRASP65 by small interference RNA (siRNA) reduced the number of cisternae in the Golgi stacks; this reduction was rescued by expressing exogenous GRASP65. These results provided evidence and a molecular mechanism by which GRASP65 stacks Golgi cisternal membranes. Further experiments revealed that inhibition of mitotic Golgi disassembly by expressing non-regulatable GRASP65 mutants did not affect equal partitioning of the Golgi membranes into the daughter cells. However, it delayed mitotic entry and suppressed cell growth; this effect was diminished by dispersing the Golgi apparatus with Brefeldin A treatment prior to mitosis, suggesting that Golgi disassembly at the onset of mitosis plays a role in cell cycle progression.
Keywords: cell cycle, Golgi stacking, GRASP65, oligomerization, phosphorylation
The Golgi apparatus is the central conduit for protein trafficking, processing and secretion in almost all eukaryotic cells. In interphase mammalian cells, the Golgi apparatus consists of stacks of parallel aligned flattened membrane cisternae, which are further linked laterally by tubules to form a ribbon-like structure (1). During mitosis, the Golgi apparatus undergoes extensive disassembly; the generated vesicles are equally partitioned into the two daughter cells, where these membranes are reassembled into new Golgi stacks (2). In vitro assays that reconstitute the cell cycle-regulated Golgi disassembly and reassembly processes showed that mitotic Golgi disassembly consists of two reactions: Golgi unstacking mediated by phosphorylation of Golgi structural proteins and vesiculation mediated by coat protein I (COPI) vesicle formation; while postmitotic Golgi reassembly requires vesicle fusion for single cisterna formation and stacking of the newly formed cisternae (3). The molecular mechanism of Golgi stacking remains unclear. Accumulating evidence suggests that stack formation requires the Golgi reassembly stacking protein GRASP65. GRASP65 was first identified as a Golgi peripheral protein that is accessible to the alkylating reagent N-ethylmaleimide (NEM) only when the Golgi stacks were disassembled (4). Further experiments showed that adding GRASP65 antibodies to the in vitro Golgi reassembly assay inhibited stacking of newly formed Golgi cisternae (4). Consistently, microinjection of GRASP65 antibodies into mitotic cells inhibited subsequent Golgi stack formation in the daughter cells (5).
Biochemical experiments revealed a possible mechanism for GRASP65 in Golgi stacking. GRASP65 forms homodimers that further oligomerize in trans to hold the adjacent Golgi membranes into stacks. Oligomerization is mitotically regulated. During mitosis, GRASP65 is phosphorylated by two mitotic kinases, cdc2 and polo-like kinase (plk), which leads to GRASP65 deoligomerization and thus Golgi unstacking (5,6). Oligomerization of GRASP65 is mediated by the N-terminal GRASP domain, while its phosphorylation occurs at the C-terminal serine/proline-rich (SPR) domain (6). Further studies showed that, when targeted to the outer membranes of mitochondria, GRASP65 is capable of tethering mitochondria into clusters (7). Finally, the expression of a caspase-resistant form of GRASP65 partially prevented Golgi fragmentation in apoptotic cells (8). GRASP65 is concentrated in the cis Golgi membranes, while its homolog, GRASP55, is primarily localized to the medial-trans cisternae (9). These studies provided evidence that GRASP65 is both necessary and sufficient to hold the cis Golgi membranes into stacks, while GRASP55 may stack the medial-trans cisternae in a similar manner (10,11). During mitosis, phosphorylation of the GRASP proteins allows unstacking of the Golgi cisternae, which may facilitate COPI vesicle formation and fragmentation of the Golgi membranes (11–14). Subsequent dephosphorylation of GRASP and other Golgi structural proteins is required for postmitotic Golgi reassembly (12,15).
However, the hypothesis that Golgi stacking is the primary role of GRASP65 has recently been challenged based on studies of GRASP65 homologs in other species (16,17) and knockdown experiments of mammalian GRASP65 (18,19). In Drosophila, knockdown of the sole GRASP protein, dGRASP, and its interacting protein GM130 resulted in at least partial disassembly of the Golgi stacks into single cisternae and vesicles in S2 cells (17). In mammalian cells, depletion of GRASP65 using small interference RNA (siRNA) led to either Golgi ribbon unlinking (19) or an arrest in cell division accompanied by irregular mitotic spindle formation and cell death, with the number of cisternae in the stacks decreased (18). Obviously, the siRNA technique has limitations (such as incomplete knockdown and off-target effects); simple knockdown experiments (18,19) and the experiments using forced ectopic targeting of GRASP65 (7) may not be able to exclude the possibility of side-effects. In addition, the roles (e.g. stacking, ribbon linking and cell cycle control) of GRASP65 in the cell may be related to each other. Therefore, a re-evaluation of GRASP65 functions in vivo is necessary and important.
The exact role of mitotic Golgi disassembly is so far unknown, but it is thought to facilitate equal partitioning of the Golgi membranes into the daughter cells (15,20). In addition, Golgi ribbon unlinking at late G2 phase may function as a new checkpoint for cell cycle progression into mitosis, as inhibition of Golgi ribbon cleavage at the onset of mitosis by blocking the activity of the Golgi fission protein BARS (21) or MEK1 kinase (22) delayed mitotic entry of the cells. This delay was rescued when the cells were treated with Brefeldin A (BFA), a fungal product that disassembles the Golgi apparatus, prior to mitosis (22,23). Interestingly, interrupting GRASP65 function by microinjection of either a C-terminal fragment of GRASP65 or antibodies to this fragment into normal rat kidney (NRK) cells prevented cell entry into mitosis (23). Further studies showed that phosphorylation of GRASP65 on serine 277 plays an important role in cell cycle regulation, as a peptide containing this site, but not its serine to alanine (S277A) mutant, delayed mitotic entry when microinjected into NRK cells (24). The underlying mechanism for GRASP65 in cell cycle control is so far unclear. One possibility is through its regulation on mitotic kinases. As GRASP65 is a substrate of cdc2 and plk, manipulation of GRASP65 level may affect the localization and activity of these kinases in cell cycle progression, as suggested by a previous study (25). Alternatively, physical disruption of the Golgi structure mediated in part by GRASP65 phosphorylation and thus deoligomerization may be required for the separation of the duplicated centrosomes for spindle formation and mitotic progression (26). If this is the case, inhibition of GRASP65 phosphorylation may delay Golgi disassembly at the onset of mitosis and thus interrupt cell cycle progression.
In this study, we have applied biochemical and cell biology approaches to study the function of GRASP65 in mammalian cells. We established stable cell lines in which endogenous GRASP65 was depleted by siRNA and exogenous wild-type (WT) GRSAP65 or its phosphorylation defective mutants were expressed under an inducible promoter. This allowed us to determine the role of GRASP65 in Golgi stacking by systematic electron microscopic (EM) analysis. Our results support that GRASP65 stacks Golgi cisternal membranes. In addition, we found that exogenous expression of the PDZ1 region, which is necessary and sufficient for Golgi targeting and promotion of cisternal stacking but lacks the entire phosphorylation domain, delays mitotic progression, indicating that Golgi disassembly, instead of GRASP65 phosphorylation itself, is important for the regulation of mitotic progression. Our study provides evidence and a molecular mechanism showing that GRASP65 directly functions as a Golgi stacking factor and that mitotic Golgi disassembly is linked to cell cycle progression.
Results
The N-terminal 112 amino acids (PDZ1) of GRASP65 are sufficient for dimerization and oligomerization
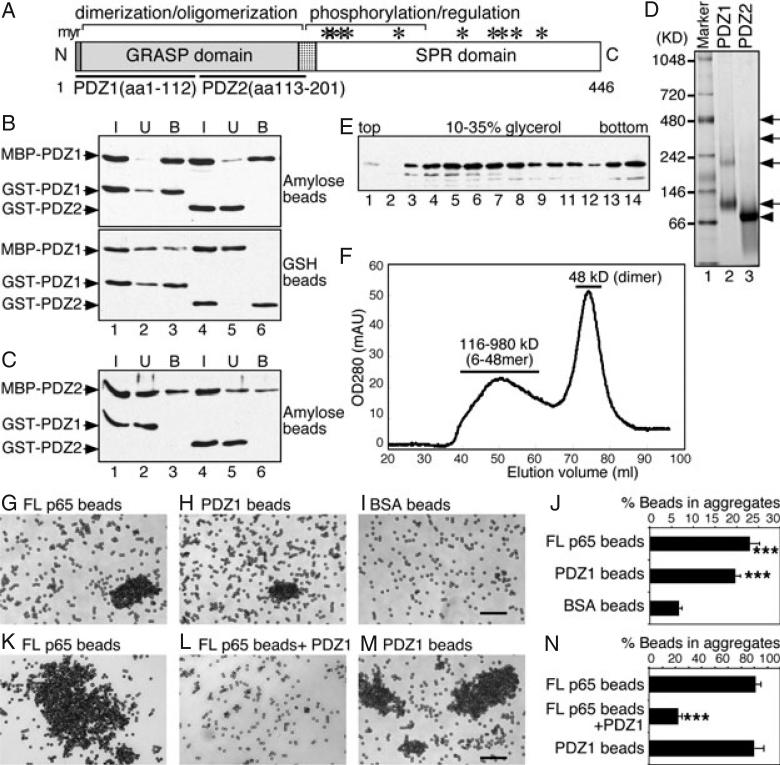
Previous results showed that GRASP65 forms both dimers and oligomers (5) and subsequent studies revealed that the N-terminal GRASP domain (aa 1–201) is both necessary and sufficient for dimerization and oligomerization (6). As the GRASP domain has two PDZ-containing domains, PDZ1 (aa 1–112) and PDZ2 (aa 113–201; Figure 1A), we first tested whether PDZ1 or PDZ2 is sufficient to mediate GRASP65 oligomerization using an established copurification assay (5,6). Recombinant maltose binding protein (MBP)- and glutathione S-transferase (GST)-tagged PDZ1 and PDZ2 were expressed in bacteria and purified separately, then mixed and incubated. Protein complexes were reisolated using amylose beads or glutathione beads and the copurified proteins were determined by western blotting. As shown in Figure 1B, only MBP-PDZ1 and GST-PDZ1 copurified, regardless of the beads that were used (Figure 1B, lane 3). However, MBP-PDZ1 was not able to interact with GST-PDZ2 (Figure 1B, lane 6), nor MBP-PDZ2 with GST-PDZ1 or GST-PDZ2 (Figure 1C, lanes 3 and 6). The results suggest that PDZ1 interacts with itself to form oligomers, while PDZ2 is not sufficient for oligomerization.
