This year marks the 25th anniversary of the discovery of NF-κB, a key regulatory molecule in the immune system and many other physiological processes. Hayden and Ghosh provide a comprehensive review that assesses the current knowledge of the activation and control of NF-κB, which has served as a paradigm for inducible transcription factors and how environmental signaling influences gene expression.
Keywords: NF-κB, IKK, signaling, inflammation, cancer
Abstract
The ability to sense and adjust to the environment is crucial to life. For multicellular organisms, the ability to respond to external changes is essential not only for survival but also for normal development and physiology. Although signaling events can directly modify cellular function, typically signaling acts to alter transcriptional responses to generate both transient and sustained changes. Rapid, but transient, changes in gene expression are mediated by inducible transcription factors such as NF-κB. For the past 25 years, NF-κB has served as a paradigm for inducible transcription factors and has provided numerous insights into how signaling events influence gene expression and physiology. Since its discovery as a regulator of expression of the κ light chain gene in B cells, research on NF-κB continues to yield new insights into fundamental cellular processes. Advances in understanding the mechanisms that regulate NF-κB have been accompanied by progress in elucidating the biological significance of this transcription factor in various physiological processes. NF-κB likely plays the most prominent role in the development and function of the immune system and, not surprisingly, when dysregulated, contributes to the pathophysiology of inflammatory disease. As our appreciation of the fundamental role of inflammation in disease pathogenesis has increased, so too has the importance of NF-κB as a key regulatory molecule gained progressively greater significance. However, despite the tremendous progress that has been made in understanding the regulation of NF-κB, there is much that remains to be understood. In this review, we highlight both the progress that has been made and the fundamental questions that remain unanswered after 25 years of study.
Inducible regulation of gene expression allows organisms to adapt to environmental, mechanical, chemical, and microbiological stresses. Owing to its amenability to experimentation and its importance in disease, NF-κB has, for the past 25 years, served as a model for inducible transcription factors. NF-κB plays its most important and evolutionarily conserved role in the immune system, and much of our understanding of NF-κB is derived from the quest to decipher and manipulate the immune response. However, besides its role in the immune system, NF-κB also acts broadly to influence gene expression events that impact cell survival, differentiation, and proliferation. As a result of such broad effects on physiology, the dysregulation of NF-κB can lead to severe consequences (Courtois and Gilmore 2006; Karin 2006), including numerous diseases.
Transcriptional programs regulated by NF-κB are essential to the development and maintenance of the immune system (Hayden and Ghosh 2011), skeletal system (Novack 2011), and epithelium (Wullaert et al. 2011). In these settings, the NF-κB pathway contributes to the control of cell survival, differentiation, and proliferation. NF-κB does the same in the various diseases with which aberrant activation has been associated: cancer, autoimmune diseases, neurodegenerative diseases, cardiovascular disease, diabetes, and many more. The contribution of NF-κB to disease is perhaps most easily understood in the context of chronic inflammatory and autoimmune diseases wherein proinflammatory cytokines drive activation of NF-κB and the activation of NF-κB drives proinflammatory cytokine production (Lawrence 2009), or in malignancies in which NF-κB is intrinsically activated, resulting in tumor survival by driving up-regulation of proliferative and anti-apoptotic transcripts. As a multifaceted regulator of fundamental aspects of cell survival and function acting downstream from numerous cell signaling pathways, NF-κB also has the capacity to link physiology to pathology. Thus, NF-κB is now appreciated to form an etiological mechanism tying obesity to inflammation (Baker et al. 2011), and inflammation to malignancy (Ben-Neriah and Karin 2011) and metabolic diseases (Baker et al. 2011; Donath and Shoelson 2011). The role of NF-κB in disease is not always the result of inflammation gone awry. A host of diseases and disease susceptibilities result from germline and/or acquired mutations in components of the NF-κB pathway (Courtois and Israel 2011).
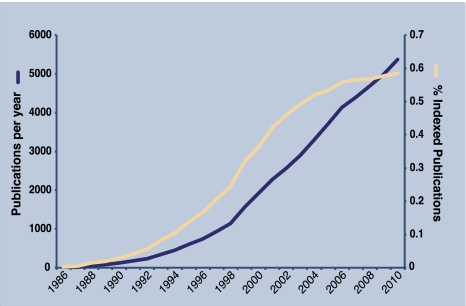
The involvement of NF-κB in so many facets of biology has resulted in a vast number of publications on this transcription factor (Fig. 1). While a limited body of work has produced the watershed events that comprise the past 25 years of NF-κB research (Supplemental Material), our current understanding of NF-κB is informed by a broad swath of literature resulting from investigations in diverse areas of the biomedical sciences. Searching PubMed with the search term “NF-κB” alone yields nearly 40,000 hits, accumulating at a rate of >3500 studies published annually. As a result, NF-κB-related publications astoundingly account for almost one-half of 1% of all publications: One out of every 200 publications indexed in PubMed relates to this one transcription factor family. The progressively expanding NF-κB literature has made writing a comprehensive review on NF-κB an untenable undertaking: One must choose areas of focus in any review dealing with such a broad subject matter. Therefore, in the current review, we discuss the most fundamental aspects of NF-κB regulation, focusing on those areas that we believe are undergoing significant progress or in which there are significant outstanding questions. We organized this review around the core components of the NF-κB pathway; namely, the IκB (inhibitor of κB) kinase (IKK) complex, the inhibitory IκB proteins, and the transcription factor NF-κB itself. Thus, the first section deals with insights and outstanding questions regarding activation of IKK, the second deals with the emerging functional complexity of IκB proteins, and the third deals with the mechanisms of transcriptional regulation by NF-κB. It goes without saying that in covering these broad areas, many primary references are not cited, and for this we refer readers to important previous reviews and collections of reviews (Lenardo and Baltimore 1989; Beg and Baldwin 1993; Baeuerle and Henkel 1994; Siebenlist et al. 1994; Verma et al. 1995; Barnes and Karin 1997; Ghosh et al. 1998; Rothwarf and Karin 1999; Karin and Delhase 2000; Ghosh and Karin 2002; Hayden and Ghosh 2004, 2008; Gilmore 2006; Karin 2009; Sun and Liu 2011).
Figure 1.
Twenty-five years of NF-κB literature. Graph indicates the total number of publications identified in PubMed using the keywords NF-κB, Rel, or IKK for each year since 1986 (shown in the left axis). Also graphed are the total publications identified with the above keywords as a percentage of all PubMed indexed publications in the same calendar year (shown in the right axis).
Overview of NF-κB
NF-κB activation is governed by a number of positive and negative regulatory elements. In the “resting” state, NF-κB dimers are held inactive in the cytoplasm through association with IκB proteins. Inducing stimuli trigger activation of the IκB kinase complex, leading to phosphorylation, ubiquitination, and degradation of IκB proteins. Released NF-κB dimers translocate to the nucleus, bind specific DNA sequences, and promote transcription of target genes. Thus, the core elements of the NF-κB pathway are the IKK complex, IκB proteins, and NF-κB dimers. Consequently, research into the regulation of NF-κB has focused on the mechanisms that activate the IKK complex, the inhibition of NF-κB by IκB proteins, and the capacity of NF-κB family members to bind to and promote transcription from the promoters of selected target genes. Inactivation of the pathway requires deactivation of the IKK complex, resynthesis of IκB proteins, and displacement of NF-κB dimers from both DNA and transcriptional coactivators. Therefore, to understand these processes, we must first introduce the families of proteins that together comprise the NF-κB signaling pathway.
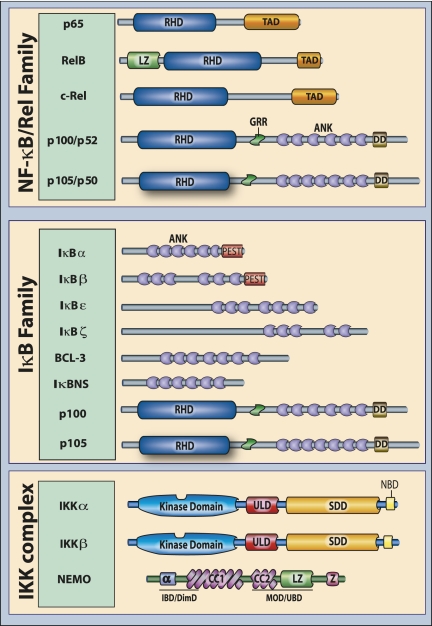
There are five NF-κB family members in mammals: RelA/p65, RelB, c-Rel, p50 (NF-κB1), and p52 (NF-κB2) (Fig. 2). NF-κB proteins bind to κB sites as dimers, either homodimers or heterodimers, and can exert both positive and negative effects on target gene transcription. NF-κB proteins are characterized by the presence of an N-terminal Rel homology domain (RHD). Functional analyses and crystal structures of NF-κB dimers bound to κB sites have shown that it is the RHD that makes contact with DNA and supports subunit dimerization. Only p65, c-Rel, and RelB possess C-terminal transactivation domains (TADs) that confer the ability to initiate transcription. Although p52 and p50 lack TADs, they can positively regulate transcription through heterodimerization with TAD-containing NF-κB subunits or interaction with non-Rel proteins that have transactivating capability. Alternatively, p50 and p52 homodimers can negatively regulate transcription by competing with TAD-containing dimers for binding to κB sites. These p50 and p52 dimers may also constitutively occupy some κB sites and thus enforce an activation threshold for certain NF-κB target genes.
Figure 2.
Components of the NF-κB pathway. The mammalian Rel (NF-κB) protein family consists of five members: p65 (RelA), RelB, c-Rel (Rel), and the precursor proteins p100 (NF-κB2) and p105 (NF-κB1), the latter giving rise to p52 and p50, respectively. The IκB family consists of eight bona fide members, IκBα, IκBβ, IκBɛ, IκBζ, BCL-3, IκBNS, p100, and p105, which are typified by the presence of multiple ankyrin repeat domains. Not pictured is the potential IκB family member IκBη, which is discussed in the text. The IKK complex consists of IKKα (IKK1 or CHUK), IKKβ (IKK2), and NEMO (IKKγ). Relevant domains typifying each protein family are indicated. (ANK) Ankyrin repeat domain; (DD) death domain; (RHD) REL homology domain; (TAD) transactivation domain; (LZ) leucine zipper domain; (GRR) glycine-rich region; (SDD) scaffolding and dimerization domain; (ULD) ubiquitin-like domain; (Z) zinc finger domain; (CC) coiled-coil domain; (NBD) NEMO-binding domain; (α) α-helical domain; (IBD/DimD) IKK-binding domain/dimerization domain; (MOD/UBD) minimal oligomerization domain/ubiquitin-binding domain; (PEST) proline-rich, glutamic acid-rich, serine-rich, and threonine-rich.
A hallmark of the NF-κB pathway is its regulation by IκB proteins. IκB proteins IκBα, IκBβ, IκBɛ, IκBζ, BCL-3 (B-cell lymphoma 3), and IκBns and the precursor proteins p100 (NF-κB2) and p105 (NF-κB1) are defined by the presence of multiple ankyrin repeat domains (Fig. 2). Activation of NF-κB is achieved through phosphorylation of IκBs on conserved serine residues, so-called destruction box serine residues (DSGXXS), leading to recognition by βTrCP proteins. Recognition of the phosphorylated destruction box induces K48-linked polyubiquitination by the Skp1–Culin–Roc1/Rbx1/Hrt-1–F-box (SCF or SCRF) family of E3 ligases acting coordinately with the E2 enzyme UbcH5. The prototypical and most extensively studied member of the family is IκBα. IκBα is rapidly degraded during activation of canonical NF-κB signaling pathways, leading to the release of multiple NF-κB dimers, although the p65:p50 heterodimer is considered the primary target of IκBα. Although IκBα:p65:p50 complexes are capable of constant shuttling between the nucleus and the cytoplasm, masking of the p65 nuclear localization signal (NLS), combined with the effect of IκBα nuclear export signal, results in steady-state cytoplasmic localization of NF-κB dimers, thus preventing DNA binding. Proteasomal degradation of IκBα thus favors nuclear localization of NF-κB.
Following removal of IκB from the NF-κB complex, NF-κB dimers are able to accumulate in the nucleus and bind to DNA κB sites with the sequence 5′-GGGRNWYYCC-3′ (where N is any base, R is purine, W is adenine or thymine, and Y ispyrimidine) in promoters and enhancers of target genes. The degenerate nature of the κB site sequence; the ability of individual NF-κB subunits to form homodimers and heterodimers, heterotypic interactions with other transcription factors; and the regulation of transcriptional activity by post-translational modifications (PTMs) targeting NF-κB subunits allow both positive and negative regulation of transcription of a wide variety of target genes (Pahl 1999; Hayden and Ghosh 2004). Termination of the transcriptional response depends on not only resynthesis of typical IκB proteins, but also removal of active NF-κB dimers from the DNA.
The NF-κB signaling pathways have been broadly classified into two types: canonical and noncanonical. The canonical, or classical, pathway is representative of the general scheme of how NF-κB is regulated. Upon recognition of ligand, cytokine receptors such as the TNF receptor (TNFR) (Fig. 3) and IL-1 receptor (IL-1R), pattern recognition receptors (PRRs) such as Toll-like receptor 4 (TLR4) (Fig. 4), and antigen receptors (Fig. 5), among many other stimuli, trigger signaling cascades that culminate in the activation of IKKβ (also known as IKK2). IKKβ exists in a complex with the closely related kinase IKKα (also known as IKK1) and the regulatory protein NEMO (also known as IKKγ). Activated IKKβ phosphorylates IκB proteins such as IκBα. The noncanonical, or alternative, NF-κB pathway is induced by specific members of the TNF cytokine family, including the CD40 ligand (Fig. 6), BAFF, and lymphotoxin-β (Sun 2011). In contrast to the canonical pathway, the noncanonical pathway depends on IKKα and is independent of NEMO. IKKα activation by these cytokines leads to phosphorylation of p100 and the generation of p52/RelB complexes. Given that IKKα can contribute to some canonical signaling pathways and that activation of canonical pathways augments noncanonical signaling through the induction of p100 expression, the most useful distinction between canonical and noncanonical signaling remains the dependence of the signaling pathway on NEMO. In addition to phosphorylation of IκB proteins, it is important to note that as the key enzymatic constituents of the NF-κB pathway, IKKα and IKKβ can mediate cross-talk with additional signaling pathways—including the p53, MAP kinase (MAPK), and IRF pathways—and directly regulate aspects of the transcriptional responses (Oeckinghaus et al. 2011).
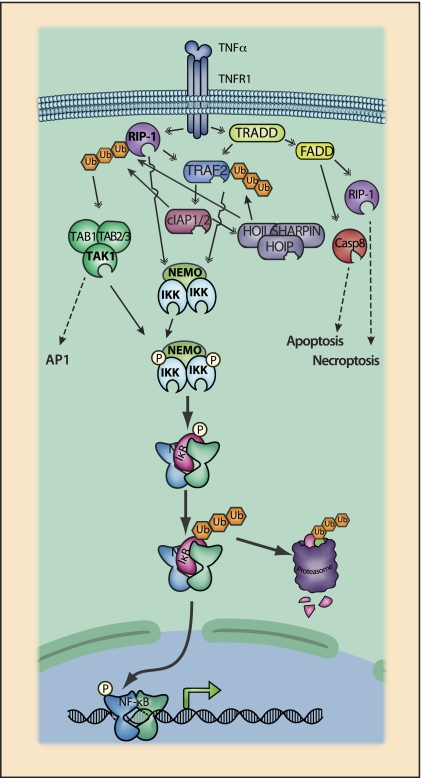
Figure 3.
TNFR1 signaling to NF-κB. TNFR1 activates multiple signaling pathways, including NF-κB, AP-1, and the apoptosis and necroptosis cell death pathways. TNF-induced activation of NF-κB is mediated by a series of intermediary adapters. The cytoplasmic tail of TNFR1 exhibits several protein-binding domains, most notably a death domain (DD) that mediates signaling events following TNF binding. Signaling events are partially organized by subcellular compartmentalization of receptor complexes, and the TNFR cytoplasmic tail contains adapter protein-binding motifs that direct trafficking following TNF binding (Schutze and Schneider-Brachert 2009). Upon ligand binding, the DD of TNFR1 binds TRADD (TNFR-associated protein with a DD) and the DD-containing kinase RIP1 (Box 3). Mechanisms coordinating binding between the DDs of TRADD, RIP1, and TNFR1 are not fully established. Nevertheless, it is clear that each of these DD-containing proteins are capable of binding to other DD-containing proteins (Wajant and Scheurich 2011). TRADD also provides an assembly platform for recruitment of another DD adapter protein, FADD (Fas receptor-associated DD). TNFR1 lacks a TRAF interaction motif, and TRAF recruitment is thus also dependent on TRADD, which has a TRAF-binding domain. Although RIP1 also has a TRAF-binding domain and may contribute to TRAF2 recruitment under some circumstances (Pobezinskaya et al. 2008), it is generally thought that TRAF2 recruitment is primarily dependent on TRADD (Chen et al. 2008; Ermolaeva et al. 2008; Pobezinskaya et al. 2008). TRAF2 recruits cIAP1 and cIAP2 (Box 2), which are essential for IKK activation (Mahoney et al. 2008; Varfolomeev et al. 2007; Vince et al. 2007). The cIAPs can function as E3 ubiquitin ligases and are also responsible for the recruitment of the linear ubiquitin chain assembly complex, which is required for efficient activation of IKK and JNK pathways (LUBAC [linear ubiquitin assembly complex]) (Box 4; Haas et al. 2009; Rahighi et al. 2009; Tokunaga et al. 2009, 2011; Gerlach et al. 2011; Ikeda et al. 2011). RIP1 and TRAF2 cooperate in the recruitment of the TAK1 and IKK kinase complexes, leading to IKK activation and activation of NF-κB.
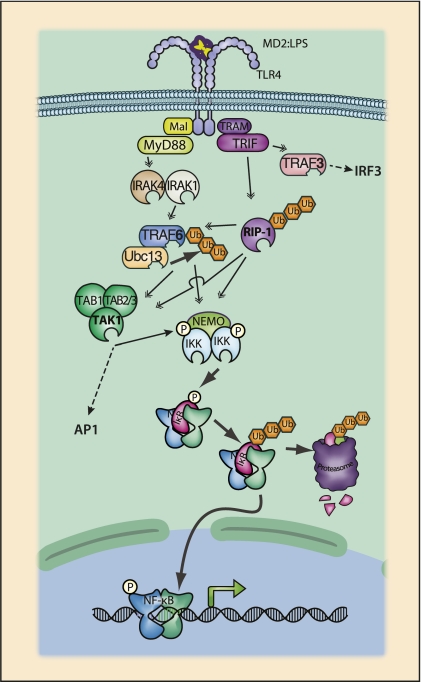
Figure 4.
TLR4 signaling to NF-κB. Both TLR and IL-1R receptor families are defined by the presence of cytoplasmic TIR (Toll IL-1R) domains. Upon ligand binding, TIR domains mediate the recruitment of TIR-containing adapter proteins such as MyD88, TRIF, Mal, or TRAM (Yamamoto et al. 2004a). TLR4, which responds to bacterial lipopolysaccharide, has a complex bifurcating signaling scheme. MyD88 is the prototypical TIR adapter and is used in all characterized TLR signaling pathways, with the exception of TLR3. TLR4 recognizes LPS bound to either LPS-binding protein or MD2, and signaling is also dependent on the glycoprotein CD14. Formation of a complex between LPS and TLR4:MD2:CD14 results in the homodimerization of TLR4 and recruitment of the TIR-containing adapters Mal and TRAM. Mal serves as an adapter to recruit MyD88 to TLR4, while TRAM is an adapter between TLR4 and TRIF. Following recruitment to the receptor complex, dimerized MyD88 recruits IL-1R-associated kinases-4 (IRAK-4) through the DD of MyD88 and IRAK-4. IRAK-4 recruits IRAK-1, and the IRAK-1:IRAK-4 complex is responsible for binding to TRAF6. TRAF6, in turn, recruits the TAK1 and IKK complexes, leading to activation of NF-κB. TRIF, recruited by TRAM, predominantly activates the interferon pathway through an N-terminal TRAF3-binding motif. TRAF3 recruits the IKK family members IKKɛ and TBK1, which phosphorylate IRF3, leading to the induction of type I interferons. TRIF may also induce NF-κB activation through a C-terminal RHIM (RIP homology interaction motif) domain capable of recruiting RIP1 and also the IKK complex (Cusson-Hermance et al. 2005).
Figure 5.
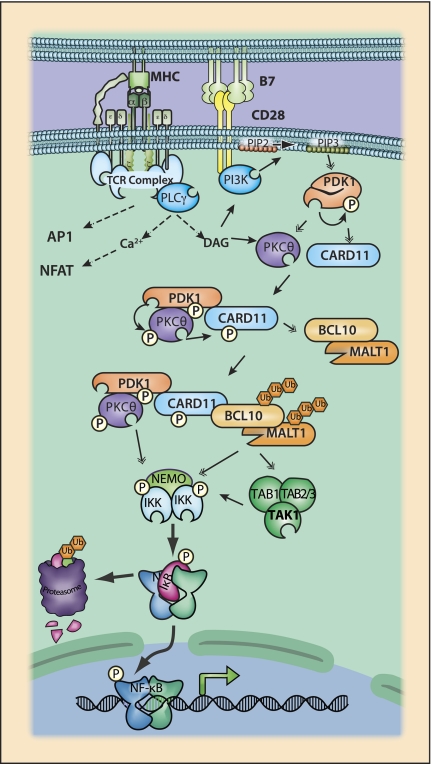
T-cell receptor (TCR) signaling to NF-κB. TCR-induced NF-κB activation requires ligation of both the TCR and the associated coreceptor CD28. Signaling involves the formation of large supramolecular clusters at the interface of the T-cell and antigen-presenting cell (APC). In vivo TCR ligation occurs upon presentation of cognate antigen in MHC-I or MHC-II for CD8 and CD4 T cells, respectively, by activated APCs expressing costimulatory molecules such as B7.1 or B7.2. Antigen:MHC complexes are engaged by T cells expressing somatically encoded antigen-specific TCRs. TCR:MHC binding is augmented by CD8:MHC-I or CD4:MHC-II interactions. ITAM motifs on CD3ζ are phosphorylated, leading to recruitment and activation of the ZAP70 kinase, which in turn activates PLCγ. Active PLCγ leads to DAG production. Coreceptor ligation involves recognition of B7 molecules on the APC surface by the TCR coreceptor CD28. CD28 ligation results in activation of PI3K and phosphorylation of PIP2 to PIP3. PDK1 binds PIP3 and undergoes autophosphorylation, revealing a PKC-binding site. DAG, in conjunction with PDK1-mediated recruitment and phosphorylation, leads to activation of the atypical PKC family member PKCθ. PKCθ (Sun et al. 2000) and perhaps also the related family members PKCɛ and PKCη (Quann et al. 2011) are selectively required for the activation of NF-κB downstream from TCR. In addition to activating PKCθ, PDK1 also facilitates the formation of a signaling complex (Lee et al. 2005; Park et al. 2009) in which PKCθ and potentially other kinases phosphorylate CARD11 (Matsumoto et al. 2005; Sommer et al. 2005; Shinohara et al. 2007). Phosphorylation of CARD11 results in a structural change that allows the formation of the CARD11, BCL10, and MALT1 (CBM) (Ruefli-Brasse et al. 2003; Ruland et al. 2003) signaling complex. Recruitment of the IKK complex to the CBM through PKCθ (Lee et al. 2005) or ubiquitinated MALT1 (Oeckinghaus et al. 2007), together with recruitment of the TAK1 complex, perhaps through TRAF6-mediated ubiquitination of BCL10 (Sun et al. 2004), results in the activation of IKK and the NF-κB signaling pathway.
Figure 6.
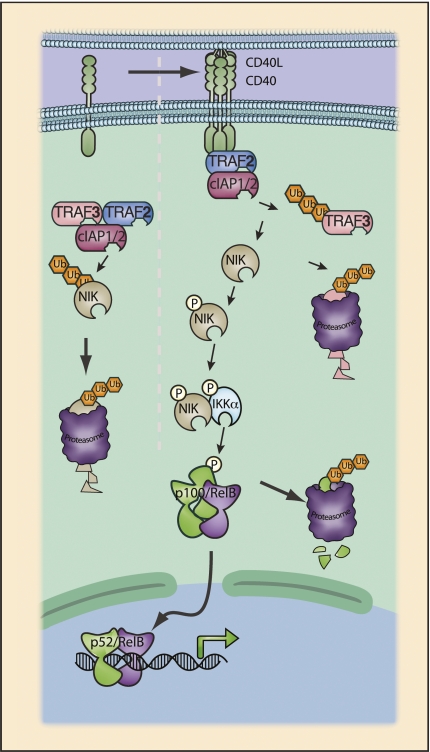
CD40 signaling to the alternative NF-κB pathway. CD40 possesses multiple TRAF interaction motifs, and binding of CD40L to CD40 triggers direct binding to multiple TRAF proteins. Noncanonical NF-κB activation requires the NF-κB-inducing kinase (NIK). NIK is constitutively active; thus, the noncanonical pathway is activated through the post-translational regulation of NIK protein levels. In the steady state, NIK is subject to constitutive ubiquitination by TRAF3 and consequent degradation. Upon CD40 ligation, TRAF2 is recruited to the receptor and, in conjunction with cIAP, targets TRAF3 for proteasomal degradation (Liao et al. 2004). CD40 ligation may also promote NIK stabilization through an “allosteric model” in which binding of NIK and binding of CD40 by TRAF proteins are mutually exclusive events (Sanjo et al. 2010). Thus, upon TRAF binding to CD40, NIK is displaced from the TRAF2:TRAF3 complex. As a result of displacement of NIK and the loss of TRAF3, active NIK accumulates. NIK binds and phosphorylates IKKα, leading to activation of IKKα kinase activity. Phosphorylation of p100 C-terminal serine residues by IKKα results in p100 ubiquitination by the SCFβTRCP complex and proteasomal processing to p52. Activation of the noncanonical pathway has also been well characterized for LTβR, BAFFR, and RANK.
Activating NF-κB
A wide range of soluble and membrane-bound extracellular ligands activate the NF-κB pathway, most notably through members of the TNFR, TLR, IL-1R, and antigen receptor superfamilies. In addition, in recent years, a growing list of signaling pathways has also been described that regulate NF-κB activity in response to changes in the intracellular environment. These intracellular NF-κB-activating pathways include the responses to DNA damage and reactive oxygen species, as well as recognition of intracellular pathogens mediated by the NOD and RIG-I-like (NLR) family of proteins. Rather than attempt to describe the many signaling pathways that activate NF-κB, we proceed through a general discussion of the mechanisms by which the IKK complex becomes activated, which remains the common upstream component of all NF-κB pathways. For the uninitiated reader with a basic familiarity with the mechanistic aspects of the regulation of the IKK complex, we provide brief overviews of prototypical NF-κB signaling pathways, with accompanying references to more detailed reviews of each pathway (Figs. 3–6).
Research on NF-κB signaling has produced an unexpected convergence in discoveries relating to signaling pathways upstream of the IKK complex. In fact, many of the signaling intermediates required for activation of IKK are shared between the different pathways. In most cases, RIP (receptor-interacting protein) and TRAF families of proteins are required in pathways that lead to IKK activation. Members of the TRAF family (Box 1) of proteins are required in canonical and noncanonical pathways, whereas RIP family members (Box 2) are selectively used in NEMO-dependent, canonical signaling pathways. The requirement for RIP family proteins highlights two entirely distinct mechanisms of initiating NF-κB signaling: dissociative and aggregative. In dissociative signaling, receptor ligation results in disruption of inhibitory complexes and, in the case of NF-κB, leads to activation of the alternative pathway. Aggregative signaling, on the other hand, relies on the assembly of large signaling platforms, and in the case of canonical NF-κB signaling pathways, it appears that RIP proteins play an important role in this process. However, before delving into how these divergent concepts may underlie IKK activation, we provide an overview of commonly studied NF-κB signaling pathways and introduce some of the general principles and common players in NF-κB signaling.