Figure 1. The N-terminal half of the GRASP domain (PDZ1) is sufficient to form dimers and oligomers.
A) Domain structure of GRASP65. Indicated are the N-terminal GRASP domain with myristoylated N-terminal glycine (myr) and the C-terminal serine/proline-rich (SPR) domain with phosphorylation sites (*). The GRASP domain contains two PDZ subdomains, PDZ1 (aa 1–112) and PDZ2 (aa 113–201). B) PDZ1 is sufficient to form oligomers as shown by a copurification assay. Separately purified recombinant MBP-tagged PDZ1 was mixed and incubated with GST-tagged PDZ1 or PDZ2. The protein complexes were isolated using either amylose or glutathione (GSH) beads, as indicated. Equal proportions of the input (I), unbound (U) or bound (B) fractions were analyzed by immunoblotting for GRASP65. Note that PDZ1 copurified with itself (lane 3), but not with PDZ2 (lane 6). C) PDZ2 does not form oligomers. As in (B) but using MBP-PDZ2 as the bait. Note that PDZ2 did not copurify with PDZ1 (lane 3) or itself (lane 6). D) Analysis of oligomerization of PDZ1 and PDZ2 by non-denaturing gels. GST-tagged PDZ1 and PDZ2 were incubated at 37°C and analyzed by non-denaturing electrophoresis and stained with Coomassie Blue. Molecular weight standards are indicated on the left. Note that PDZ1 formed dimers and oligomers (arrows), while PDZ2 formed dimers (arrowhead) only. E) Analysis of PDZ1 oligomerization by velocity gradient. Purified recombinant MBP-tagged PDZ1 was sedimented in 10–35% glycerol gradients. Shown is the western blot for GRASP65 in each fraction. PDZ1 is found in both the upper (4–7; dimers) and deep fractions (13–14; higher-order structures). F) Gel filtration analysis of PDZ1. Purified His-tagged PDZ1 was analyzed by gel filtration using a Sephacryl S-300 column. The protein was separated into two peaks: a broad peak of 116–980 kDa (6-48mer) and a sharp peak of 48 kDa (dimer). Sizes were calculated according to the calibration with a gel filtration molecular weight standard (Sigma). G–I) Bead aggregation assay showing that PDZ1 is sufficient to form oligomers. Beads coated with His-tagged full-length (FL) GRASP65 (G), PDZ1 (H) or BSA (I) were incubated with BSA at 37°C for 60 min. After incubation, the beads were placed on glass slides and random fields were photographed. A representative image of each condition is shown. Bar, 50 μm. Note that FL and PDZ1-coated beads formed aggregates. J) Quantitation of the percentage of beads in aggregates in (G–I) from three independent experiments. ***p < 0.001, compared with BSA-coated beads. K–M) Bead aggregation assay showing that soluble PDZ1 inhibits oligomerization of FL GRASP65. Beads coated with His-tagged FL GRASP65 (K and L) or PDZ1 (M) were incubated with interphase cytosol at 37°C for 60 min. In (L), purified recombinant PDZ1 was added into the reaction. After incubation, the beads were placed on glass slides and random fields were photographed. A representative image of each condition is shown. Bar, 50 μm. Note that PDZ1 can inhibit the aggregation of GRASP65-coated beads. N) Quantitation of the percentage of beads in aggregates in (K–M). Results in (J) and (N) expressed as the mean ± SEM from three sets of independent experiments. ***p < 0.001, compared with FL GRASP65-coated beads in the absence of PDZ1 added to the reaction.
We then analyzed PDZ1 oligomerization by non-denaturing electrophoresis. As shown in Figure 1D, GST-PDZ1 exhibited four bands on the gel, as indicated by arrows (lane 2). The lowest band of 82 kDa was about twice the size of the estimated molecular weight of monomeric GST-PDZ1 (41 kDa); the upper bands exhibited molecular weights of 185, 307 and 424 kDa. These results suggest that PDZ1 exists as dimers and higher-order oligomers, consistent with the oligomeric properties of GRASP65 shown in previous studies (5,6). GST-PDZ2 (Figure 1D, lane 3), however, exhibited only one band of 64 kDa on the gel, which was about twice the molecular weight of GST-PDZ2 (33 kDa), indicating that PDZ2 forms dimers but not oligomers. All these bands were excised from the gel and further confirmed by denaturing electrophoresis and western blotting. As GST itself forms homodimers (27), this result was confirmed using His-tagged PDZ1 and PDZ2 (not shown). We also analyzed PDZ1 dimerization and oligomerization using other biochemical techniques. In a sedimentation gradient (Figure 1E), MBP-tagged PDZ1 was concentrated in two groups of fractions, dimers in fraction 4–7 and oligomers in heavier fractions including 13–14; in gel filtration (Figure 1F), His-tagged PDZ1 was eluted as two peaks, one sharp peak of 48 kDa (dimer) and another broad one of 116–980 kDa (6-48mer). As PDZ1 was the smallest fragment that is capable of oligomerization, it was further characterized in the following experiments.
We employed a well-established bead aggregation assay (5) to test whether PDZ1 is sufficient to link adjacent surfaces. When incubated with BSA, a significant amount (19.7 ± 1.1%) of the Dynal beads coated with His-tagged PDZ1 formed aggregates, similar to those coated with full-length GRASP65 (23.1 ± 1.7%). In contrast, only 6.7 ± 0.6% of the beads coated with BSA formed aggregates under the same condition (Figure 1I,J). Incubation with interphase cytosol (IC) greatly enhanced the aggregation of Dynal beads coated with PDZ1; 80.2 ± 3.2% of the beads were in aggregates, similar to those coated with full-length GRASP65 (81.0 ± 1.7%; Figure 1, M versus K, N). In addition, an excess amount of His-PDZ1, when added into the reaction, inhibited the aggregation of beads coated with full-length GRASP65 (Figure 1L and N). In agreement with our results, a recent study showed that PDZ1, when expressed and targeted to the outer membrane of mitochondria, linked mitochondria to form clusters (7). Taken together, these results show that PDZ1 of GRASP65 is both necessary and sufficient to hold surfaces together through oligomerization.
Expression of non-regulatable GRASP65 mutants enhances Golgi stacking in interphase cells
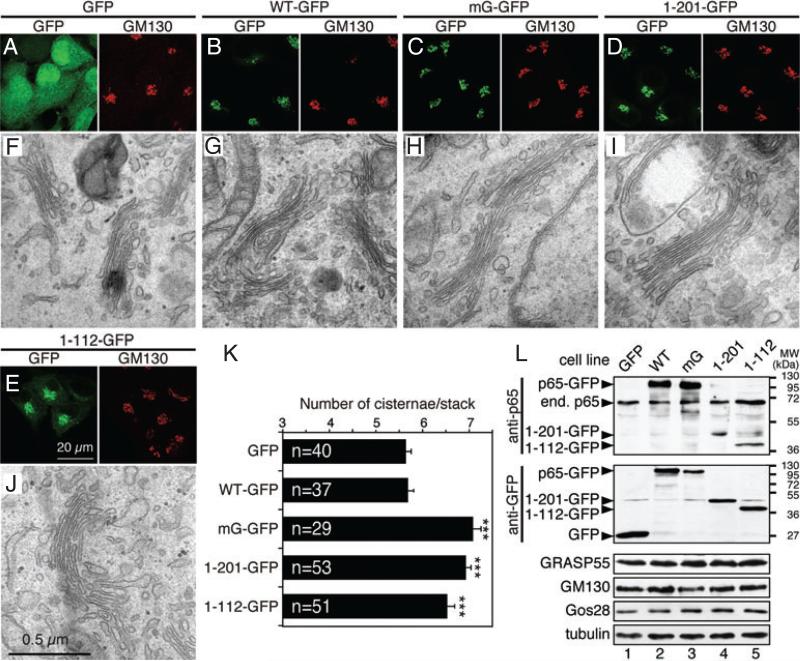
To test the function of GRASP65 in the cell, we expressed the WT protein or its mutants using a retroviral tet-inducible mammalian expression system (tet-on) (11,28). The coding sequence was integrated into the genome but the exogenous protein is not expressed unless induced by doxycycline. This is particularly important, as continuous overexpression of GRASP65 or its mutants may affect cell growth (18). Establishment of stable cell lines expressing the exogenous protein in an inducible manner also allowed a systematic analysis by EM. The expression level of the exogenous proteins was well controlled, thus non-specific effects caused by protein overexpression were not a concern in our study. Five lines of HeLa cells were generated that express (i) GFP (enhanced green fluorescent protein) and GFP-tagged, (ii) WT GRASP65, (iii) non-phosphorylatable GRASP65 mutant, mG, with seven phosphorylation sites mutated to alanines (S216A/S217A/T222A/T224A/S277A/S367A/S376A) (6,29), (iv) the GRASP domain (aa 1–201) and (5) PDZ1 (aa 1–112) of rat GRASP65. The last three mutants form oligomers, but their oligomerization cannot be regulated through phosphorylation (6), and thus they are termed as non-regulatable mutants. The GRASP domain has been examined previously (6) and was used as a positive control in this study. Other constructs, such as PDZ2 (Figure 1) and the SPR domain (6), do not form oligomers and thus were not examined in this experiment. The cytoplasmic soluble mutant (G2A) of the protein was originally included in this study, but did not show a significant effect on the Golgi structure (6) and thus was not further characterized. The exogenous proteins were well expressed when induced by doxycycline, as shown by western blot analysis (Figure 2L). Fluorescence microscopy (Figure 2A–E) showed that the expressed GRASP65 WT-GFP, mG-GFP and GRASP domain-GFP localized to the Golgi in interphase cells, indicated by colocalization with the Golgi marker GM130 (Figure 2B–D); while PDZ1-GFP (Figure 2E) was partially Golgi localized and GFP (Figure 2A) was found in the nucleus and cytosol. Expression of GRASP65 and its mutants did not significantly affect the overall Golgi organization at the light microscopic level when using GM130 (Figure 2A–E), α-mannosidase II and Gos28 (not shown) as Golgi markers.
Figure 2. Expression of non-regulatable GRASP65 mutants enhances Golgi stacking in interphase cells.
A–E) Confocal images of interphase HeLa cells stably expressing indicated GRASP65 constructs using an (tet-on) inducible retroviral expression system. Cells were treated with 1 μg/mL doxycycline for 72 h, fixed and stained for GM130. F–J) Representative EM images of interphase cells expressing indicated GRASP65 constructs. Bar, 0.5 μm. Note that the number of cisternae in the Golgi stacks was increased in cells expressing the mG mutant (H), the GRASP domain (aa 1–201, I) and PDZ1 (aa 1–112, J) compared with cells expressing GFP (F) and wild-type GRASP65 (G). K) Quantitation of (F–J). Results expressed as the mean ± SEM. The number of stacks quantified for each cell line was indicated. Statistical significance was assessed by comparison to the GFP cell line. ***p < 0.001. Bar charts indicating the actual frequency of each value of cisternal numbers are shown in Figure S1. L) Western blot images of cells described in (A–E). Cells were lysed in SDS buffer followed by western blotting for GFP and other indicated proteins. ’End. p65’, endogenous GRASP65.
When analyzed under the EM, cells expressing non-regulatable GRASP65 showed improved cisternal alignment in the Golgi stacks in comparison with cells expressing GFP or WT GRASP65 (Figures 2F–J and S1). The number of cisternae per stack significantly increased in mG (7.1 ± 0.2), GRASP domain (6.9 ± 0.1) or PDZ1 (6.5 ± 0.2) expressing cells compared with control GFP-expressing cells (5.6 ± 0.1), but not in cells expressing WT GRASP65 (5.7 ± 0.1; Figure 2K). Further analysis of the EM micrographs showed that most Golgi stacks in cells expressing the non-regulatable GRASP65 mutants contained 7–8 cisternae, a significant increase from 5 to 6 cisternae per stack in cells expressing GFP or WT GRASP65 (Figure S1). These results provided direct evidence that GRASP65 plays a critical role in Golgi stacking in vivo. In addition, as WT GRASP65 did not enhance stacking as its non-regulatable mutants, it suggests that phosphorylation of GRASP65 may regulate stacking in interphase, which may be important for Golgi remodeling under certain conditions such as cell migration in wound healing (26).
Expression of non-regulatable GRASP65 mutants inhibits mitotic Golgi disassembly
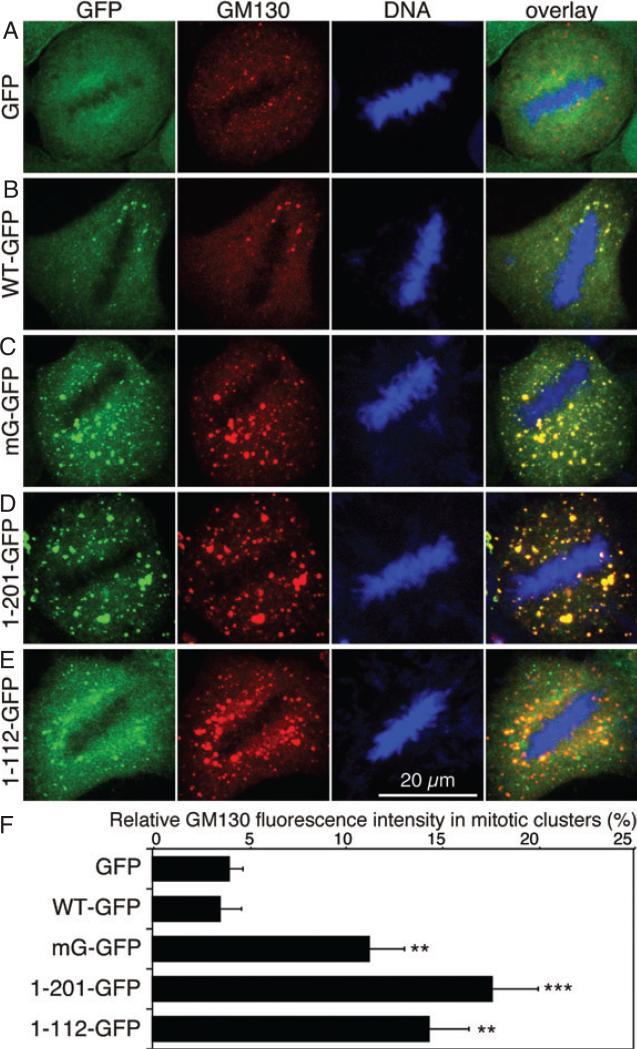
Previous reports showed that GRASP65 phosphorylation at the onset of mitosis is required for Golgi membrane unstacking, which facilitates subsequent vesiculation of the membranes (3,5). Consistently, the expression of the GRASP domain of GRASP65 inhibited Golgi fragmentation in mitosis (6), suggesting that the expressed non-regulatable GRASP65 mutants may inhibit mitotic Golgi disassembly as they remain as oligomers during mitosis. We therefore examined mitotic Golgi fragmentation in the cell lines described above. After 72 h doxycycline induction, cells were fixed, stained for DNA to select metaphase cells and GM130 to reveal the Golgi membranes. As shown in Figure 3, the Golgi membranes were extensively fragmented in metaphase cells expressing GFP or GRASP65-GFP. However, in cells expressing either the GRASP65 mG mutant, the GRASP domain or PDZ1, the Golgi was observed as large dots, or mitotic Golgi clusters, indicated by the Golgi marker GM130. Quantitation of the images showed that a significant higher amount of the Golgi membranes remained as clusters in cells expressing non-regulatable GRASP65 mutants compared with that in cells expressing GFP or WT GRASP65-GFP (Figure 3F). The exogenous GRASP65 mG-GFP and the GRASP domain-GFP colocalized with the endogenous GM130 on the mitotic Golgi clusters, suggesting that the reduced Golgi disassembly was caused by the expressed exogenous proteins. Although the PDZ1 was only partially localized to the Golgi, it inhibited Golgi fragmentation to the same extent as the mG mutant or the GRASP domain (Figure 3).
Figure 3. Expression of non-regulatable GRASP65 mutants inhibits mitotic Golgi disassembly at the light microscopy level.
A–E) Confocal images of mitotic HeLa cells stably expressing the indicated GRASP65 proteins. Cells were induced by doxycycline for 72 h and fixed, and stained for DNA and GM130. Note that the cells expressing the mG mutant (C), the GRASP domain (aa 1–201, D) and PDZ1 (aa 1–112, E) had more mitotic clusters compared with those expressing GFP (A) and wild-type GRASP65 (B). Scale bar, 20 μm. F) Relative distribution (%) of the Golgi membranes in mitotic Golgi clusters in the indicated cell lines. The fluorescence intensity of GM130 was quantified using the ImageJ software for the total fluorescent intensity and a threshold was used to filter the fluorescence in Golgi haze to quantify the fluorescent intensity in mitotic Golgi clusters. The average percentage of fluorescent intensity in Golgi clusters (mean ± SEM) from 10 cells in each cell line is shown. Statistical significance was assessed by comparison to the GFP cell line. **p < 0.005; ***p < 0.001.