Box 1. TRAFs.
The TRAF family of proteins is defined by the presence of the eponymous C-terminal coiled-coil domain responsible for homotypic and heterotypic protein–protein interactions. There are seven mammalian TRAF proteins (TRAF1–7), of which TRAF2, TRAF3, and TRAF6 have been best characterized as regulators of signaling to NF-κB (Dempsey et al. 2003; Hacker et al. 2011). TRAF family members are obligate signaling intermediates in nearly all NF-κB signaling pathways (Hacker et al. 2011). All TRAF family members except TRAF1 have an N-terminal RING finger domain. In general, TRAF2 leads to NF-κB activation downstream from TNFR superfamily members, while TRAF6 has been shown to induce NF-κB activation in response to a wide variety of stimuli, including activation of TLRs, NLRs, and antigen receptors. Research focused on the role of ubiquitination in NF-κB signaling initially suggested an important role for TRAF E3 ubiquitin ligase activity that was demonstrated for TRAF2 and TRAF6 (Chen 2005). It remains unclear whether TRAF3, TRAF4, TRAF5, and TRAF7 can also act as E3 ubiquitin ligases. However, it is important to note that E3s come in multiple forms. The RING domain of TRAF proteins is not an E3 ligase with catalytic activity like that of HECT domain E3 ligases. Instead, TRAF proteins act as E3 ligase by serving as adaptors linking substrates with E2 enzymes, rather than by directly mediating the transfer of ubiquitin to substrate lysines. It was initially thought that TRAF2 and TRAF6, both of which have been shown to facilitate K63-linked ubiquitination of various substrates, did so through the recruitment of the heterodimeric E2, consisting of Ubc13 and Uev1A. There is particularly strong support for cooperation between TRAF6 and Ubc13/Uev1A in mediating K63 linkages. However, the genetic evidence for such a model for TRAF2 has been unsatisfying. Instead, with the identification of IAPs (inhibitor of apoptosis proteins) as ubiquitin ligases (see Box 4), it now appears that the picture is significantly more complex.
Box 2. Receptor-interacting (RIP) kinases.
The RIP family consists of seven serine/threonine kinases (RIP1–7), several of which have been shown to be crucial for signaling to NF-κB as well as in the regulation of cell death (Meylan and Tschopp 2005). RIP1 proteins share homologous serine/threonine kinase domains (KDs) but divergent protein:protein interaction motifs. RIP1 and RIP3 have RIP homology interaction motifs (RHIMs), RIP2 contains a caspase activation and recruitment domain (CARD) (Box 6), and RIP1 has a death domain (DD). These protein:protein interaction motifs and the consequent ability of RIPs to function as adapters are crucial for the roles of RIPs in canonical NF-κB signaling. Thus, while RIP1 is needed for activation of IKK by TNFR1, at least in most cell types, the kinase activity of RIP1 is not required (Hsu et al. 1996; Ting et al. 1996; Devin et al. 2000; Lee et al. 2004). RIP1 and RIP2 have been most thoroughly studied with regard to NF-κB activation, although there has also been increasing interest in RIP3 as a regulator of programmed necrosis (necroptosis) in conjunction with RIP1 (D Zhang et al. 2010; Liu 2005; Meylan and Tschopp 2005). RIP family members have been implicated in most NF-κB signaling pathways. As discussed in the review, it is apparent that RIP proteins generally serve as scaffolds in the IKK activation pathway; that is, RIPs function to recruit the IKK complex and also serve as a scaffold onto which ubiquitination is anchored, leading to IKK activation. In general, the linkage of ubiquitination onto RIPs is carried out in a TRAF-dependent manner. Thus, in TNFR1 signaling, TRAF2, in conjunction with cIAPs, mediates K63-linked ubiquitination of RIP1 (Karin 2009). Yet when a knockdown and replacement strategy was used to prevent K63 linkages from being formed during signaling, activation of IKK by TNF was largely unaffected. Conversely, activation of IKK downstream from IL-1βR, which acts independently of RIPs, was significantly blunted (Xu et al. 2009). These findings may suggest the contribution of other ubiquitin linkages. For example, there is recent evidence to suggest that linear head-to-tail ubiquitin may be important in TNF and IL-1β signaling. Furthermore, K11 linkages have been detected on RIP1. The ability of RIP proteins to interact with NEMO during signaling is consistent with the requirement of RIPs for canonical but not noncanonical signaling pathways.
In the case of a typical signaling pathway, such as in response to TNF, activation of NF-κB is initiated following binding of the ligand to the receptor present at the plasma membrane. Interestingly, activation of NF-κB generally occurs in the absence of receptor enzymatic activity. Instead, for most NF-κB pathways, signaling proceeds through the binding of a series of adapter proteins, which possess protein:protein interaction domains. Key interaction domains that participate in NF-κB pathways include death domains (DDs), caspase activation and recruitment domains (CARDs) (Box 3), RIP homotypic interaction motifs (RHIMs), and Toll IL-1R (TIR) domains. Conformational changes in the receptor or changes in receptor stoichiometry triggered by ligand binding facilitate adapter protein binding, which provides links to downstream components. Thus, the early events of the signaling cascade are based on conformational change, protein:protein interaction, and assembly of large protein complexes, rather than on enzymatic cascades or PTMs. One or several steps removed from ligand binding, kinases are recruited to the receptor complex. The activation of these kinases appears to be dependent on two factors: induced proximity resulting from the dense organization of the receptor signaling complex, and conformational changes resulting from adapter protein binding. As is the case for the IKK complex itself, kinases in NF-κB signaling pathways exist in multisubunit complexes consisting of both kinase and nonenzymatic regulatory subunits. These regulatory kinase complex subunits—e.g., NEMO or TAB proteins—function as adapters that mediate kinase recruitment and are therefore required for kinase activation following binding to the receptor signaling complex. Once activated, these regulated kinase complexes can instigate traditional kinase cascades. In the case of the NF-κB pathway, the kinase “cascades” are often exceptionally short and primarily achieve signal amplification, leading to robust IκB phosphorylation and culminating in NF-κB transcriptional programs.
Box 3. Caspase recruitment domains (CARDs).
CARD-containing proteins function in multiple NF-κB signaling pathways. The role of CARDs in NF-κB signaling first came to light in antigen receptor signaling pathways. CARD-containing proteins most often act through TRAF family proteins in NF-κB signaling pathways. Both BCL10 and CARD11 (CARMA1) are CARD-containing proteins that are crucial for IKK activation downstream from the T-cell receptor (TCR) and B-cell receptor (BCR). CARD–CARD interactions mediate the formation of large signaling complexes. In addition to antigen receptor signaling, members of the NOD-LRR family, and components of RIG-I-like receptor (RLR) signaling proteins are CARD-containing proteins. In NOD signaling, the CARD-containing kinase RIP2 (RICK/CARDIAK) binds to NEMO, TAK1, and TRAFs to activate the IKK complex by proximity-induced mechanisms (Inohara et al. 2000; Abbott et al. 2007; JY Kim et al. 2008). RIP2 is also ubiquitinated in the process, which is thought to be necessary for recruitment of TAK1, rather than NEMO (Hasegawa et al. 2008). The PIDDosome, another cytoplasmic signalosome, uses RIP-associated ICH-1/CED-3 homologous protein with a DD (RAIDD), a CARD, and DD-containing protein to promote IKK activation in response to genotoxic stress (Janssens et al. 2005; Hacker and Karin 2006). Aggregative induction of the NF-κB signaling pathways is most strikingly illustrated by the recent description of the ability of the RLR adapter MAVS to form large fibril-like aggregates upon viral infection (Hou et al. 2011). Although the CARD domain of MAVS is sufficient for the formation of these fibrils both in vivo and in vitro, it appears that fibril formation and downstream signaling are augmented or stabilized by the formation of K63-linked ubiquitin chains (Zeng et al. 2010; Hou et al. 2011).
Structure and regulation of IKK
A cursory view of the diverse signaling pathways shown in Figures 3–6 reveals several common features, some of which we discussed in detail previously (Hayden and Ghosh 2008). Here we try to present a more generalizable and historical narrative on the mechanisms by which the NF-κB signaling pathway is initiated. The identification of the kinase complex responsible for phosphorylating IκBα was a watershed event in NF-κB research. Given the diversity of stimuli leading to NF-κB activation, and the existence of multiple members of the IκB family, one might have predicted that there would be multiple IKKs. Instead, multiple groups identified a common IKK complex with the same protein constituents.
The IKK was initially observed to exist as a high-molecular-weight kinase activity that was purified and characterized by multiple groups as a stimulus-dependent kinase (DiDonato et al. 1997; Mercurio et al. 1997; Regnier et al. 1997; Woronicz et al. 1997; Zandi et al. 1997). The first identified component of this 550- to 900-kDa complex was IKKα (IKK1), a serine/threonine kinase, previously known as CHUK. IKKβ (IKK2) was subsequently identified based on both sequence homology and biochemical purification. IKKα and IKKβ, 85 and 87 kDa, respectively, are serine/threonine kinases that share a homologous N-terminal kinase domain (KD) and are able to phosphorylate multiple members of the IκB family in vitro (Fig. 2; Zandi et al. 1997, 1998). NEMO (also known as IKKγ, IKKAP1, or Fip-3) was identified by multiple groups through complementation of an NF-κB-unresponsive cell line, by affinity purification, or as a factor binding an adenoviral inhibitor of NF-κB (Rothwarf et al. 1998; Yamaoka et al. 1998; Y Li et al. 1999; Mercurio et al. 1999). NEMO, which does not posses kinase activity, is a 48-kDa protein that is not related to IKKα and IKKβ (Fig. 2).
There is ample and unambiguous genetic evidence for the preeminent role of IKKα, IKKβ, and NEMO in NF-κB signaling pathways (Gerondakis et al. 2006). Loss of IKKβ results in a phenotype mimicking p65 knockouts, confirming the importance of IKKβ in activation of canonical p65-containing dimers (Q Li et al. 1999b; ZW Li et al. 1999; Tanaka et al. 1999). The ability to rescue embryonic lethality through deletion of TNFR1 supports a crucial role for IKKβ in TNF signaling to NF-κB (Q Li et al. 1999b; ZW Li et al. 1999; Senftleben et al. 2001b). Unlike IKKβ-deficient animals, IKKα-deficient mice die perinatally with multiple morphological defects (Hu et al. 1999; Q Li et al. 1999a; Takeda et al. 1999). While initial studies demonstrated little role for IKKα in classical NF-κB activation, subsequent reports revealed the requirement for IKKα in multiple noncanonical NF-κB signaling pathways and perhaps some canonical signaling pathways as well (Takaesu et al. 2003; Solt et al. 2007). NEMO is required for canonical NF-κB pathways, and as a result, NEMO-deficient mice succumb to embryonic hepatocyte apoptosis, and cells without NEMO fail to activate NF-κB through all canonical pathways (Yamaoka et al. 1998; Rudolph et al. 2000; Schmidt-Supprian et al. 2000).
Following the identification of the IKK complex, it has been clearly shown that most NF-κB signaling pathways proceed through the IKK complex (Li et al. 2000). Many of the upstream signaling components that regulate IKK activation in these diverse pathways have been identified and suggest significant overlap in the mechanisms that regulate IKK activity, even between functionally divergent pathways. Nevertheless, significant questions about IKK regulation remain unanswered. Even the most basic questions—e.g., how IKK is activated, how IKK is inactivated, or how IKK substrate specificity is determined—remain to be answered satisfactorily. Here we discuss some of the significant progress that has occurred in each of these areas, but also draw attention to the work that remains to be done to fully address these important, outstanding questions.
Given the large number of publications in this area, it is surprising that 14 years after the description of the IKK complex, it remains unclear exactly how the kinase is activated. Most likely this is because the many receptors that lead to IKK activation lack kinase activity. Most, for that matter, lack any described enzymatic activity. Thus, at some point, a kinase activity must be recruited into the pathways. In the case of the alternative pathway, this is accomplished by regulating protein levels of a kinase that is otherwise constitutively active: NIK (NF-κB-inducing kinase). Elegant genetic proof of this regulatory mechanism came from the rescue of TRAF3 knockout mice through the genetic inactivation of NIK or deletion of p100 (He et al. 2006) NIK directly phosphorylates and activates IKKα, as demonstrated by analyses of NIK−/− mice and aly mice, which bear a point mutation in the NIK KD (Regnier et al. 1997; X Lin et al. 1998; Ling et al. 1998; Uhlik et al. 1998; Shinkura et al. 1999). However, for canonical NF-κB pathways, although this issue is not always appreciated, it remains unclear whether IKK is activated by an upstream kinase or autophosphorylation. Although many IKK kinases (IKK-Ks) have been implicated in canonical pathways over the years, many of them have subsequently fallen by the wayside. At present, only TAK1 remains as a generally accepted canonical pathway IKK-K. However, the mechanism of initial kinase activation in the NF-κB pathway remains a fundamentally important unsolved question. The answer is likely to lie in the numerous reports demonstrating the assembly of large oligomeric signaling complexes in nearly all canonical NF-κB signaling pathways. Therefore, whether IKK is activated directly or through TAK1, the kinase-independent higher-order organization of receptor signaling complexes suggests a mechanism for initiation of the kinase activity. Hopefully, recently described structural information on the IKK complex may help to clarify its mechanism of activation.