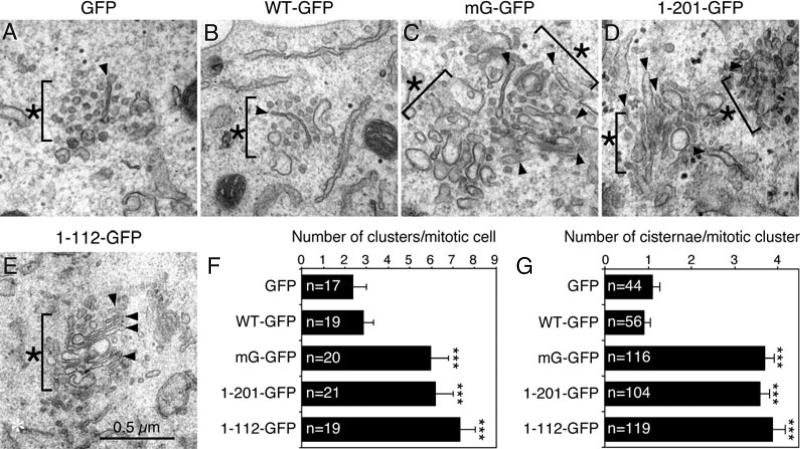
We then examined these cells under the EM. Mitotic HeLa cells were collected by shake-off after double thymidine block and release. Unlike mitotic cells expressing GFP or WT GRASP65 in which the Golgi complex was extensively disassembled and diffused, considerable amounts of Golgi membranes remained as Golgi clusters in cells expressing non-regulatable GRASP65 mutants (Figure 4C–F, asterisks). These clusters consisted of short cisternae (arrowheads), tubular structures or even ministacks that were surrounded by vesicles. Quantitation of the EM images showed that the number of mitotic clusters per mitotic cell profile (with condensed chromosomes but no nuclear envelope) was significantly increased in cells expressing mG (6.0 ± 0.8), the GRASP domain (6.2 ± 0.8) or PDZ1 (7.3 ± 0.7) compared with cells expressing GFP (2.4 ± 0.6) or WT GRASP65 (2.8 ± 0.4; Figure 4F). The mitotic clusters in cells expressing the non-regulatable mutants were also larger in size and more compact. In addition, each mitotic Golgi cluster contained more remaining Golgi cisternae in cells expressing mG (3.7 ± 0.2), the GRASP domain (3.6 ± 0.2) and PDZ1 (3.9 ± 0.3) compared with those expressing GFP (1.1 ± 0.2) and WT GRASP65 (0.9 ± 0.2; Figure 4G). These results suggest that the expression of the non-regulatable GRASP65 mutants inhibited the disassembly of Golgi membranes during mitosis.
Figure 4. Expression of non-regulatable GRASP65 mutants inhibits mitotic Golgi disassembly when examined by EM.
A–E) Representative EM images of Golgi clusters (asterisks) in mitotic cells expressing indicated GRASP65 constructs. Mitotic cells were collected by shake-off of cells released from a double thymidine block and processed for EM. Bar, 0.5 μm. Note that the mitotic Golgi clusters were larger and more compact in cells expressing the mG mutant (C), the GRASP domain (D) and PDZ1 (E), with an increased number of remaining Golgi cisternal structures (arrowheads) in the clusters. F and G) Quantitation results of (A–E) to determine the number of mitotic clusters per mitotic cell profile (F) and the number of cisternae per mitotic cluster (G). Only cells with condensed chromosomes but no detectable nuclear envelope were counted. The number of cells (F) and clusters (G) quantified for each cell line was indicated and results expressed as the mean ± SEM. Statistical significance was assessed by comparison to the GFP cell line. ***p < 0.001.
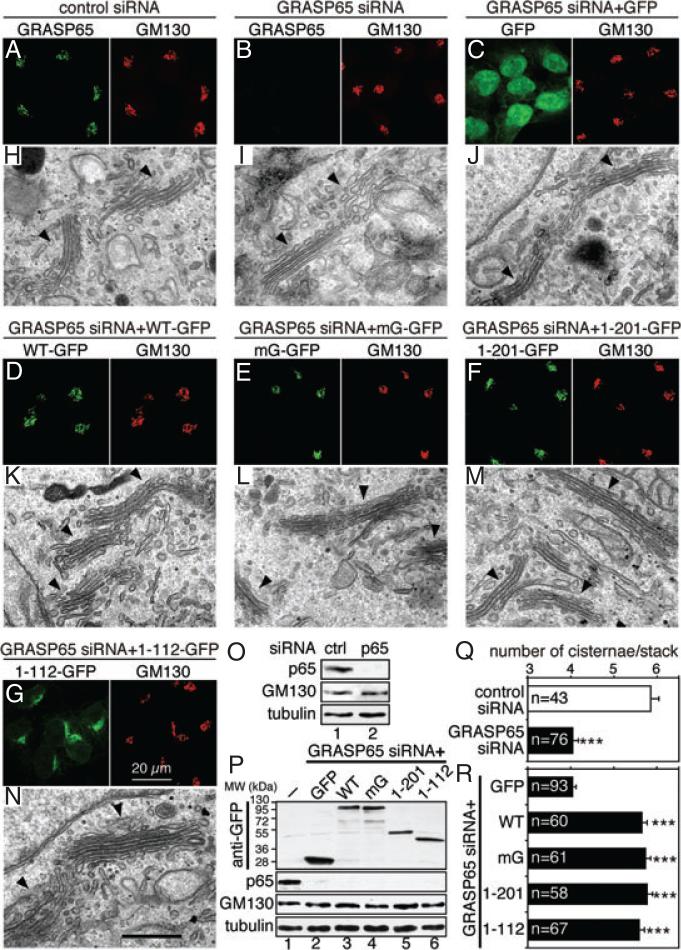
Depletion of GRASP65 reduced the number of cisternae per Golgi stack, which was rescued by expressing exogenous GRASP65
If GRASP65 plays a direct role in Golgi stacking, depletion of GRASP65 would reduce the number of cisternae in the stack. Indeed, it has been reported that knockdown of GRASP65 reduced the number of cisternae per stack from 6 to 3 (18). However, in another report, GRASP65 depletion in the same cell line using identical siRNA oligos led to Golgi ribbon unlinking (19). It is not clear whether the discrepancy was caused by different knockdown efficiencies between the two groups, off-target effects of the siRNA, or a limitation of the methods employed to analyze the effects. Therefore, we have re-evaluated the possible phenotypes in cells in which GRASP65 was efficiently depleted by both immunofluorescence microscopy and EM. Both western blotting (Figure 5O) and immunofluorescence microscopy (Figure 5B) showed that endogenous GRASP65 was efficiently depleted (94.1 ± 2.9% depletion, quantified from western blot results) 96 h after transfection with GRASP65 siRNA oligos. At the fluorescence microscopy level, no obvious change in the Golgi morphology was detected (Figure 5, B versus A). However, when analyzed under the EM, the number of cisternae per Golgi stack in GRASP65-depleted cells (4.0 ± 0.1) was significantly reduced compared with cells transfected with control siRNA (5.9 ± 0.2; Figure 5, I versus H, Q; Figure S2A), in agreement with a previous report (18). As GRASP65 is concentrated to the cis Golgi (9), this result indicates that GRASP65 is essential for stacking the cis Golgi membranes.
Figure 5. Depletion of GRASP65 reduces the number of cisternae per stack, which can be rescued by the expression of exogenous GRASP65.
A and B) Confocal fluorescence images of GRASP65 knockdown cells. HeLa cells transfected with indicated siRNA oligos were fixed and immunostained for GRASP65 and GM130. C–G) Confocal fluorescence images of HeLa cells in which endogenous GRASP65 was replaced by exogenous GRASP65 or its mutants. HeLa cells expressing indicated proteins were transfected with siRNA oligos for human GRASP65. Doxycycline was added 24 h after transfection. Cells were fixed 96 h after transfection and immunostained for GM130. H–N) Representative EM micrographs of cells described in (A–G). Arrowheads indicate Golgi stacks. Note that the number of cisternae per stack was reduced in GRASP65-depleted cells (I), which was restored by the expression of rat GRASP65 or its mutants (K–N), but not by GFP (J). O) Immunoblots of HeLa cells described in (A) and (B). The knockdown efficiency of GRASP65 was 94.1 ± 2.9%. P) Immunoblots of HeLa cells described in (C–G), in which endogenous GRASP65 was replaced by rat GRASP65 or its mutant. Lane 1, control cells with neither GRASP65 depleted nor exogenous protein expressed; lanes 2–6, cells with GRASP65 knocked down and indicated exogenous protein expressed. The knockdown efficiency for GRASP65 was 95.4 ± 0.6% for the GFP cell line and was similar in other cell lines, while GM130 expression level was not significantly affected. Q) Quantitation of the EM images in (H and I) in one representative experiment from three sets of independent experiments. Only stacks with three or more cisternae were counted. The number of Golgi stacks quantified was indicated. Results expressed as the mean ± SEM. Note that GRASP65 depletion reduced the average number of cisternae per stack. ***p < 0.001. R) Quantitation of the EM images in (J–N). Note the increase in cisternal number per stack by expression of GRASP65 and its mutants in GRASP65 knockdown cells. Statistical significance was assessed by comparison to the GFP cell line. ***p < 0.001. Bar charts indicating the actual frequency of each value of Golgi cisternae are shown in Figure S2.
To ensure that the observed effect was specific for GRASP65 depletion, we expressed exogenous rat GRASP65 in cells in which the endogenous GRASP65 was knocked down. We took advantage of the HeLa cell lines described earlier that stably express rat GRASP65 or its mutant in an inducible system. These cells were first transfected with siRNA targeted to human but not rat GRASP65; 24 h after transfection, doxycycline was added into the tissue culture medium followed by an additional 72 h incubation to induce the expression of exogenous GRASP65 proteins. As shown by the western blot results (Figure 5P), 95.4 ± 0.6% of the endogenous GRASP65 was depleted in the GFP cell line, while the exogenous GFP or GFP-tagged GRASP65 and its mutants were well expressed. At the fluorescence microscopy level using GM130 (Figure 5) and Gos28 (not shown) as Golgi markers, the overall Golgi structure did not exhibit any obvious differences between GFP- and GRASP65-expressing cells in which the endogenous GRASP65 was depleted (Figure 5, D versus C). When examined under the EM, cells expressing GRASP65-GFP had an increased number of cisternae per stack (5.7 ± 0.1) compared with those expressing GFP (4.0 ± 0.1; Figures 5R and S2B). A similar result was obtained in cells expressing GFP-tagged mG (5.7 ± 0.1), the GRASP domain (5.8 ± 0.1) or PDZ1 (5.6 5R ± 0.1; Figures and S2B). These results showed that the reduction of Golgi stacking in cells transfected with GRASP65 siRNA oligos was a direct consequence of GRASP65 depletion.
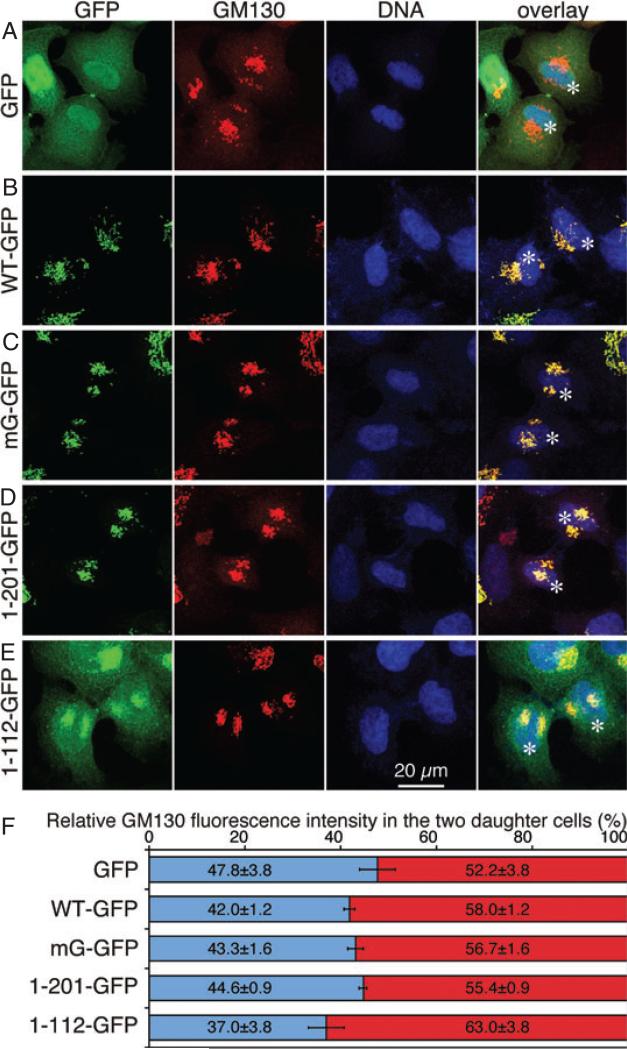
Expression of GRASP65 mutants does not affect equal distribution of Golgi membranes into the daughter cells during cell division
Golgi disassembly during mitosis in mammalian cells is generally thought to facilitate its equal partitioning into the two daughter cells (15,30). However, it has also been suggested that mitotic Golgi clusters may undergo ordered inheritance (31). Therefore, it is necessary to evaluate whether inhibition of mitotic Golgi disassembly affects Golgi partitioning. We took advantage of the cell lines expressing non-phosphorylatable GRASP65 mutants in which mitotic Golgi disassembly is inhibited (Figure 3) and quantified the amount of the Golgi membranes in the two daughter cells using GM130 as the marker. At least 10 pairs of daughter cells in cytokinesis in each cell line were observed under confocal microscopy and the fluorescence intensity in each daughter cell was quantified (Figure 6). No statistical difference in GM130 distribution in the two daughter cells was detected between the cell lines examined. This result showed that although mitotic Golgi fragmentation is inhibited in the cells that express GRASP65 mutants, Golgi partitioning into the two daughter cells is as accurate as control GFP-expressing cells. Similar results were obtained using other Golgi markers such as Gos28 (not shown). Consistent with this observation, it has been shown that inhibition of mitotic Golgi fragmentation by microinjection of a non-phosphorylatable p47 mutant did not affect equal distribution of Golgi membranes into the daughter cells (32). These results imply that either partial disassembly of the Golgi in mitosis (e.g. fragmentation of the Golgi ribbon into stack) is sufficient for its equal partitioning into the daughter cells, in which case an ordered partitioning mechanism may be involved, or the primary role of mitotic Golgi disassembly is not for equal Golgi membrane distribution.
Figure 6. Expression of non-regulatable GRASP65 mutants does not significantly affect Golgi membrane distribution into the two daughter cells.
A–E) Confocal images of indicated HeLa cell lines in cytokinesis. Cells were induced by doxycycline for 72 h and fixed, stained for DNA and GM130. Asterisks indicate the two daughter cells. Scale bar, 20 μm. F) Relative distribution of the Golgi membranes in the two daughter cells using GM130 as a marker. The fluorescence intensity of GM130 in 10 pairs of daughter cells in each cell line was quantified using the NIH ImageJ software. The average (mean ± SEM) intensity in the daughter cells with relatively lower intensity compared with their pairs is shown in blue, and the average intensity in the cells with relatively higher GM130 signal is shown in red. The distribution of GM130 in the two daughter cells is statistically insignificant between all the cell lines (p > 0.05).
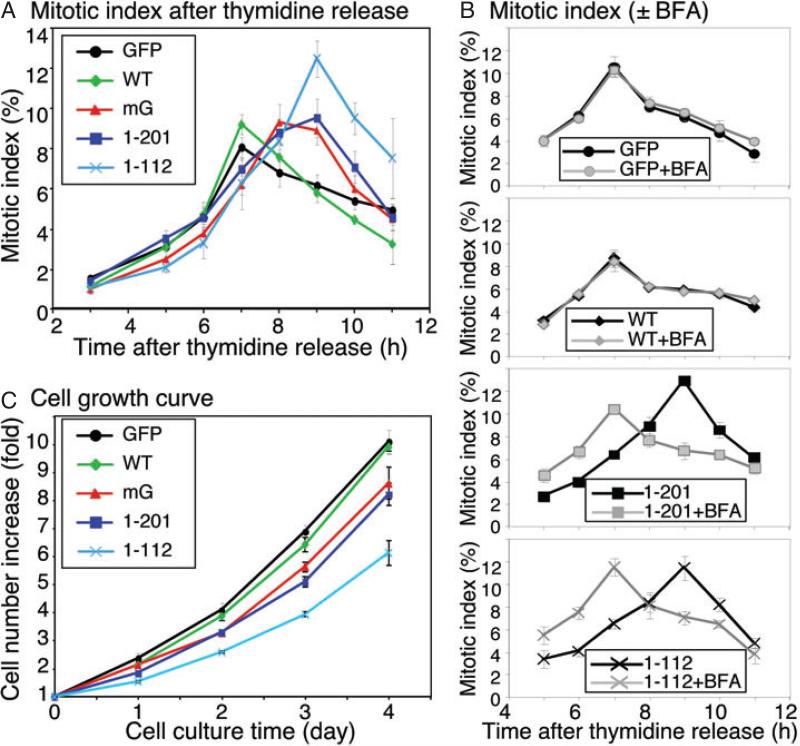
Inhibition of mitotic Golgi disassembly delays cell cycle progression
Previous reports suggested that the Golgi structure (and/or its mitotic disassembly) plays a role in cell cycle control (22,33). As expression of the non-regulatable GRASP65 mutants inhibited mitotic Golgi fragmentation, these cell lines provided us with excellent tools to determine the relationship between mitotic Golgi fragmentation and cell cycle progression. To evaluate whether inhibition of mitotic Golgi fragmentation affects mitotic entry, cells expressing GFP or GFP-tagged GRASP65 constructs (induced by doxycycline) were synchronized to G1/S phase by double thymidine block and the mitotic index of each cell line was determined over time after thymidine release. As shown in Figure 7A, the mitotic index of the GFP-expressing cells peaked at 7 h after thymidine release. Cells expressing WT GRASP65 showed a similar cohort of mitotic index, showing that the expression of WT GRASP65 protein did not affect mitotic entry. However, when the mG mutant, the GRASP domain or PDZ1 was expressed, the peak mitotic index was delayed for 1–2 h, suggesting that inhibition of mitotic Golgi fragmentation delayed cell cycle progression, which is consistent with previous reports (22,33).
Figure 7. Inhibition of mitotic Golgi fragmentation by expression of non-regulatable GRASP65 mutants delays mitotic entry and inhibits cell proliferation.
A) Inhibition of mitotic Golgi fragmentation delayed cell cycle progression. The percentage of cells in mitosis (mitotic index) at the indicated time-points of release from double thymidine block was determined for cells expressing GFP or GRASP65 constructs as indicated. Note that the cells expressing GFP or wild-type GRASP65 passed through mitosis in a cohort that peaks at 7 h. This peak was significantly delayed in the cells expressing the mG mutant, the GRASP domain or PDZ1 of GRASP65. B) BFA-induced Golgi disruption in cells expressing non-regulatable GRASP65 mutants rescued mitotic progression. As in (A), except that 5 μg/mL BFA was added to the tissue culture medium 4 h after the release from the double thymidine block. Note that the BFA treatment suppressed the mitotic delay in cells expressing the GRASP domain or PDZ1. C) Expression of non-regulatable mutants of GRASP65 reduced the cell growth rate. The cell number of each cell line was measured by staining the cells with crystal violet at indicated time-points and normalized with the cell number at the starting point. Note that the expression of the mG mutant, the GRASP domain or PDZ1 reduced the cell growth rate compared with expression of GFP or wild-type GRASP65.