The discrepancy between the molecular weight of the constituents of the IKK complex and the observed molecular weight of the complex by gel filtration has, for more than a decade, led to considerable speculation about both the existence of additional components and the stoichiometry of known components in the complex. Available evidence, reviewed briefly here, now strongly suggests that the IKK complex consists only of IKKα1IKKβ1NEMO2. The evidence for a dimer of dimers is as follows: (1) Recombinant NEMO—in this case, trimers—with a predicted molecular weight of 150 kDa elutes at an apparent molecular weight of 550 kDa upon gel filtration, demonstrating a particularly elongated structure (Agou et al. 2004). (2) Recombinant NEMO with IKKα or IKKβ assembles into a complex with an apparent molecular weight that is similar to the purified complex (Krappmann et al. 2000; Miller and Zandi 2001). (3) Recombinant NEMO and IKKβ associate in a 2:2 molar ratio, as do the minimum interaction domains of the same proteins (Drew et al. 2007). (4) The recent IKKβ crystal structure, discussed below, suggests that IKKβ would interact with NEMO dimers as a dimer (Xu et al. 2011), and it is anticipated that IKKβ:IKKα heterodimers would do the same. Thus, the large Stoke's radius of NEMO and the end-to-end assembly of IKK with NEMO predict that a complex consisting of an IKK dimer in complex with a NEMO dimer is sufficient to account for the observed molecular weight of the IKK complex (Agou et al. 2004; Xu et al. 2011).
Recently, after many years of effort by numerous groups, an IKK protein has finally been crystallized (Xu et al. 2011). This work offers important additional insight into both the potential mechanisms of IKK activation and substrate specificity. The structure obtained, of IKKβ from Xenopus in complex with the kinase inhibitor Cpmd1 or Cpmd2, demonstrates a trimodular composition differing significantly from the predicted modular organization. Predicted leucine zipper and helix–loop–helix domains were not seen, and instead these regions formed an α-helical scaffold/dimerization domain (SDD). The other structural domains that were identified were the previously described ubiquitin-like domain (ULD) (May et al. 2004) and the KD. The SDD, and in particular the portion previously thought to form a leucine zipper domain, mediates IKK dimerization, as had previously been predicted (Mercurio et al. 1997; Woronicz et al. 1997; Zandi et al. 1997). While dimerization was necessary for inducible activation of IKKβ, it was not necessary for kinase activity per se (Xu et al. 2011). The ULD is necessary for kinase activity (May et al. 2004) and also appears to cooperate with the SDD in binding to IκBα such that the appropriate serine residues within the destruction box are targeted for phosphorylation (Xu et al. 2011). The resulting structural insights will hopefully lead to an improved understanding of substrate targeting by IKK. Nevertheless, despite the information provided by elucidation of the structure, the mechanism of IKK complex activation remains ambiguous.
IKK activation depends on phosphorylation of activation loop serines. IKKα and IKKβ can each be phosphorylated on two serines: Ser 176 and Ser 180 or Ser 177 and Ser 181, respectively. Activation loop phosphorylation is crucial for inducible kinase activity: Activity is lost upon treatment with phosphatases in vitro, mutation of the activation loop serines to glutamic acid yields constitutively active IKK, and mutation to alanines abrogates signal responsiveness (DiDonato et al. 1997; Mercurio et al. 1997; Ling et al. 1998; Delhase et al. 1999; Hacker and Karin 2006). Interestingly, the IKKβ structure suggests that a dimer of IKKβ would not be capable of phosphorylating itself because the active site of IKKβ within a dimer is not in proximity to the activation loop of the other IKKβ in the same dimer (Xu et al. 2011). These results are consistent with the finding that IKK complexes in which one kinase lacks activity retain the ability to activate the active kinase subunit (Zandi et al. 1997). On the other hand, in higher-order structures predicted to form during IKK activation, the activation loop and active site of IKKβ in adjacent dimers are closely juxtaposed (Xu et al. 2011). As a result, IKKβ would likely not undergo cis-autophosphorylation; however, two dimers brought into proximity could readily be activated by trans-autophosphorylation. These structural insights correlate well with a growing body of evidence that suggests that the formation of higher-order, oligomeric signaling complexes plays a key role in the activation of NF-κB signaling.
Oligomerization in activation of IKK
Our overview of NF-κB signaling pathways (Figs. 3–6) highlights the importance of adapter proteins in NF-κB activation. Adapter proteins facilitate the assembly of complex signaling networks by mediating the activation of multiple transcription factor families. The requirement for DD-, RHIM-, and CARD-containing adapters highlights the importance of the assembly of complex platforms, sometimes referred to as signalosomes, in IKK activation. Although assembly of adapter protein complexes seems most consistent with an associative signaling model in which IKK activation occurs through trans-autophosphorylation, signalosome formation could also promote activation of an IKK-K and proximity between IKK-K and IKK. Indeed, these are not mutually exclusive means of IKK activation. Induced proximity secondary to adapter oligomerization may mediate trans-autophosphorylation of TAK1, TAK1 phosphorylation of IKK, and IKK trans-autophosphorylation. However, given that TAK1 is not universally required for IKK activation, we first focus on potential mechanisms whereby oligomerization of IKK itself could induce IKK-K activity.
IKK dimerization is necessary and sufficient for activation of overexpressed IKK (Zandi et al. 1997; McKenzie et al. 2000; Tang et al. 2003). We discussed above that NEMO is in all cases required for inducible canonical IKK activation. NEMO can form dimers, trimers, and tetramers in vitro and can oligomerize in vivo (Agou et al. 2002, 2004; Tegethoff et al. 2003; Drew et al. 2007). Inducible oligomerization of NEMO by RIP1 was initially speculated to activate IKK, and it was indeed found that forced induction of NEMO oligomerization activates IKK (Inohara et al. 2000; Poyet et al. 2000). Forced oligomerization of IKK also directly activates NF-κB (Inohara et al. 2000). Consistent with these results, NEMO with a mutated oligomerization domain functions as a dominant negative, and the oligomerization domain alone can prevent IKK activation (Tegethoff et al. 2003; Agou et al. 2004). In addition to the C-terminal minimal oligomerization domain (MOD), NEMO contains both a dimerization domain and an IKK-binding domain within the N-terminal half of the protein (Fig. 2). Overexpression of a truncated NEMO, consisting of the dimerization and IKK interaction domains, alone can activate co-expressed IKK; however, reconstitution of knockout cells demonstrates that both N-terminal IKK binding and dimerization domains and the C-terminal oligomerization domain are needed to reconstitute inducible IKK activation in the absence of full-length (FL) NEMO (Marienfeld et al. 2006). Therefore, both formation of a dimer of dimers and ability to form higher-order structures are thought to be necessary for induced IKK activation. In further support of the requirement for the formation of higher-order IKK structure, modification of the dimerization domain may be an important mechanism of negative feedback regulation. NEMO becomes phosphorylated at Ser 68 within the dimerization/IKK-binding domain following stimulation, which disrupts formation of NEMO dimers and interaction between IKK and NEMO, thereby terminating signaling (Palkowitsch et al. 2008). IKK binding to NEMO may be further augmented by phosphorylation of the IKK NEMO-binding domain (NBD) (Palkowitsch et al. 2008). In addition to this evidence for positive and negative regulation of IKK activity through manipulation of the formation of higher-order IKK complexes during signaling, exogenous factors have also been reported to manipulate NF-κB signaling by altering NEMO oligomerization. Viral proteins capable of activating NF-κB have also been shown to trigger oligomerization of NEMO (Poyet et al. 2001; Huang et al. 2002). Given that higher-order IKK complexes would facilitate trans-autophosphorylation, these data suggest that on its own, formation of higher-order IKK complexes would be sufficient for IKK activation. However, to date, there is little direct evidence for the formation of higher-order IKK complexes in vivo in the course of signaling to NF-κB.
How IKK undergoes oligomerization upon signal induction remains unsettled. One model holds that oligomerization of upstream adapters RIPs, TRAFs, or CARD-containing proteins forms a higher-order structure through which IKK can become oligomerized via direct one-to-one binding to these adapter proteins. Thus, for example, RIP or BCL10 oligomerization and binding to NEMO would directly induce proximity of IKK complexes (Inohara et al. 2000). Another model posits that ubiquitin chains provide the oligomeric structure to which TAK1 and/or IKK complexes bind and become activated. Within the ubiquitin model, it has been proposed that ubiquitin chains may either directly activate kinase complexes, facilitate proximity-induced trans-autophosphorylation, or stabilize otherwise activated signaling complexes. There is further subdivision of the ubiquitin model according to the nature (K63 versus linear head-to-tail) (Box 5, below) of the ubiquitin linkage and whether the ubiquitin chains are free or attached to a substrate. Here we explore the evidence for these nonexclusive models of how the regulated formation of oligomeric signaling complexes may facilitate IKK activation.
Box 5. Ubiquitin.
Ubiquitin is a highly conserved, ubiquitously expressed, multifunctional 76-amino-acid protein encoded, in mammals, by four genes. The cell uses ubiquitin for PTM of proteins by their covalent attachment to target protein lysines through an enzyme cascade (Kerscher et al. 2006). The ubiquitination process requires an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase. E1 enzymes “activate” ubiquitin usng ATP to generate a ubiquitin AMP that allows ubiquitin conjugation to the catalytic cysteine of E1 itself through a thioester bond; ubiquitin is then transferred to the catalytic cysteine residue of the E2 enzyme. There are two mammalian E1 enzymes, >35 E2 enzymes, and >600 predicted E3s. The E3 ubiquitin ligase may possess enzymatic activity or may serve primarily as an adapter between E2 and target proteins. Ubiquitin linkage occurs when an isopeptide bond is formed between the C-terminal glycine of ubiquitin and the ɛ-amino group of the target protein lysine. Ubiquitin chains are then assembled through the attachment of C-terminal glycine of ubiquitin to lysine residues within the initially conjugated ubiquitin. These chains may be assembled using K6, K11, K27, K33, K48, or K63 in ubiquitin and also via the N-terminal methionine (M1). In addition, numerous reports have now described ubiquitin linkage to nonlysine residues of target proteins (Verhelst et al. 2011). As a result, ubiquitination of substrates can occur in both variable and specific fashions.
Linear ubiquitin assembly complex (LUBAC). While most ubiquitin chains involve linkage of the C-terminal glycine of ubiquitin to lysine residues on the substrate-linked ubiquitin (e.g., K48 or K63), recent work has demonstrated the existence of head-to-tail linear ubiquitin chains assembled by a unique LUBAC (Kirisako et al. 2006). The LUBAC complex consists of heme-oxidized iron regulatory protein 2 ubiquitin ligase 1 (HOIL-1, also known as RBCK1) and HOIL-1-interacting protein (HOIP, also known as RNF31 and Zibra) (Kirisako et al. 2006). In this early work, it was noted that overexpression of the LUBAC complex could drive an NF-κB-dependent transcriptional response. Subsequently, the LUBAC complex was copurified with the active TNF/TNFR1 signaling complex (Haas 2009), and it was demonstrated that LUBAC was required for stabilization of the TNFR1 signaling complex and full activation of NF-κB by TNF and CD40 (Haas 2009; Rahighi et al. 2009; Tokunaga et al. 2009). More recently, SHARPIN has been identified as a third component of LUBAC (Gerlach et al. 2011; Ikeda et al. 2011; Tokunaga et al. 2011). From these initial reports, it appeared that SHARPIN is required for NF-κB activation downstream from TNFR1, IL-1R, and CD40. In TLR signaling pathways, it appears that SHARPIN is selectively required for the phosphorylation of p105 but is dispensable for phosphorylation or degradation of IκBα (Zak et al. 2011). Furthermore, a mutation in NEMO, which abrogates binding of the LUBAC complex, also does not affect TLR-induced phosphorylation and degradation of IκBα (Zak et al. 2011). While several studies have demonstrated contributions of LUBAC to the stabilization of TNFR1 family signaling complexes, outstanding questions remain for this recently identified component of inflammatory signaling pathways (see the text).
To address these models, we first look at the role of RIP1 in IKK activation. RIP1 has been thoroughly studied in the TNFR1 signaling pathway and is generally accepted to be essential for TNF-induced canonical IKK activation (Hsu et al. 1996; Kelliher et al. 1998; Zhang et al. 2000). Although recent data have suggested that RIP1 may not be universally required in TNF signaling (Wong et al. 2010; Zhang et al. 2011), other studies suggest that the residual NF-κB activation in the absence of RIP1 may be the result of a noncanonical pathway (Gentle et al. 2011). That this residual NF-κB activity is augmented by FADD (Fas receptor-associated DD) deletion is consistent with a noncanonical mechanism of NF-κB activation in the absence of RIP1 (Zhang et al. 2011). RIP1 knockout animals (Kelliher et al. 1998) survive longer than IKKβ or p65 knockouts, but this difference may be attributable to NF-κB-independent contributions of RIP1 to prodeath signaling pathways (Zhang et al. 2011) such as necroptosis (Degterev et al. 2008). In addition to TNFR1 signaling and IKK activation via other DD-containing TNFR family members, RIP1 has also been reported to be required for TRIF-dependent NF-κB activation via TLR3 and TLR4, NF-κB activation via RIG-I (Meylan et al. 2004; Cusson-Hermance et al. 2005), and DNA damage-induced activation of IKK (Hur et al. 2003; Janssens et al. 2005). RIP1 binds directly to NEMO and always acts through NEMO in activating IKK. Thus, RIP1 is not required for activation of the alternative pathway through CD40 or LTβR (Vivarelli et al. 2004). Substantial data exist to support a key role for RIPs as a substrate for ubiquitination and as a platform for canonical IKK complex activation. However, it is not yet clear that these properties of RIP proteins are one and the same; namely, whether RIP1 ubiquitination is necessary for RIP1-dependent IKK activation.
The idea that RIP1 could nucleate the assembly of a signaling complex that activates IKK by proximity-induced trans-autophosphorylation was proposed not long after the identification of IKK (Delhase et al. 1999; Inohara et al. 2000). This model preceded the recognition of RIP1 as an ubiquitin substrate. Hence, the question was whether induced RIP1 oligomerization, in the absence of upstream signaling events, was sufficient for IKK activation. Indeed, induced oligomerization of RIP1 or RIP2 does induce IKK activation (Inohara et al. 2000). Although these forced oligomerization studies strongly suggest direct effects on the IKK complex, the potential role of an intervening kinase, such as TAK1, was not investigated. RIP1 can interact with the TAK1 complex and can act as a mediator of induced proximity between kinase and substrate; e.g., between TAK1 and IKK or IKK alone. The interaction between RIP and other kinases must, therefore, be a regulated part of the signaling pathway. Forced oligomerization experiments suggest that oligomerization itself could be the regulated event. A competing but nonexclusive model is that ubiquitination of RIP1 may generate the substrate for TAK1 or IKK recruitment and activation (Bhoj and Chen 2009). In such a model, a single RIP molecule modified with oligomeric ubiquitin chains could mediate oligomerization of the IKK complex.