The fact that disrupting GRASP65 function by expression or microinjection of a C-terminal fragment of the protein leads to a delay in cell mitotic entry has been described previously (23,24); however, the mechanism remains unknown. One possible mechanism is that GRASP65 regulates the function of mitotic kinases cdc2 and plk and therefore delays mitosis progression. It has been shown that the expression of the WT GRASP65 C-terminal SPR domain, but not the phosphorylation defective mutant, caused a delay in cell cycle progression (25). However, the non-regulatable GRASP65 mutants used in our study were not substrates of the mitotic kinases, because the phosphorylation sites were either mutated or deleted. In addition, these proteins are localized on the Golgi when expressed in the cell, which is different from the cytosolic SPR domain used in previous studies (23,25). Expression of these constructs also showed no effect on the expression level and localization of cdc2, cyclin B1 and plk in the cell (not shown). Therefore, it is unlikely that the delay in mitotic entry observed in this study occurred through inhibition or modulation of the mitotic kinases.
Another possible explanation for the cell cycle progression delay is that Golgi disassembly is required for mitotic entry, as suggested by previous reports (23,26). In this model, GRASP65 has no direct role in cell cycle control, but inhibition of mitotic Golgi fragmentation by interrupting GRASP65 function may delay mitotic entry. If this is the case, physical disruption of the Golgi by BFA before mitosis (22) may abolish the mitotic delay caused by the expression of non-regulatable GRASP65 mutants. To test this possibility, we added BFA into the tissue culture medium 4 h after final release from the double thymidine block and measured the mitotic index at later time-points. As shown in Figure 7B, BFA treatment abrogated the 2 h mitotic delay caused by the expression of the GRASP domain or PDZ1. In contrast, disruption of the Golgi structure by BFA treatment did not affect mitotic entry of cells expressing GFP or WT GRASP65-GFP. This result indicated that the cell cycle delay caused by the expression of non-regulatable GRASP65 mutants was through inhibition of Golgi disassembly at the onset of mitosis.
Next, we examined the growth of cells that express non-regulatable GRASP65 mutants. Equal numbers of cells were plated into 24-well plates. After 2 days of doxycycline induction to express the exogenous proteins, the number of cells was measured over time using crystal violet, a DNA-binding dye. As shown in Figure 7C, the increase in the cell number was reduced in cells expressing the mG mutant, the GRASP domain or PDZ1 compared with those expressing GFP or GRASP65 WT protein at all time-points examined. The effect was most significant on day 3, with p < 0.01 when comparing cells expressing mG, the GRASP domain or PDZ1 with those expressing GFP (Figure 7C). It became less significant on day 4, possibly because the cells were confluent on the dish. The effect was observed only when the exogenous proteins were induced, but not in the absence of doxycycline. These results suggest that inhibition of mitotic Golgi disassembly delays cell cycle progression and slows down cell growth. To summarize, our study shows that oligomerization of GRASP65 through its N-terminal 112 amino acids is essential for Golgi stacking in tissue culture cells, and inhibition of mitotic Golgi disassembly by expressing GRASP65 mutants delays cell cycle progression.
Discussion
In this study, we have examined the role of GRASP65 in Golgi stacking using a combination of biochemical approaches, an inducible expression system, the siRNA technique and systematic EM analysis. We first showed that its first PDZ domain oligomerizes with itself, which is sufficient to hold surfaces together. Our results further showed that the expression of non-regulatable GRASP65 mutants enhances Golgi stacking and inhibits Golgi fragmentation in mitotic cells. GRASP65 is required for full stacking as GRASP65 depletion reduces the number of cisternae in the stack; this effect can be rescued by expression of exogenous GRASP65. Furthermore, using non-regulatable GRASP65 as a tool, we showed that inhibition of Golgi fragmentation at the onset of mitosis does not significantly affect the accuracy of Golgi inheritance, but it delays mitotic entry and slows down cell growth, supporting a model in which Golgi fragmentation is needed for proper cell cycle progression. Our results revealed the mechanism for GRASP65 as a Golgi stacking factor, which holds the adjacent cisternal membranes by forming trans-oligomers through the N-terminal PDZ1 domain. Although GRASP65 does not seem to play a direct role in cell cycle control, physical disruption of the Golgi is required for mitotic entry.
GRASP65 has multiple functions that are possibly carried out by different domains. The first PDZ domain interacts with itself and plays a significant role in Golgi stacking. The second PDZ domain interacts with GM130, a protein that plays a role in protein trafficking (34–37). Therefore, GRASP65 may also coordinate with GM130 through the second PDZ domain and function in membrane trafficking (38). The C-terminal SPR domain contains all the phosphorylations sites and thus is important for mitotic regulation of GRASP65 oligomerization and Golgi disassembly (6). In addition, different phosphorylation sites may be regulated by different pathway. Mutants that affect one function but not the others may serve as useful tools to further dissect the functions of the GRASP65 as well as the biological significance of Golgi stacking in mammalian cells.
One interesting observation concerns PDZ1 targeting to the Golgi membranes. GRASP65 is myristoylated, which serves as the anchor to the Golgi membranes. Deletion of this myristoylation site by mutating the N-terminal glycine to alanine (G2A) led to the loss of Golgi membrane targeting and thus the protein became cytosolic (6,39). GRASP65 function requires correct Golgi localization, as the G2A mutant of the protein did not exhibit significant effect on Golgi stacking in our study (6). GRASP65 and GM130 form a stable complex, which is directly targeted to the Golgi membranes right after their biosynthesis. It was originally proposed that GRASP65 targets GM130 to the Golgi membranes, as it contains a membrane anchor (39,40). However, GM130 is found on the Golgi membranes in GRASP65-depleted cells (Figure 5), consistent with a previous observation (19). Furthermore, PDZ1 (aa 1–112, tagged with GFP), which lacks the GM130 binding site, was largely concentrated on the Golgi (Figure 2E). Interestingly, the Golgi localization of PDZ1 did not depend on (the oligomerization with) the endogenous GRASP65, as it was found enriched on the Golgi membranes in cells in which the endogenous GRASP65 was depleted (Figure 5G). In general, one myristoylation modification may not be sufficient to target the protein to certain membranes. However, as GRASP65 forms oligomers, multiple myristic acid moieties on the protein complex may be sufficient for Golgi membrane localization. In this case, the interaction with GM130 may enhance and specify the Golgi targeting of GRASP65, but may not be absolutely required for its Golgi localization.
Another interesting observation concerns the different effects caused by the expression of GRASP65 WT protein versus its non-regulatable mutants. Expressing the non-regulatable GRASP65 mutants enhanced Golgi stacking during interphase and inhibited Golgi fragmentation during mitosis, while the expression of WT GRASP65 had little, if any, effect. These results suggest that GRASP65 may be regulated during both mitosis and interphase, consistent with the report that GRASP65 is phosphorylated by the mitogen-activated protein (MAP) kinase extracellular signal-regulated kinase (ERK) on S277, when the Golgi structure is remodeled for cell migration in a wound healing assay (26). From an evolutionary point of view, it is not known what controls the number of cisternae in each stack. For example, the budding yeast Saccharomyces cerevisiae has a GRASP65 homolog but lacks stacked Golgi under normal conditions; while in mammalian cells, the number of cisternae per stack is highly conserved in different cell types, at least in the cell lines and tissues that we have examined. In this study, the number of cisternae per stack (7.1 ± 0.2) in cells expressing the non-phosphorylatable GRASP65 was significantly increased compared with that of 5.6 ± 0.1 in control cells expressing GFP (Figure 2K), while depletion of the endogenous GRASP65 reduced the number to 4.0 ± 0.1, suggesting that studies on GRASP65 and its homolog protein, GRASP55 (11), may shed light on the molecular control of the cisterna number in the Golgi stacks. Future EM analysis will determine the identity of the additional cisternae in cells expressing GRASP65 mutants and the lost cisternae in GRASP65-depleted cells.
Although GRASP65 was originally discovered as a Golgi stacking factor and many studies support this idea, it has also been shown that GRASP65 depletion in HeLa cells led to Golgi ribbon unlinking (19). However, as knockdown of GRASP65 affects the integrity of the Golgi stacks, it is possible that Golgi ribbon unlinking in GRASP65-depleted cells was caused, at least in part, by disassembly of the Golgi stacks. In all the conditions examined in this study, we observed by both light and electron microscopy that the Golgi membranes were concentrated in the perinuclear region and thus we were unable to obtain evidence that GRASP65 depletion leads to scattered Golgi as previously reported (19), although the number of cisternae in the stacks was reduced (Figure 5), which is consistent with a different report (18). In addition, the observed phenotype was rescued by expression of rat GRASP65, which validated the specificity of the effect. A recent report showed that GRASP65, when targeted to the outer membrane of the mitochondria, caused mitochondria clustering (7). This result was used to support the role of GRASP65 in Golgi ribbon formation. However, it can also be interpreted as evidence for Golgi stacking, as it indicates oligomerization of the protein. In general, a protein involved in Golgi ribbon linking should localize (and possibly concentrate) on the Golgi rims where Golgi ribbon linking occurs, as suggested by the Golgi ribbon-linking protein GMAP210 (41). However, GRASP65 was discovered as a Golgi membrane protein that is accessible to the alkylating reagent, NEM, only when the Golgi membranes were disassembled by mitotic cytosol (4), suggesting that the protein is not concentrated on the rims of the Golgi cisternae. Instead, immuno-EM showed that GRASP65 is distributed on the body of the Golgi cisternae (9). Therefore, it is more likely that GRASP65 is primarily involved in Golgi stacking. Indeed, results from the expression and knockdown experiments in this study provided strong evidence that GRASP65 is directly involved in Golgi stacking. Whether GRASP65 plays a direct role in Golgi ribbon linking requires further investigation.
Golgi disassembly is related to cell cycle progression, as inhibition of Golgi fragmentation at the onset of mitosis delays mitotic entry (22,23,33). So far it is unclear what role GRASP65 plays in cell cycle control, nor the biological significance of mitotic Golgi disassembly. It has been suggested that Golgi dispersal during mitosis may aid in equal partitioning of Golgi into the new daughter cells (20). In our study, the inhibition of Golgi fragmentation during mitosis by expression of the non-regulatable mutants of GRASP65 did not lead to an unequal distribution of the Golgi membranes (Figure 6), suggesting that extensive Golgi fragmentation may not be required for accurate Golgi inheritance.
An alternative possible role for mitotic Golgi disassembly is to allow the separation of the duplicated centrosomes for mitotic spindle formation, as an intact Golgi complex may physically block centrosome separation during prometaphase. Consistent with this speculation, our results showed that inhibition of Golgi fragmentation by expression of non-regulatable GRASP65 mutants delayed mitotic entry, while disruption of the Golgi by BFA treatment diminished this effect. Previous studies showed that microinjecting recombinant GRASP65 or its antibodies, or expressing its C-terminal fragment, caused a delay in mitotic entry (23,24), possibly through modulation of the related mitotic kinases (25). In contrast to the previous studies, the expression of Golgi-localized non-regulatable GRASP65 does not affect mitotic kinase expression or subcellular localization, as these constructs are not substrates of the mitotic kinases. Therefore, our results suggest that the observed mitotic delay may occur through inhibition of mitotic Golgi disassembly, which is generally regulated through GRASP65 phosphorylation and deoligomerization. Furthermore, the mitotic index of cells expressing non-regulatable mutants of GRASP65 exhibited broader peaks. This may be because there are more mitotic cells when the mutants were expressed or these cells require a longer period to complete mitosis. In the latter case, an extended time period required for complete Golgi disassembly may contribute largely to the delay in cell cycle progression. Alternatively, the cells may need a longer time period to equally distribute the mitotic clusters (than completely disassembled Golgi vesicles) into the two daughter cells. In summary, our results show that GRASP65 stacks Golgi cisternae by oligomerization through its N-terminal PDZ1 domain. The disassembly of GRASP65 trans-oligomers caused by phosphorylation is essential for Golgi unstacking and fragmentation, which in turn regulates the cell cycle progression and cell growth. Future directions include determination of the effects caused by the expression of GRASP65 with single point mutation of the phosphorylation sites (e.g. S277), the dynamics of mitotic Golgi disassembly and postmitotic Golgi reassembly in cells expressing GRASP65 mutants, the signaling pathways that regulate Golgi membrane dynamics during the cell cycle, and especially the effect of Golgi stacking/unstacking on protein trafficking, modifications and secretion.
Materials and Methods
Reagents
All reagents were from Sigma Co, Roche or Calbiochem, unless otherwise stated. The following antibodies were used: monoclonal antibodies against Gos28 and GM130 (Transduction Laboratories), and α-tubulin (K. Gull); polyclonal antibodies against GFP (J. Seemann), GM130 (N73, J. Seemann), human GRASP65 (J. Seemann), rat GRASP65 (6) and human GRASP55 (Proteintech Group Inc).
Preparation of GRASP65 fusion proteins
GRASP65 cDNAs (4–6) were cloned into pMAL-c2X (New England Biolabs), pGEX (GE Healthcare) or pET30a (Novagen) vectors by polymerase chain reaction (PCR) or restriction digestion and confirmed by DNA sequencing. GRASP65 point mutations were introduced using the QuikChange mutagenesis kit (Stratagene) and confirmed by DNA sequencing. Proteins were expressed in BL21-CodonPlus(DE3)RILP bacteria and purified on amylose (New England Biolabs), nickel (Qiagen) or glutathione Sepharose (GE Healthcare) beads.
GRASP65 dimerization and oligomerization
To test the oligomerization of GRASP65 fragments, separately expressed and purified MBP- or GST-tagged GRASP65 fragments were mixed and incubated in the presence of an ATP-regenerating system (10 mm creatine phosphate, 0.1 mm ATP, 20 μg/mL creatine kinase, 20 μg/mL cytochalasin B) at 37°C for 1 h. The protein complex was isolated using glutathione or amylose beads and analyzed by western blotting.
For sedimentation analysis, purified MBP-tagged PDZ1 (120 μg) was incubated at 37°C for 1 h in gradient buffer (25 mm HEPES–KOH, pH 7.4, 150 mm KCl, 5 mm MgCl2, 1 mm DTT) in a final volume of 150 μL. Top-loaded glycerol gradients (10–35% w/v) were centrifuged for 4 h at 260 000 × g in a VTI65.1 rotor and portions of the 1 mL fractions were analyzed by western blotting.
For non-denaturing electrophoresis, purified GST-tagged PDZ1 or PDZ2 domain was incubated with KHM buffer [20 mm HEPES–KOH, pH 7.4, 0.2 m sucrose, 60 mm KCl, 5 mm Mg(OAc)2, 2 mm ATP, 1 mm GTP, 1 mm glutathione with protease inhibitors] at 37°C for 1 h, mixed with non-denaturing sample buffer and loaded onto a 4–12% NuPAGE Novex Bis-Tris mini gel (Invitrogen) (11,42). After electrophoresis, the gel was stained with Coomassie Blue. Major bands were excised from the gel and further analyzed by denaturing electrophoresis and western blotting.
For gel filtration, His-tagged PDZ1 was incubated with an ATP regeneration system at 37°C with KHM buffer for 1 h and analyzed by a Sephacryl S-300 column. Fractions of 2 mL were collected at a flow rate of 0.5 mL/min. Proteins in the peak fractions were confirmed by western blotting.
Bead aggregation assay was performed as previously described (5,6). Briefly, His-tagged recombinant full-length GRASP65 and PDZ1 were subjected to centrifugation at 21 130 g (15 000 rpm) for 30 min in a tabletop centrifuge at 4°C. The proteins in the supernatant were then cross-linked to M500 magnetic beads (Dynal Biotech). The beads were incubated for 60 min at 37°C with either BSA or IC prepared from HeLa S3 cells (5), in KHM buffer to allow the aggregation. In some experiments, an excess amount (0.5 μg/μL) of purified His-PDZ1 was included in the reaction. For quantitation of the beads, 15 random phase contrast digital images of each reaction were captured with a 20× lens and a Spot Slider2 Camera (Diagnostic Instruments). Images were analyzed with the MATLAB7.4 software to determine the surface area of objects, which was used to calculate the number of beads in the clusters. Aggregates were defined as those with ≥6 beads. For large aggregates, only visible surface beads were counted; therefore, the number of beads in these aggregates was underestimated. Results were expressed as the mean ± SEM from three independent experiments; statistical significance was assessed by Student's t-test.
Cell culture, siRNA and establishment of stable cell lines
HeLa cells were transfected with Stealth siRNA oligos (Invitrogen) prepared according to the reported sequence for human GRASP65 (5′-CCTGAAGGCACTACTGAAAGCCAAT-3′) (18) using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instructions. Control non-specific siRNA oligos were purchased from Ambion. Assays were performed 96 h after transfection. The knockdown efficiency of endogenous GRASP65 was analyzed using the ImageJ software from western blotting results and normalized with tubulin as a loading control.
To establish stable cell lines, rtTAm2 HeLa cells were infected with retroviral particles encoding indicated exogenous genes cloned in the pRevTRE2 vector. Wild type rat GRASP65, the mG mutant in which seven phosphorylation sites were mutated to alanines (S216A/S217A/T220A/T224A/S277A/S367A/S376A) (6) and the GRASP domain (aa 1–201) were first cloned into the pEGFP-N2 vector via the EcoRI and ApaI sites. GFP (enhanced green fluorescent protein), GRASP65-GFP, GRASP65 mG-GFP and GRASP domain-GFP cDNA were then excised by Hind III and NotI and religated into the pRevTRE2 vector. For GRASP65 PDZ1 (aa 1–112)-GFP, GFP cDNA was first cloned into the pRevTRE2 vector through the HindIII and NotI sites. The PDZ1 coding sequence was inserted in frame into the BamHI and HindIII sites at the N-terminus of GFP. Procedures for viral preparation, infection and cell selection were previously described (28). GFP-positive cells were enriched by flow cytometry. The expression of exogenous proteins was induced with 1 μg/mL doxycycline for 72 h. For the gene replacement experiment, GRASP65 or GFP cell lines were transfected with GRASP65 siRNA for 24 h followed by induction with doxycycline for 72 h.
Microscopy and quantitation
Immunofluorescence microscopy and collection of mitotic cells were previously reported (14). Pictures were taken with a Leica SP5 confocal laser scanning microscope using a 100× oil lens. For metaphase cells, images were captured using fixed parameters with 0.3-μm intervals at the z-axis and then processed for maximum value projection. Images were analyzed using the NIH ImageJ software, and a threshold was introduced to exclude cytoplasmic staining and the Golgi haze (43). All pixels above this threshold value were counted as Golgi remnants and summed up as the fluorescence intensity in Golgi clusters. The percentage of fluorescence intensity in Golgi clusters was calculated versus the total fluorescence intensity in the cell above background. Cells in cytokinesis were defined as two daughter cells linked with a thin connection (contractile ring); chromosomes are concentrated in the center of each daughter cell although the DNA may not decondensed; the Golgi has started to reform but the process has not completed. Fluorescence intensity for indicated Golgi proteins in each daughter cell (above the background of the image) was measured; the percentage of the Golgi membrane inherited by one daughter cell was calculated against the total fluorescence intensity of the cell pair. More than 10 cells or cell pairs were quantified for each cell line in each experiment. The average (mean ± SEM) percentage of fluorescence intensity in the daughter cells with relative lower fluorescence level compared with their pairs was calculated and presented in Figure 6F.