There is abundant evidence that RIP1 is ubiquitinated upon signaling from multiple pathways. Following TNF stimulation or the induction of DNA damage, SDS-PAGE analysis reveals slower-migrating forms of RIP1 (Lee et al. 2004). This laddering effect is greatly enhanced if only TNF-bound receptor complexes are analyzed (Wu et al. 2006; Haas et al. 2009). Analyses by multiple groups revealed that this modified RIP1 was in fact ubiquitinated through either K63 (Cusson-Hermance et al. 2005; Ea et al. 2006; Li et al. 2006; Wu et al. 2006) or linear (Haas et al. 2009) ubiquitin linkages. Thus, it was proposed that ubiquitin chains branching from RIP1 might mediate either recruitment or stabilization of the IKK and TAK1 complexes. It remains unclear how either the TAK1 or IKK complex might distinguish polyubiquitin chains associated with RIP1 from those that are bound to other proteins or even floating freely within the cell. One possibility is that coordinate binding of IKK to both adapter proteins—e.g., TRAF or RIP—and ubiquitin chains through the NEMO ubiquitin-binding domain (UBD) could provide specificity. Ubiquitination mediated by either cIAPs, with TRAF2 in TNFR1 signaling and TRAF6 in DNA damage (Hinz et al. 2010), or the linear ubiquitin assembly complex (LUBAC), in TNFR1 signaling, would thus provide a model of inducible scaffold formation.
Ubiquitination of RIP1 and the role of ubiquitination in IKK activation were originally attributed to the E3 ligase activity of TRAFs. The requirement for TRAF6 and TRAF2/5 in signaling by the TIR and TNFR superfamilies, respectively, is well established. Ligation of TNFR1 by TNF results in the formation of a proinflammatory, multisubunit signaling structure, termed complex I (Micheau et al. 2001), in which TRAF2 is a key component. TRAF2 is recruited to complex I via an induced interaction with TRADD (Hsu et al. 1996). High-affinity interaction between TRADD and TRAF2 mediates robust activation of NF-κB and AP-1 pathways (Ayabe et al. 2000). However despite deficiencies in JNK and AP-1 activation, TRAF2-deficient cells have relatively intact TNF-induced activation of NF-κB (Yeh et al. 1997). TRAF5 is also part of the TNFR1 signaling complex, and although TRAF5 knockouts activate NF-κB normally, TRAF2/5 double-knockout cells are defective in IKK activation (Yeh et al. 1997; Nakano et al. 1999; Tada et al. 2001). Although deletion of the RING finger domain of TRAF2 inhibits IKK activation, it may also prevent TRAF2-mediated recruitment of IKK to the receptor complex (Devin et al. 2000). Finally, knockdown of the E2 UBC13 or deletion of UBC13 in macrophages prevents TRAF2 ubiquitination with minimal effects on NF-κB activation (Habelhah et al. 2004; Yamamoto et al. 2006). TRAF6 is necessary for MyD88-dependent activation of NF-κB (Cao et al. 1996; Wesche et al. 1997; Ye et al. 2002), and TRAF6-deficient cells fail to activate NF-κB in response to IL-1 and LPS (Lomaga et al. 1999; Naito et al. 1999). However, like TRAF2, the importance of the E3 ligase activity of TRAF6 remains controversial. Reconstitution of TRAF6-deficient cells with a TRAF6 mutant lacking the RING finger motif completely restored IL-1-induced activation of NF-κB and JNK in vitro (Kobayashi et al. 2001). In addition, UBC13 knockouts also failed to show significant defects in TRAF6-mediated activation of NF-κB downstream from LPS, IL-1, CD40, or BAFF despite impaired MAPK activation (Yamamoto et al. 2006). However, ubiquitin replacement with K63R abrogates MyD88-mediated NF-κB activation by LPS and IL-1, suggesting that another E2/E3 pair may have important functions in this process (Xu et al. 2009).
The E3 function of cIAP1/2 has also been implicated in multiple NF-κB pathways (Box 4). The LUBAC complex may also serve to ubiquitinate relevant substrates in TNFR and TLR signaling pathways (Box 5). It should be noted, however, that there are significant outstanding questions regarding the LUBAC complex and, in particular, the recently implicated component SHARPIN with regard to its role in the activation of NF-κB. First, the phenotype of SHARPIN-deficient cpdm/cpdm mice is difficult to reconcile with the phenotypes of other essential components of the canonical NF-κB pathway. While NEMO, IKK, or p65-deficient mice exhibit embryonic lethality, the SHARPIN-deficient mice are born normal but exhibit an IL-1-dependent inflammatory phenotype later in life (Gerlach et al. 2011; Ikeda et al. 2011; Tokunaga et al. 2011). Indeed, SHARPIN-deficient mice exhibit a robust activation of an NF-κB gene signature, and their phenotype can be ameliorated with bortezomib, a proteasome inhibitor that is thought to function in part as an NF-κB inhibitor (Liang et al. 2011). These findings seem directly at odds with the proposed role of SHARPIN and accepted biological functions of the NF-κB pathway. The proinflammatory phenotype could be the result of the recently demonstrated role of SHARPIN as an inhibitor of integrin signaling in fibroblasts, leukocytes, and keratinocytes (Rantala et al. 2011). However, while IL-1β could certainly be produced in an NF-κB-independent manner, the ability of IL-1β to mediate inflammation would be expected to depend on activation of NF-κB. Furthermore, others have shown that canonical IKK activation by TLR stimulation of macrophages from SHARPIN-deficient mice is normal (Zak et al. 2011), suggesting that the role of SHARPIN in NF-κB activation may be less direct than initially anticipated. Nevertheless, the implication of additional ubiquitin ligases fails to account for the lack of correlation between ubiquitination of proposed substrates—e.g., RIP1 in TNFR1 signaling—and the activation of IKK.
Box 4. Inhibitor of apoptosis proteins (IAPs).
IAPs are an evolutionarily conserved family of proteins that have been of significant interest primarily because of functions in cancer (Gyrd-Hansen and Meier 2010). IAPs are defined by the presence of a baculovirus IAP repeat (BIR) domain that mediates protein:protein interactions. Mammalian cIAP1 and cIAP2 are also notable for possessing RING finger, ubiquitin-associated, and CARD domains that mediate E3 ubiquitin ligase, ubiquitin binding, and protein:protein interactions, respectively. Although originally characterized primarily as inhibitors of caspase activity, particularly caspase 3, recent findings have established important roles for IAPs in the regulation of both canonical and noncanonical NF-κB pathways. In canonical TNFR signaling, cIAP1 and cIAP2 are required for TRAF2-dependent K63-linked ubiquitination of RIP1 (Yin et al. 2009), while in the noncanonical pathway, cIAPs are required for TRAF2-mediated ubiquitination and degradation of NIK (NF-κB-inducing kinase). Thus, current data suggest that cIAP1 and cIAP2 may generally function with TRAF2, analogously to the role of Ubc13/Uev1A with TRAF6. Finally, XIAP, which appears to primarily function as a regulator of caspase 3 activation, also binds to the TAK1 complex through the TAB1 protein (Lu et al. 2007).
Thus, there are several complications in what at first seems a well-supported model. In induced oligomerization experiments, ubiquitination of RIP1 was not noted, and coimmunoprecipitated RIP1/RIP2 and NEMO did not appear to be ubiquitin-modified (Inohara et al. 2000). If RIP1 or NEMO ubiquitination was necessary for IKK activation under these artificial circumstances (Inohara et al. 2000), ubiquitin modification should have been readily apparent in the experiments performed, although the presence or absence of such species was not tested directly. The strongest evidence for a role for RIP1 ubiquitination in IKK and TAK1 recruitment comes from studies in which cells lacking RIP1 were reconstituted with RIP1 lacking Lys 377, which was shown to be essential for RIP1 ubiquitination (Ea et al. 2006; Li et al. 2006). However, while this mutant fails to be ubiquitinated, it also, apparently, does not bind to the activated receptor (Ea et al. 2006). As a result, the implications of these findings for the mechanistic role of RIP1 K63 ubiquitination are unclear. Furthermore, several studies have now failed to observe correlations between RIP1 ubiquitination and IKK activation. For example, elegant inducible ubiquitin knockdown and replacement strategies indicate that K63-linked ubiquitination is not required for IKK activation by TNFR1 (Xu et al. 2009), and there is a modest effect on activation of IKK in Ubc5 knockdown cells in which RIP1 ubiquitination is almost completely abolished (Xu et al. 2009). Finally, reconstitution of TRAF2/5 double-knockout cells with TRAF2 RING mutants abrogates RIP1 ubiquitination but not IKK activation (L Zhang et al. 2010). Therefore, while ubiquitination of multiple substrates does occur at the ligand-bound receptor complex, and the role of multiple deubiquitinases (DUBs) as negative regulators of NF-κB activity is well established (Harhaj and Dixit 2011), it remains unclear exactly how nondegradative ubiquitination activates IKK.
Based on the lack of receptor kinase activity in multiple NF-κB signaling pathways, there is inherent appeal in the model of oligomerization-mediated IKK activation. While unclear in terms of the mechanistic details, ubiquitination may well have a role in regulating this process. We envision four potential mechanisms through which nondegradative ubiquitination might be required for IKK activation: (1) direct activation, (2) induced proximity, (3) stabilization, and (4) nonsignaling functions. While it was initially posited that ubiquitination might play a role analogous to that of phosphorylation, the evidence in support of such a model is unconvincing. Direct activation could, alternatively, occur through conformational change in the kinase complex resulting from either ubiquitination of NEMO or NEMO binding to ubiquitin. Although NEMO has been shown to be ubiquitinated in some pathways, the requirement for ubiquitination of a single lysine residue for activation of the IKK has not been established. Although both events occur in the course of signaling, there is insufficient evidence to conclude that they directly mediate kinase activation. If direct activation were to occur, then short ubiquitin oligomers should activate IKK or TAK1 complexes as efficiently as longer polymers. While recent work has shown that K63-linked ubiquitin chains can activate TAK1 or IKK complexes in vitro (Xia et al. 2009), it appears that this is not the result of conformational changes that directly activate the kinase complex. Instead, in vitro kinase complex activation by ubiquitin chains suggests that activation of IKK or TAK1 by ubiquitin chains is primarily the result of induced proximity and trans-autophosphorylation (Xia et al. 2009). This work suggested that unanchored ubiquitin chains might mediate kinase activation in cells (Xia et al. 2009). It was proposed that unanchored ubiquitin chains might function analogously to second messenger systems; however, it remains unclear how such a process could be regulated and why the mechanisms of IKK regulation and signaling outcomes would differ between signaling pathways. Alternatively, nondegradative ubiquitination could function by stabilizing signaling complexes through one of two mechanisms: (1) by preventing K48 ubiquitination and degradation of signaling intermediates, as has been demonstrated for RIP1 (Harhaj and Dixit 2011), and (2) by physically stabilizing complexes through “cross-linking” of UBD-containing proteins. In fact, both A20 and, more recently, CYLD (cylindromatosis protein) (Ahmed et al. 2011) are reported to negatively regulate signaling not merely by removing K63 linkages, but also by facilitating replacement of these linkages with K48 linkages that target signaling components for proteasomal degradation. These data suggest that removal of K63 linkages is not sufficient for signal termination and that DUBs may primarily suppress signaling by targeting active signaling complexes for degradation, rather than by removing K63. Yet another hypothesis that we believe has been poorly explored is the idea that nondegradative ubiquitination could serve a role that is separate from, but necessary for, signaling to occur. In this regard, we hypothesize that non-K48 linkages may serve to target previously activated signaling complexes to cellular machines, such as the HSP90/cdc37 complex, that are capable of re-establishing signaling pathway competence. If the formation of large oligomeric signaling complexes, including those that appear to be highly stable (Hou et al. 2011), is a necessary part of IKK activation, then one must predict that there are active mechanisms required for complex disassembly and re-establishment of the inducible signaling pathway. This latter hypothesis is one for which there are little data, but we speculate that analogous to the targeting of proteins for proteasomal degradation by K48 ubiquitin, alternative linkages would be an efficient means of targeting signalosomes to appropriate chaperones in order to disassemble these multiprotein complexes and reset signaling pathways.
To summarize, there are still significant gaps in our understanding of IKK regulation. The most fundamental aspects of the signaling pathway—namely, activation of IKK and determination of substrate specificity—remain inadequately understood. In some ways, however, there has been progress toward a more unified model of kinase activation. The basic issue remains the activation of kinases downstream from receptors lacking inherent kinase activity. Despite increasing numbers of players in pathways leading to IKK activation, particularly with regard to ubiquitin ligase and DUB complexes, the model of kinase activation by trans-autophosphorylation continues to hold sway. The idea that nondegradative ubiquitin linkages might function analogously to phosphorylation and through direct target conjugation alter kinase activity no longer appears to be embraced. Similarly, the idea that K63 linkages on RIP1 and other upstream adapters serve as recognition motifs, analogous to binding to phosphorylated SH3 domains, for IKK and TAK1 recruitment is not fully supported. Instead, it is now argued that nondegradative ubiquitin chains can mediate proximity-induced trans-autophosphorylation, although it remains to be robustly established in vivo. The alternative, but nonexclusive, viewpoint that K63 and linear linkages may prolong signaling through stabilization of signaling complexes is also insufficiently understood in vivo. Thus, it is imperative to making progress in understanding IKK signaling that efforts be directed at understanding mechanistically whether and how nondegradative ubiquitination functions in IKK activation and regulation of IKK activity. Finally, as a more complex picture evolves about the role of IκB proteins in NF-κB signaling, future efforts must also address how targeting of IκB proteins by IKK complexes is regulated.
The IκB family: multifaceted NF-κB regulators
The classical role of IκB family proteins is to sequester NF-κB complexes in the cytoplasm, thus inhibiting binding of NF-κB dimers to κB DNA sequences. It is readily apparent, however, that the functions of individual IκB family members are quite heterogeneous and are not limited to this particular role in regulating NF-κB signaling. IκB family proteins are best thought of as NF-κB cofactors or chaperones, which can promote the formation of otherwise unstable NF-κB dimers in both the cytoplasm and the nucleus. IκB binding to DNA-bound transctiption factors, including but not limited to NF-κB, can influence the recruitment of coactivators and, therefore, the transcriptional response. Finally, through interactions with other proteins, IκBs can mediate cross-talk between NF-κB and heterologous signaling pathways. Thus, it has become increasingly clear that IκB proteins contribute immensely to mechanisms that allow NF-κB to participate in diverse biological processes. Therefore, it continues to be important to address the significant gaps in our understanding of IκB regulation.
Among the classical IκBs, IκBα and IκBβ are broadly expressed in all tissues, whereas IκBɛ is expressed only in hematopoietic cells. The precursor proteins p100 and p105 can be either processed to form p52 and p50, respectively, or degraded, resulting in the release of NF-κB complexes. The atypical IκB proteins BCL-3, IκBζ and IκBNS (also known as NF-κBδ and TA-NF-κBH) exhibit a far more limited expression pattern. These atypical IκB proteins are up-regulated following NF-κB activation and, therefore, generally mediate their effects late in the transcriptional response or during secondary responses. Here we review some of the emerging diversity of IκB protein functions (Fig. 7).
Figure 7.
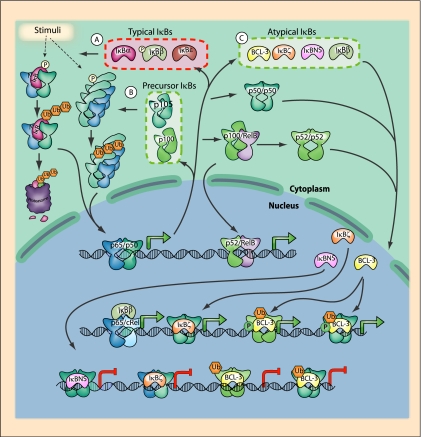
IκB functions. (A) Typical IκB proteins function by promoting cytosolic sequestration of NF-κB dimers. Upon stimulation of the canonical signaling pathway, such IκB proteins are phosphorylated by IKK and targeted for proteasomal degradation. Activation of NF-κB results in resynthesis of IκB proteins. (B) The precursor IκB proteins p100 and p105 have multiple functions. Their constitutive processing results in the generation of p50 and p52 subunits. Unprocessed precursor proteins may, alternatively, form complexes with other NF-κB proteins. Proteasomal degradation or processing, in the case of p100:RelB complexes, is induced by activation of the noncanonical pathway. Proteasomal degradation of p105-containing complexes is mediated by the canonical pathway and can result in the activation of both NF-κB and ERK via release of p105-bound Tpl2. (C) Atypical IκB proteins are induced by various stimuli, including NF-κB activation, and exert both positive and negative effects on NF-κB-mediated transcription. Atypical IκB proteins function by binding to nuclear DNA-associated NF-κB dimers. In the case of IκBβ, newly synthesized hypophosphorylated IκBβ protein acts to augment transcription of p65:c-Rel dimers. BCL-3 may promote or inhibit transcription, depending on various PTMs. IκBζ promotes transcription, whereas IκBNS inhibits transcription, by binding p50 dimers.
IκBα: the prototype
IκBα remains the prototypical member of the IκB family, exhibiting the classical traits that characterize IκB proteins. IκBα is chiefly responsible for the regulation of the prototypical NF-κB complex, the p65/p50 heterodimer. In the resting state, p65/p50 dimers are predominantly sequestered by IκBα (Urban and Baeuerle 1990; Haskill et al. 1991; Kerr et al. 1991; Nolan et al. 1991), and signal-induced release (Ghosh and Baltimore 1990; Haskill et al. 1991) and degradation (Henkel et al. 1993; Mellits et al. 1993; Lin et al. 1995) of IκBα via the proteasome (Palombella et al. 1994; Alkalay et al. 1995; Chen et al. 1995) are necessary for nuclear import and DNA binding by NF-κB. The nuclear NF-κB drives IκBα expression, generating a negative feedback loop (Brown et al. 1993; de Martin et al. 1993; Le Bail et al. 1993; Sun et al. 1993). Therefore, in the absence of IκBα, the termination of NF-κB activation in response to stimuli such as TNFα is significantly delayed (Beg et al. 1995; Klement et al. 1996). The duration of the NF-κB response depends heavily on the kinetics of the feedback pathway (Hoffmann et al. 2002). As a result, the kinetics of NF-κB inactivation can be restored by placing a different IκB (e.g., IκBβ) under the control of the IκBα promoter (Cheng et al. 1998).
IκBα, IκBβ, and IκBɛ are considered traditional IκB proteins; that is, they sequester NF-κB dimers away from κB elements in unstimulated cells, thus inhibiting transcription. Although IκBβ knocked into the genome replacing IκBα can serve analogously to IκBα, there are enough differences between these inhibitors such that it is unlikely that IκBα and IκBβ are truly interchangeable. Indeed, analyses of NF-κB responses in mouse embryonic fibroblasts (MEFs) lacking one, two, or all three IκB proteins demonstrate that they have unique functions, even within a given signaling pathway. The functional characteristics of IκBα, IκBβ, and IκBɛ are somewhat due to temporal differences in their degradation and resynthesis (Hoffmann et al. 2002). More recently, using cells deficient in all three traditional IκB proteins, it has been shown that the long-standing model of cytoplasmic sequestration by IκB proteins is only partially true (Tergaonkar et al. 2005). In particular, cells that lack all three subunits show relatively normal nuclear/cytoplasmic p65 distribution but significantly increased basal NF-κB-dependent gene expression, suggesting that regulation of NF-κB transcriptional activity by IκB proteins is partly independent of cytoplasmic sequestration. This work also definitively confirmed that stimulus-induced activation of canonical NF-κB requires the three typical IκB proteins (Tergaonkar et al. 2005).
IκBɛ: slow starter
Like IκBα, IκBɛ is degraded in an IKK-dependent manner and, following phosphorylation of Ser 157 and Ser 161, undergoes β-TrCP-dependent proteasomal degradation (Whiteside et al. 1997; Shirane et al. 1999). However, IκBɛ degradation and resynthesis occur with considerably delayed kinetics compared with that of IκBα. The difference in the kinetics of IκBɛ and IκBα degradation has profound effects on the nature of the transcriptional response to TNF (Kearns et al. 2006). IκBɛ is thought to selectively regulate p65 homodimers and c-Rel:p65 heterodimers (Simeonidis et al. 1997; Whiteside et al. 1997). IκBɛ is induced slowly, and hence it is thought to suppress late NF-κB gene activation by p65:c-Rel. The origin of the differential kinetics of IκBα and IκBɛ degradation is not clear. One possibility is that the canonical IKK complex exhibits substrate preference for IκBα. Alternatively, active mechanisms may delay IκBɛ degradation. For example, IκBɛ constitutively associates with the PP6 phosphatase holoenzyme through the PP6R1 subunit (Bouwmeester et al. 2004). PP6R1 knockdown results in accelerated degradation of IκBɛ (Stefansson and Brautigan 2006). It is tempting to speculate that ongoing dephosphorylation of IκBɛ S157 and S161 may delay IκBɛ degradation relative to IκBα.
Probably due to the presence of a noncanonical NES, IκBɛ undergoes less nuclear–cytoplasmic shuttling than IκBα and is therefore more restricted to the cytoplasm (Simeonidis et al. 1997; Tam et al. 2000; Lee and Hannink 2002). IκBɛ is primarily expressed in hematopoietic cells, and loss of IκBɛ results in selective defects in hematopoietic lineages. However, in initial analyses, it was shown that IκBɛ loss is largely compensated for by the presence of IκBα (Goudeau et al. 2003; Samson et al. 2004). More recently, it has been observed that B cells lacking IκBɛ have augmented basal and B-cell receptor (BCR)-induced nuclear c-Rel (Clark et al. 2011). IκBɛ is differentially expressed during B-cell development and has been proposed to regulate both p65- and c-Rel-containing NF-κB complexes in B cells (Doerre and Corley 1999; Doerre et al. 2005). The temporal and cell type-specific degradation and expression of IκBɛ support the hypothesis that different IκBs play unique functions in NF-κB responses and indicate that a more detailed analysis of IκBɛ function in vivo is needed.
IκBβ: still mysterious
Like IκBα and IκBɛ, IκBβ is phosphorylated by the IKK complex on two N-terminal serine residues (Box 6); however, the kinetics and pattern of IκBβ degradation are remarkably different (Kerr et al. 1991; Thompson et al. 1995; Tran et al. 1997; Weil et al. 1997). It has long been appreciated that IκBβ behaves quite differently than IκBα, but the physiological relevance of this observation has been unclear. IκBβ undergoes slow degradation and resynthesis, similar to IκBɛ, yet deletion of IκBβ does not dramatically affect the kinetics of the NF-κB responses, as seen upon deletion of IκBα or IκBɛ (Hoffmann et al. 2002; Kearns et al. 2006). However, both the nuclear/cytoplasmic localization and PTM of IκBβ seem to be unique, and IκBβ is capable of associating with NF-κB dimers that are bound to DNA (Thompson et al. 1995; Suyang et al. 1996; Phillips and Ghosh 1997).
Box 6. The IκB degron.
Phosphorylation of the conserved destruction box serine residues (DSGXXS) in IκB proteins induces recognition by βTrCP, followed by K48-linked polyubiquitination by the Skp1–Culin–Roc1/Rbx1/Hrt-1–F-box (SCF or SCRF) family of E3 ligases (Yaron et al. 1998; Fuchs et al. 1999; Kroll et al. 1999; Spencer et al. 1999; Suzuki et al. 1999; Winston et al. 1999; Wu and Ghosh 1999) coordinately with the E2 UbcH5 (Yaron et al. 1998; Spencer et al. 1999). IκBβ (Wu and Ghosh 1999), p100 (Fong and Sun 2002; Fong et al. 2002; Amir et al. 2004; Xiao et al. 2004), and p105 (Orian et al. 2000; Heissmeyer et al. 2001) also possess similarly defined degrons.
Upon stimulation by LPS, IκBβ is degraded with slow kinetics followed by resynthesis and accumulation of a hypophosphorylated form of IκBβ that can be detected in the nucleus (Thompson et al. 1995; Suyang et al. 1996; Phillips and Ghosh 1997; Weil et al. 1997). The crystal structure of IκBβ bound to p65 homodimers suggests that this association might occur in conjunction with binding to DNA (Malek et al. 2003). In vitro, hypophosphorylated IκBβ does not mask the NLS of p65 and can bind DNA with p65 and c-Rel (Suyang et al. 1996; Phillips and Ghosh 1997; Tran et al. 1997). DNA-bound NF-κB:IκBβ complexes are resistant to newly synthesized IκBα, suggesting that hypophosphorylated, nuclear IκBβ may prolong the expression of certain genes (Suyang et al. 1996). Conversely, phosphorylated IκBβ present in unstimulated cells masks the p65 NLS and inhibits binding to DNA in vitro (Suyang et al. 1996; Phillips and Ghosh 1997; Tran et al. 1997).
Recently, two groups, including our own, have reported the characterization of IκBβ knockout mice (Rao et al. 2010; Scheibel et al. 2010). These animal models support the notion that IκBβ has both cytoplasmic and nuclear functions. In the cytoplasm, it appears that IκBβ acts as a traditional IκB by sequestering p65- and c-Rel-containing complexes (Rao et al. 2010; Scheibel et al. 2010). IκBβ may not only inhibit, but also allow, the formation of p65:c-Rel heterodimers (Rao et al. 2010). Stimulus-induced IκBβ degradation allows nuclear translocation and transcriptional regulation by these dimers. As we and others have previously hypothesized, newly synthesized, hypophosphorylated IκBβ binds with NF-κB complexes to DNA. In particular, IκBβ binds p65:c-Rel heterodimers and, based on the effects of the knockout, promotes continued binding to specific κB sites and augments late transcription of select target genes, such as TNF (Rao et al. 2010) and IL-1β (Scheibel et al. 2010). As a result, mice lacking IκBβ show decreased transcription of these important proinflammatory genes and are resistant to LPS-induced septic shock and collagen-induced arthritis (Rao et al. 2010; Scheibel et al. 2010). Thus, both biochemical and genetic evidence support a complex role for IκBβ as both a positive and negative regulator of NF-κB transcriptional responses.
BCL-3—a multipurpose protein
BCL-3 was identified as a proto-oncogene that is overexpressed in B-cell chronic lymphocytic leukemia (Ohno et al. 1990). A clearly delineated role for BCL-3 in NF-κB signaling has remained elusive, although several unique properties have been described. It appears that BCL-3 may both inhibit and facilitate NF-κB-dependent transcription in a context-specific manner. The most striking characteristics of BCL-3, compared with other IκB proteins, is that it has a well-defined TAD and is mainly localized to the nucleus, characteristics consistent with a role as a direct regulator of transcription (Nolan et al. 1993). In the nucleus, BCL-3 is detected bound to p50 and p52 homodimers (Bours et al. 1993; Fujita et al. 1993; Nolan et al. 1993), and the nuclear localization of BCL-3 depends on these interactions. Thus, PTMs affecting association of BCL-3 with DNA-binding proteins can alter the nuclear/cytoplasmic localization of BCL-3. As p50 and p52 lack TADs, it is likely that BCL-3 confers transactivating potential to what are otherwise transcriptional repressors. Alternatively, BCL-3 might facilitate transcription by displacing p50 and p52 homodimers and allowing occupancy by transcriptionally active NF-κB (Hatada et al. 1992; Inoue et al. 1992; Wulczyn et al. 1992; Franzoso et al. 1993; Naumann et al. 1993). BCL-3 may also repress transcription by stabilizing p50 and p52 dimers bound to κB sites.