For EM analysis, Golgi stacks and clusters were identified using morphological criteria and quantified using standard stereological techniques (6). For interphase cells, the profiles had to contain a nuclear profile with an intact nuclear envelope; only the obvious stacked structures with three or more cisternae were counted. For mitotic cells, the profile had to contain one or more profiles of condensed chromosomes and lack a nuclear envelope; very often multiple condensed chromosomes were aligned in the center of the cell. A low magnification (normally 1600×) image and serial images of higher magnification (normally 11 000×) were taken to cover the entire cell. About 20 cells were quantified. To quantify the cisternae number in each mitotic Golgi cluster, a cisterna was defined as a membrane-bound structure in the Golgi cluster whose length is at least 4× its width, normally 20–30 nm in width and longer than 150 nm (44).
Mitotic index and growth rate analysis
The mitotic index analysis of cells after double thymidine block and release was performed as previously described (22). Briefly, cells of about 40% confluence were incubated in growth medium with 2 mm thymidine for 24 h, then washed with PBS and incubated with growth medium for 14 h. Cells were then maintained in growth medium with thymidine for an additional 20 h before the final release. At different time-points after release, cells were assayed for mitotic index by tallying the number of cells displaying clear mitotic (rounded perimeter, discernible mitotic plate) and interphase characteristics using an inverted microscope and a 20× phase contrast objective (22). About 300 cells were counted at each time-point within 5 min. For the stable cell lines, 1 μg/mL doxycycline was added at the starting time of the first thymidine block. In some experiments, 5 μg/mL BFA from a 5 mg/mL stock, or an equal volume of ethanol, was added 4 h after the final thymidine release to disperse the Golgi structure. The experiment was repeated at least 3× and the results were expressed as the mean ± SEM.
To analyze the growth rate of the stable cell lines, 5000 cells were added into each well of 24-well plates and incubated in growth medium with 1 μg/mL doxycycline to induce the expression of the exogenous proteins. Two days later, cell numbers were measured every 24 h for 4 days by crystal violet staining (45). Briefly, cells were washed with prewarmed PBS, fixed in 4% formaldehyde for 30 min and stained with 0.5% crystal violet (in 30% methanol) for 15 min. After extensively washing with H2O, DNA-bound dye was extracted with 1 mL of 10% acetic acid and measured for optical density (OD) at 590 nm. The cell numbers at different days were normalized to the cell number from the first measurement. The results were shown as the mean ± SEM, n = 3.
Supplementary Material
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Expression of non-regulatable GRASP65 mutants enhances Golgi stacking in interphase cells. Bar charts of Figure 2K indicating the actual frequency of each value of number of cisternae per stack in HeLa cells expressing GFP or indicated GFP-tagged GRASP65 constructs. Note that the expression of non-regulatable GRASP65 mutants, but not the wild-type protein, increased the number of cisternae in the stacks.
Figure S2: Knockdown of GRASP65 reduces the number of cisternae per stack, which can be rescued by expression of exogenous GRASP65. A) Bar charts of Figure 5Q showing the actual frequency of Golgi cisternal numbers in the stacks in HeLa cells transfected with control or GRASP65 siRNA. Note the shift of the bars to lower numbers when GRASP65 was depleted (lower panel) compared with the control siRNA-transfected cells (upper panel). B) Bar charts of Figure 5R showing the actual frequency of Golgi cisternal numbers in the stacks in HeLa cells in which endogenous GRASP65 was replaced by GFP, rat GRASP65 or its mutants, as indicated. Note that the expression of GRASP65 and its mutant increased the number of cisternae in the stacks compared to the GFP cell line.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgments
We thank Joachim Seemann and Graham Warren for antibodies and cDNA constructs, Blanche Schwappach for the retroviral vector and the rtTA HeLa cell line, Hana Popelkova for technical help with gel filtration, Majan Varedi and Xiaoxia Lin for developing the Matlab program to quantify beads in aggregates and Yi Xiang and other members of the Wang Lab for suggestions and reagents. This work was supported by the Pardee Cancer Research Foundation, the National Institute of Health (GM087364), the American Cancer Society (RGS-09-278-01-CSM), the University of Michigan Rackham Faculty Research Grant, the NIH-funded Michigan Alzheimer's Disease Research Center (P50 AG08761) and an anonymous donation to Y. W.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucocq JM, Warren G. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J. 1987;6:3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186:41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ, Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeffer SR. Constructing a Golgi complex. J Cell Biol. 2001;155:873–875. doi: 10.1083/jcb.200109095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle regulated Golgi disassembly and reassembly in a cell free system. Nat Protocols. 2010;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi cisternal unstacking stimulates COPI vesicle budding and protein transport. PLoS ONE. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang Y, Seemann J, Bisel B, Punthambaker S, Wang Y. Active ADP-ribosylation factor-1 (ARF1) is required for mitotic Golgi fragmentation. J Biol Chem. 2007;282:21829–21837. doi: 10.1074/jbc.M611716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y. Golgi apparatus inheritance. In: Mironov A, Pavelka M, Luini A, editors. The Golgi Apparatus State of the art 110 Years After Camillo Golgi's Discovery. Springer-Verlag Gmbh; Wien-New York: 2008. pp. 580–607. [Google Scholar]
- 16.Behnia R, Barr FA, Flanagan JJ, Barlowe C, Munro S. The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J Cell Biol. 2007;176:255–261. doi: 10.1083/jcb.200607151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondylis V, Spoorendonk KM, Rabouille C. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol Biol Cell. 2005;16:4061–4072. doi: 10.1091/mbc.E04-10-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 20.Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 21.Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimura S, Yoshioka K, Barr FA, Lowe M, Nakayama K, Ohkuma S, Nakamura N. Convergence of cell cycle regulation and growth factor signals on GRASP65. J Biol Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- 25.Preisinger C, Korner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, Yoshimura S, Nakamura N, Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan W, Husler P, Klump H, Erhardt J, Sluis-Cremer N, Dirr H. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: a detoxification enzyme and fusion-protein affinity tag. Protein Sci. 1997;6:399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heusser K, Yuan H, Neagoe I, Tarasov AI, Ashcroft FM, Schwappach B. Scavenging of 14-3-3 proteins reveals their involvement in the cell-surface transport of ATP-sensitive K+ channels. J Cell Sci. 2006;119:4353–4363. doi: 10.1242/jcs.03196. [DOI] [PubMed] [Google Scholar]
- 29.Vielemeyer O, Yuan H, Moutel S, Saint-Fort R, Tang D, Nizak C, Goud B, Wang Y, Perez F. Direct selection of monoclonal phosphospecific antibodies without prior phosphoamino acid mapping. J Biol Chem. 2009;284:20791–20795. doi: 10.1074/jbc.M109.008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 31.Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchiyama K, Jokitalo E, Lindman M, Jackman M, Kano F, Murata M, Zhang X, Kondo H. The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J Cell Biol. 2003;161:1067–1079. doi: 10.1083/jcb.200303048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colanzi A, Carcedo CH, Persico A, Cericola C, Turacchio G, Bonazzi M, Luini A, Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seemann J, Jokitalo EJ, Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle Cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 36.Weide T, Bayer M, Koster M, Siebrasse JP, Peters R, Barnekow A. The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2001;2:336–341. doi: 10.1093/embo-reports/kve065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-golgi traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- 38.D'Angelo G, Prencipe L, Iodice L, Beznoussenko G, Savarese M, Marra P, Di Tullio G, Martire G, De Matteis MA, Bonatti S. GRASP65 and GRASP55 sequentially promote the transport of C-terminal valine bearing cargoes to and through the golgi complex. J Biol Chem. 2009;284:34849–34860. doi: 10.1074/jbc.M109.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura SI, Nakamura N, Barr FA, Misumi Y, Ikehara Y, Ohno H, Sakaguchi M, Mihara K. Direct targeting of cis-Golgi matrix proteins to the Golgi apparatus. J Cell Sci. 2001;114:4105–4115. doi: 10.1242/jcs.114.22.4105. [DOI] [PubMed] [Google Scholar]
- 41.Cardenas J, Rivero S, Goud B, Bornens M, Rios RM. Golgi localisation of GMAP210 requires two distinct cis-membrane binding mechanisms. BMC Biol. 2009;7:56. doi: 10.1186/1741-7007-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 43.Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Lucocq JM, Berger EG, Warren G. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J Cell Biol. 1989;109:463–474. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz M, Oren YS, Bester AC, Rahat A, Sfez R, Yitzchaik S, de Villartay JP, Kerem B. Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS One. 2009;4:e4516. doi: 10.1371/journal.pone.0004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Expression of non-regulatable GRASP65 mutants enhances Golgi stacking in interphase cells. Bar charts of Figure 2K indicating the actual frequency of each value of number of cisternae per stack in HeLa cells expressing GFP or indicated GFP-tagged GRASP65 constructs. Note that the expression of non-regulatable GRASP65 mutants, but not the wild-type protein, increased the number of cisternae in the stacks.
Figure S2: Knockdown of GRASP65 reduces the number of cisternae per stack, which can be rescued by expression of exogenous GRASP65. A) Bar charts of Figure 5Q showing the actual frequency of Golgi cisternal numbers in the stacks in HeLa cells transfected with control or GRASP65 siRNA. Note the shift of the bars to lower numbers when GRASP65 was depleted (lower panel) compared with the control siRNA-transfected cells (upper panel). B) Bar charts of Figure 5R showing the actual frequency of Golgi cisternal numbers in the stacks in HeLa cells in which endogenous GRASP65 was replaced by GFP, rat GRASP65 or its mutants, as indicated. Note that the expression of GRASP65 and its mutant increased the number of cisternae in the stacks compared to the GFP cell line.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.