The ability of BCL-3 to function as a transactivating protein or inhibitor of NF-κB is in part determined by PTMs. BCL-3 is subject to several phosphorylation events that regulate its ability to bind to p50 or p52 homodimers and possibly confer transactivating potential (Nolan et al. 1993; Caamano et al. 1996; Bundy and McKeithan 1997). Thus, the formation of transcriptionally active BCL-3–p52–p52 or BCL-3–p50–p50 complexes may require phosphorylation and ubiquitination of BCL-3 (Bours et al. 1993; Massoumi et al. 2006). The important NF-κB-regulated cell proliferation gene cyclin D1 is known to be regulated by BCL-3. Regulation of cyclin D1 expression depends on the binding of BCL-3–p52–p52 complexes to a κB site in the cyclin D1 promoter (Westerheide et al. 2001). However, when the tumor suppressor transcription factor p53 is active, BCL-3 levels decrease, and instead p52 homodimers bind to the same κB site within the cyclin D1 promoter, thereby recruiting histone deacetylase 1 (HDAC1) to repress transcription (Rocha et al. 2003).
Transcriptional activation by BCL-3 depends on its localization to the nucleus, which has recently been shown to depend on ubiquitination. Furthermore, ubiquitination of BCL-3 is negatively regulated by CYLD, a DUB and known negative regulator of NF-κB signaling (Massoumi et al. 2006). Because CYLD is itself an NF-κB-regulated gene, these findings may help to clarify some of the confusion surrounding BCL-3 function: To form transcriptionally active complexes, BCL-3 must be induced either in the absence of CYLD or under conditions in which BCL-3 resists targeting by CYLD. When CYLD and BCL-3 are coexpressed, repressive p50 or p52 homodimers are displaced by deubiquitinated BCL-3 (Massoumi et al. 2006).
BCL-3 may also function as an inhibitor of NF-κB activity by stabilizing repressive NF-κB dimers in a DNA-bound state and preventing the binding of transcriptionally active dimers. Stabilization of repressive complexes by induction of BCL-3 has therefore been proposed to function in processes such as LPS tolerance (Wessells et al. 2004; Carmody et al. 2007). BCL-3 may prevent ubiquitination of p50 homodimers and their replacement with transcriptionally active NF-κB dimers (Carmody et al. 2007). Inhibition of NF-κB by this mechanism would occur at κB sites that exhibit a strong binding preference for p50 dimers. BCL-3 expression is regulated by NF-κB, which may result in termination of the late stage p50-dependent transcriptional responses (Brasier et al. 2001; Ge et al. 2003). Expression of BCL-3 can also be regulated independently of NF-κB. For example, IL-4 acting through AP1 (Rebollo et al. 2000), and IL-6 acting through Stat3 (Brocke-Heidrich et al. 2006) can induce BCL-3 expression. However, it is the interplay of the anti-inflammatory cytokine IL-10 with BCL-3 that has received the most attention. IL-10 treatment induces the expression of BCL-3, and this leads to inhibition of LPS-induced TNF production (Kuwata et al. 2003). In IL-10-deficient mice, BCL-3–p50–p50 complexes are decreased, and p65-dependent cytokine production (e.g., IL-23) is enhanced (Muhlbauer et al. 2008). Finally, BCL-3 may also be a cofactor for other transctiption factors. BCL-3 binds TORC3 (transducer of regulated CREB activity 3) and Tax/pCREB complexes and represses transcription by preventing p300 recruitment to the HTLV promoter (Hishiki et al. 2007; YM Kim et al. 2008). A recent report identified ERRα and PPARα as BCL-3-binding partners (Yang et al. 2009). BCL-3 appears to act with the coactivator PGC1α (PPARγ coactivator 1α) in the regulation of a subset ERRα- and PPARα-dependent transcriptional responses (Yang et al. 2009).
Thus, BCL-3 can mediate a complex set of functions by regulating the function of NF-κB and non-NF-κB transctription factors. Although loss of BCL-3 reveals a relatively mild phenotype (Franzoso et al. 1997), BCL-3 has multiple described roles in vivo, particularly in lymphocyte development and function. BCL-3 knockout mice have well-described defects in T-cell responses and fail to develop antigen-specific responses (Franzoso et al. 1997; Schwarz et al. 1997). Induction of BCL-3 in T cells during infection or immunization promotes the survival of activated T cells (Mitchell et al. 2001; Valenzuela et al. 2005; Bauer et al. 2006). BCL-3 also exhibits anti-inflammatory function by limiting granulopoiesis under settings of acute inflammation (Kreisel et al. 2011). It remains to be determined whether the effects of BCL-3 on granulopoiesis—or, for that matter, similar effects on T-cell differentiation and survival (Rangelova et al. 2008)—are due to repression of p50- or p52-dependent transcriptional responses. Thus, BCL-3 restricts inflammation by suppressing proinflammatory gene expression and inducing anti-inflammatory gene expression, as well as through regulation of granulocyte responses.
IκBζ—an induced transcription activator
IκBζ (MAIL and INAP) was identified through its sequence similarity to BCL-3 (Kitamura et al. 2000; Haruta et al. 2001; Yamazaki et al. 2001), with which it also exhibits some functional homology. IκBζ, like BCL-3, can augment transcription in association with p50 NF-κB dimers. Unlike BCL-3, it is thought that expression levels rather than PTMs primarily regulate the contribution of IκBζ to NF-κB responses. IκBζ expression is regulated by NF-κB; however, an increase in IκBζ protein levels occurs only in response to select NF-κB signaling pathways; in particular, those depending on MyD88 (Kitamura et al. 2000; Haruta et al. 2001; Yamazaki et al. 2001; Eto et al. 2003; Muta et al. 2003). It was subsequently shown that stabilization of the IκBζ transcript and IκBζ protein expression depends on additional signaling provided by TLR/IL-1R, but not TNF (Yamazaki et al. 2005). The mechanism of IκBζ regulation by IL-1 is unclear. IL-17 stimulation also stabilizes IκBζ mRNA; however, because IL-17 alone does not induce transcription of IκBζ, costimulation with TNF and IL-17 is required to induce IκBζ protein up-regulation (Yamazaki et al. 2005). It has recently been shown that signaling through IL-17R can activate NF-κB (Bulek et al. 2011; Sun et al. 2011). Surprisingly, stabilization of IκBζ mRNA is independent of TRAF6 (Hartupee et al. 2009), and the components upstream of TRAF proteins are not shared between the Toll/IL-1R and IL-17R pathways (Gaffen 2009). Therefore, the common signal between IL-17R and IL-1R that leads to IκBζ protein expression is not obvious. Recent work suggests that IκBζ mRNA contains both transcriptional silencing and destabilizing elements and implicates the kinase IRAK-1 (IL-1R-associated kinases-1), downstream from IL-1R, in the regulation of IκBζ expression (Hartupee et al. 2007, 2009; Dhamija et al. 2010). Although IL-17 seems to act via the same mechanism in regulating IκBζ translation (Dhamija et al. 2010), IRAK-1 is not required for IL-17R signaling (Gaffen 2009). However, IL-17R does signal through Act1, and IL-17 fails to stabilize IκBζ in the absence of Act1 (Chang et al. 2006; Hartupee et al. 2009). Therefore, IL-1 and IL-17 induce IκBζ expression through distinct pathways, depending on IRAK-1 and Act1, respectively, which likely culminate in the same mechanism of IκBζ mRNA stabilization and translation.
Once induced, IκBζ acts to selectively augment NF-κB-dependent transcriptional responses (Yamamoto et al. 2004b). Like BCL-3, IκBζ binds p50 dimers at κB sites, most notably on the IL-6 promoter. Although IκBζ does not have a well-defined TAD, there is some evidence to suggest that, at least upon overexpression, IκBζ can impart transactivating capacity to p50 dimers (Motoyama et al. 2005). Consequently, IL-6 is not induced by LPS in IκBζ-deficient cells (Yamamoto et al. 2004b). In contrast to its role with p50, IκBζ may negatively regulate p65-containing NF-κB complexes (Motoyama et al. 2005). Indeed, a splice variant of IκBζ, IκBζ-D, lacking the N-terminal region that has been implicated in transactivation, may selectively act as a transcriptional inhibitor (Motoyama et al. 2005). Consistent with the proposed inhibitory role of IκBζ, a slight elevation of some p65-dependent transcripts is observed in cells from IκBζ-deficient mice (Yamamoto et al. 2004b). Perhaps as a consequence of up-regulation of p65-regulated genes, IκBζ-deficient mice exhibit a proinflammatory phenotype. IκBζ-deficient mice exhibit atopic dermatitis with elevated cytokine production in skin (Shiina et al. 2004) and increased serum TNF levels following LPS treatment (Yamamoto et al. 2004a). Finally, like BCL-3, IκBζ may also regulate transcription in an NF-κB-independent manner. IκBζ knockout mice are deficient in the development and induction of proinflammatory TH17 cells (Okamoto et al. 2010). This failure of TH17 development is due to impaired transcription of the IL-17A gene. However, neither IκBζ-mediated induction of IL-17A nor TH17 development requires p50. Instead, it appears that IκBζ cooperates with, but may not directly bind to, the nuclear orphan receptors RORα and RORγ (Okamoto et al. 2010). It remains to be seen how IκBζ is recruited to the il17a promoter and through what mechanism the IκBζ TAD acts to drive transcription.
IκBNS—an induced transcription repressor
IκBNS was identified as an inducibly expressed IκB protein in T cells undergoing negative selection (Fiorini et al. 2002) but was subsequently shown to be inducibly expressed in multiple cell types. Unlike typical IκB proteins, IκBNS overexpression only marginally inhibits NF-κB (Fiorini et al. 2002). Like BCL-3, IκBNS is strongly induced by the anti-inflammatory cytokine IL-10 and appears to mediate some of the repressive effects of IL-10 on proinflammatory cytokine production during TLR stimulation (Hirotani et al. 2005). Given that IκBNS shows a preference for binding to p50 (Fiorini et al. 2002), it has been assumed that it inhibits NF-κB-dependent proinflammatory gene expression by stabilizing p50 homodimers at κB sites. This effect is selective, as in IκBNS knockout cells, IL-6 and IL-12p40 are increased following LPS stimulation, but not TNF (Kuwata et al. 2006). Recently, it has also been shown that in “regulatory” dendritic cells (DCs), a subset of DCs that produce high levels of IL-10 in response to LPS stimulation, both BCL-3 and IκBNS are induced following stimulation with LPS (Fujita et al. 2006). However, despite the initial characterization of IκBNS in thymocyte selection, knockout mice have normal T-cell development and function (Kuwata et al. 2006; Touma et al. 2007), except for mildly decreased IL-2 production and, consequently, decreased proliferative capacity (Touma et al. 2007). Therefore, whereas IκBNS bound to p50 homodimers on DNA is an inducible negative regulator of NF-κB activity, it might also increase the expression of a small subset of genes, although the mechanism by which it acts in this capacity remains to be defined.
p100—NF-κB and multifunctional IκB
The precursor protein p100 adds yet another level of complexity to the IκB family. The protein p100 is encoded by NFKB2 and, like other IκB proteins, is induced in response to NF-κB stimuli. Both p100 and p52 can bind other NF-κB subunits, while only p52 can bind to DNA (Naumann et al. 1993; Scheinman et al. 1993). p100 can function as an IκB, although in unstimulated cells, p100 can undergo constitutive processing to yield p52 (Xiao et al. 2004; Qing and Xiao 2005). IKKα and NIK (Senftleben et al. 2001a; Xiao et al. 2001, 2004) can phosphorylate p100 on a C-terminal degron with sequence homology with Lys 22 of IκBα, leading to either degradation or processing to p52 (Fong et al. 2002; Amir et al. 2004; Liang et al. 2006). NIK both phosphorylates and activates IKKα and augments binding between IKKα and p100 (Vallabhapurapu et al. 2008; Zarnegar et al. 2008). Full-length p100 binds several NF-κB family members, but its function is most closely intertwined with that of RelB. RelB appears to associate primarily with p100 or p52 and, in fact, requires p100 binding for stability (Solan et al. 2002; Fusco et al. 2008). As a result of selective binding of RelB to p100, constitutive processing of p100 in certain tissues may result in constitutive production of RelB/p52 dimers and basal NF-κB activity (Qing and Xiao 2005). Such activity has been demonstrated in lymphoid organs, and as a consequence, Relb−/− mice show decreased NF-κB activity in these tissues (Weih et al. 1995). Induction of p100 expression by NF-κB can also facilitate the exchange of p65-containing dimers for RelB/p52 dimers (Saccani et al. 2003). Because induced RelB/p52 dimers are resistant to IκBα, they may promote late transcription by displacing p65-containing dimers, thereby overcoming negative feedback mechanisms at late stages of the response (Saccani et al. 2003). However, at other promoters, including those of Il1b and Tnf, RelB may act as a transcriptional repressor, and because RelB is induced at later time points, it may have a functional role in the maintenance of LPS tolerance or termination of transcriptional responses(Saccani et al. 2003; Yoza et al. 2006; El Gazzar et al. 2007).
Although stabilization of RelB dimers may be regulated exclusively by p100, p100 itself can act more broadly in inhibiting NF-κB dimers. The p100 protein can regulate a fraction of canonical, p65-containing NF-κB dimers through pathways that signal through IKKα (Basak et al. 2007). Thus, induced p100 may act in a more typical IκB feedback loop, in that newly expressed p100 may regulate p65-containing dimers (Basak et al. 2007; Shih et al. 2009). An increase in p100 and, consequently, p52 levels will promote RelB:p52 complexes, whereas increased p100 binding to “canonical” NF-κB complexes would facilitate activation of these complexes by signals that activate IKKα. For example, following T-cell receptor stimulation, p100 is up-regulated in T cells and forms complexes with p65-containing dimers (Ishimaru et al. 2006). Presumably, formation of such complexes would fundamentally alter the consequences of activating the noncanonical pathway in that NIK/IKKα-dependent phosphorylation of p100 would liberate both p65- and RelB-containing NF-κB dimers. Thus, p100 can exhibit properties that are representative of several important IκB functions: p100 may act as a typical IκB by sequestering NF-κB-containing complexes—both p65- and RelB-containing—in the cytoplasm; p100 can shape the pool of available NF-κB dimers, not only through processing to p52, but also through stabilization of RelB; and p100 can function as an induced IκB due to the high level of p100 transcriptional regulation.
p105—from IKK to ERK
The precursor protein p105, encoded by NFKB1, like p100, serves as both a NF-κB subunit precursor and an IκB protein. In unstimulated cells, p105 undergoes a constitutive rate of proteasomal processing to p50 (Fan and Maniatis 1991; Palombella et al. 1994; L Lin et al. 1998). Constitutive p105 processing to p50 may occur both cotranslationally (L Lin et al. 1998) and post-translationally (Fan and Maniatis 1991; Moorthy et al. 2006). Limited proteolysis of the precursor protein, which generates p50, is independent of ubiquitination and is likely mediated by the 20S proteasome (Moorthy et al. 2006). The p105 glycine-rich region (GRR; amino acids 376–404) serves as a stop signal for proteolysis, perhaps by acting as a poor proteolytic substrate (Lin and Ghosh 1996; Orian et al. 1999; Moorthy et al. 2006). Thus, rather than undergoing complete degradation, p105 is processed from the C terminus to the GRR, resulting in liberation of p50.
In addition to giving rise to the p50 NF-κB subunit, several additional functions have been ascribed to p105. First, p105 can act like a typical IκB protein (Rice et al. 1992; Mercurio et al. 1993; Sriskantharajah et al. 2009) by binding NF-κB dimers and undergoing signal-induced phosphorylation and degradation. This function is dependent on the generation of p105 bound to other NF-κB proteins. Indeed, the formation of heterodimeric and homodimeric complexes with other Rel proteins likely inhibits constitutive processing of p105 (Harhaj et al. 1996; Cohen et al. 2001; Moorthy et al. 2006; Shih et al. 2009). Mature p105 bound to another NF-κB family member may be processed to p50: For example, if bound to p65, p50:p65 heterodimers would be produced. If p105 is not rapidly processed to p50, then the remaining C-terminal domain will mediate the formation of stable higher-order NF-κB complexes; e.g., p105:p65:p105:p65 (Shih et al. 2009). The IκB activity mediated by this p105 pool is likely to be both heterogenous and highly dependent on the relative translation of p105 and other NF-κB proteins (Shih et al. 2009). As p105 will undergo rapid dimerization upon synthesis, the nature of the dimer depends on the pool of available binding partners, the NF-κB milieu. As different dimers may well have differing sensitivity for 20S proteasomal processing, the relative level of p105 versus p50 generated may vary depending on cell type and status.
Activation of stable p105-inhibited NF-κB complexes likely occurs through activation of the canonical NF-κB pathway. Multiple reports have demonstrated that IKKβ-dependent phosphorylation of p105 C-terminal serines, including Ser 923 and Ser 927, is followed by inducible degradation of p105 (Fujimoto et al. 1995; MacKichan et al. 1996; Heissmeyer et al. 1999, 2001; Orian et al. 2000; Salmeron et al. 2001; Lang et al. 2003; Cohen et al. 2004). Phosphorylation of p105 Ser 923/927 leads to SCFβ-TrCP binding, polyubiquitination at multiple lysine residues, and 26S proteasome-mediated degradation of the entire protein (Harhaj et al. 1996; Orian et al. 1999; Cohen et al. 2004).
Through interactions with several non-NF-κB pathway proteins, p105 has been associated with other signaling pathways. The best studied of these is TPL-2, an upstream kinase in the MKK1/2–Erk1/2 pathway (Gantke et al. 2011). Through this interaction and IKK, in particular IKKβ, p105 can regulate the activation of ERK. IKKβ-induced degradation of p105 leads to the release and activation of TPL2, rendering IKKβ and p105 critical for both NF-κB and MAPK activation (Beinke et al. 2004; Waterfield et al. 2004). Additional regulatory functions of p105 may be mediated through other interaction partners, such as the helix–loop–helix transcription factor LYL1, c-FLIP, coatomer-β subunit protein COPB2, JNK-interacting leucine zipper protein (JLP), and ABIN proteins (Ferrier et al. 1999; Li et al. 2003; Bouwmeester et al. 2004; Zhang et al. 2004).
Other potential IκBs
In addition to the IκB proteins described above, several additional potential IκB-like proteins have been identified in humans and mice. These include IκBR (Ray et al. 1995), IκBn (Yamauchi et al. 2010), and IκBL (Albertella and Campbell 1994). Despite the naming of these proteins as IκBs, IκBR and IκBL are not established IκB family members. IκBR (NFKBIL2) is encoded in the human MHC complex and was initially reported to inhibit p50-containing dimers (Ray et al. 1995). However, analysis of the IκBR gene suggests no relationship to the IκB family (Norman and Barton 2000). Recent work has implicated IκBR as a regulator of DNA replication and genomic stability (Duro et al. 2010; O'Connell et al. 2010; O'Donnell et al. 2010; Piwko et al. 2010). While it is tempting to speculate that IκBR could participate in DNA damage-induced NF-κB activation, the data do not support any function connected directly to regulation of NF-κB. Therefore, IκBR/NFKBIL2 is not an IκB family member. In contrast, an IκBL (NFKBL1) transgenic mouse model affects NF-κB-dependent gene expression, suggesting that this ankyrin repeat-containing protein may have an IκB-like function (Chiba et al. 2011). However, IκBL has been shown not to bind NF-κB proteins or alter NF-κB-driven reporter activity (Greetham et al. 2007). Thus, existing evidence also does not support a role for IκBL as an IκB family member. IκBn, the most recently reported IκB-like protein, is primarily nuclear in localization and appears to augment the transcription of certain target genes by binding to NF-κB p50 (Yamauchi et al. 2010). Thus, while IκB-R and IκB-L are not IκB family members, more work remains to be done to better determine whether IκBn is a bona fide IκB.
Thus, the IκB family is now appreciated to function in multiple aspects of the NF-κB response (Fig. 7). In the cytoplasm, IκB proteins can both inhibit, through sequestration, and facilitate, through stabilization of otherwise unstable complexes, specific aspects of the NF-κB transcriptional response. In the nucleus, IκB proteins also exert pleiotropic effects. IκBs may act to remove NF-κB complexes from the DNA, stabilize NF-κB complexes bound to DNA, and alter the recruitment of coactivators to DNA-bound NF-κB complexes. Future research will need to more fully address the differential regulation of these distinct functions, particularly in instances where individual IκB have been shown to exert divergent biological effects.
Transcription factor NF-κB
The classical model of NF-κB regulation by cytoplasmic sequestration is inherently satisfying. Perhaps as a result, the mechanistic basis of how NF-κB dimers actually mediate expression of target genes has not been explored in sufficient detail. NF-κB family members are capable of binding to multiple transcriptional coregulators, including both HDACs and histone acetyltransferases (HATs). As NF-κB lacks intrinsic enzymatic activity, it is through recruitment of HATs, including CBP and p300, and HDACs, including HDAC1 and HDAC3, that this transcription factor family modifies transcriptional responses. However, it is not yet clear that coregulator recruitment to κB sites is the only mechanism through which NF-κB family members can regulate transcription. What is clear is that release of NF-κB dimers from IκBs is not sufficient to explain the complex regulation of NF-κB responses. Instead, the regulation of transcriptional responses by NF-κB exhibits several additional layers of complexity (Box 7).
Box 7. Regulation of transcriptional specificity.
NF-κB, not unlike other transcription factor families, recognizes a degenerate DNA-binding site that is widely dispersed throughout the genome. However, under many circumstances, activation of inducible transctiption factors leads only to the expression of a subset of the genes that they are capable of activating. Therefore, in addition to binding site specificity, there are additional mechanisms that determine which target genes are transcribed when NF-κB is activated (Hoffmann et al. 2006).
Transctiption factor dimerization. NF-κB family proteins, like members of many other transctiption factor families, are capable of homodimerization and heterodimerization that can alter both their mechanisms of regulation and their DNA-binding site specificity (Smale 2011). Thus, cells expressing a distinct complement of NF-κB dimers will exhibit distinct transcriptional specificity, as will signaling pathways that activate only a subset of NF-κB dimers.
Transcription factor PTMs. Phosphorylation, SUMOylation, ubiquitinylation, acetylation, and other modifications can affect the localization, stability, and ability of NF-κB proteins to interact with DNA and transcriptional cofactors. Induction of these modifications can be differentially regulated by specific signaling pathways.
Heterologous transctiption factors. When multiple transctiption factors are activated together, as is often the case, transctiption factor cross-talk in the form of both coordinated recruitment and competition for cofactors results in either transcriptional synergy or antagonism. Genes with promoters requiring the simultaneous binding of NF-κB and another transctiption factor will thus be specifically transcribed only in response to signals that activate both transctiption factors. NF-κB can also directly interact with heterologous transctiption factors, again requiring the activation of both transctiption factors for the transcription of specific genes.
Cell state. During cellular differentiation, durable changes in chromatin structure that selectively alter DNA accessibility are acquired, resulting in cell type-specific transcriptional profiles. Less permanent changes in chromatin structure also accompany altered cellular status brought about by signaling events. Therefore, the kinetics of transctiption factor activation, both homologous and heterologous transctiption factors, can determine which genes NF-κB can subsequently activate.
As there are five members of the NF-κB family capable of forming various homodimers and heterodimers, consequently, there are a variety of potential, functional NF-κB units that may regulate distinct transcriptional programs. Binding to heterologous transcription factors either directly or in the context of DNA-associated enhanceosomes further broadens the range of NF-κB target genes. Finally, multiple PTMs targeting NF-κB subunits that affect function and stability; subcellular and subnuclear localization; and binding to DNA, cofactors, and heterologous transcription factors have all been described. Although much of what we know about the regulation of transcription by NF-κB has been based on analyses of the p65:p50 heterodimer, specific functions for individual NF-κB subunits and specific dimer pairs are increasingly being described (Smale 2011; Siggers et al. 2012). Such inherent complexity of the NF-κB system may explain some of the variety in the biological functions that are dependent on NF-κB. Although there has been great progress in understanding the regulation of specific gene expression programs by NF-κB, this is an area in which much remains to be understood. There are outstanding questions about some of the most fundamental aspects of how NF-κB controls transcription of target genes. A detailed analysis of the biological contribution of individual NF-κB complexes to transcriptional programs is well beyond the scope of this review. Instead, we focus here on a more general discussion of what is known and what remains as yet unanswered about how NF-κB regulates transcription.
It is generally thought that NF-κB induces transcription through the recruitment of coactivators that, in turn, alter the chromatin environment to allow transcription to occur. Transcription in eukaryotes classically depends on the assembly of the preinitiation complex (PIC) at the core promoter sequence flanking the transcription start site (Kornberg 2007). The core promoter consists of ∼80 base pairs (bp) and may contain a TATA element located ∼27 bp upstream of the transcriptional start site. Recognition of the core promoter is the first step of transcription and is therefore thought to be the primary event regulated by NF-κB. In the classic scenario of a TATA-containing core promoter leading to RNA polymerase II-mediated transcription, the TATA-binding protein (TBP) binds to the TATA box, leading to formation of the TFIID-containing general transcription factor complex. Other components of the TFIID complex, such as TAF subunits, also make contact with DNA at the downstream core, promoter, and initiator elements and can mediate assembly of the complex at TATA-less promoters. For example, TFIIA augments binding of TFIID to the core promoter elements. Recruitment of RNA polymerase II, along with the other general transcription factors TFIIB, TFIIE, TFIIF, and TFIIH, which includes DNA helicase activity, and the large mediator coactivator complex, consisting of co-activators and corepressors, completes the PIC and is sufficient for transcription to begin. However, in vivo, transcription also requires additional modifications of both the PIC and the chromatin. Therefore, recruitment of transcriptional activators, chromatin-modifying enzymes, and elongation factors by transctiption factors is essential for triggering transcription even after PIC assembly. Indeed, it is now increasingly appreciated that for many inducibly transcribed genes, the PIC is preassembled and poised at core promoter elements (Kim et al. 2005; Bernstein et al. 2006; Guenther et al. 2007). Thus, it appears that for many rapidly activated genes, inducible transcription depends on effects that target post-PIC steps of transcription: transcription initiation, release of paused polymerase II, and transcriptional elongation. While NF-κB was historically thought to act primarily by promoting PIC assembly, recent data have demonstrated a role for NF-κB in both pre- and post-PIC regulation of transcription. While selective regulation of these activities is clearly crucial for transcriptional specificity, our current understanding of the regulation of transcription by NF-κB discriminates poorly between pre- and post-PIC functions. This failure can be traced in part to a lack of detailed understanding of coregulator recruitment by NF-κB dimers. Therefore, it is worth discussing what is known about this process and its regulation and what remains to be done to better understand the regulation of transcription by NF-κB.
NF-κB recruits coregulators to DNA
NF-κB family members are capable of recruiting HATs and HDACs to the enhancers and promoters of target genes. HATs, including PCAF, CBP, and p300, can promote open chromatin structure through histone acetylation and allow PIC assembly and transcription. However, this is not the sole mechanism through which these coregulators affect transcription, as HAT activity has been shown to be dispensable for some p300 functions (Kimbrel et al. 2009). NF-κB subunits have intrinsic differences in their ability to bind to HATs and HDACs and, therefore, different capacities to induce or repress transcription. NF-κB p50 and p52, which lack a TAD, bind only to HDACs and, therefore, when binding to DNA alone, act primarily as transcriptional repressors.
PTMs influence coregulator recruitment
Numerous regulatory PTMs of NF-κB have been reported and shown to have effects on transcriptional responses (Box 8). PTMs targeting NF-κB can be mediated by components of both the NF-κB or heterologous signaling pathways. By providing an additional layer of regulation to transcriptional responses, it is thought that NF-κB PTMs prevent inadvertent induction of target gene transcription and also provide an additional means of generating specificity in transcriptional programs. In addition, it is increasingly apparent that PTMs function in the termination of NF-κB transcriptional responses.
Box 8. P65 PTMs. Phosphorylation.
Several kinases, including IKKα and IKKβ, are reported to phosphorylate p65 within the TAD region at S536 (Sizemore et al. 2002; Sakurai et al. 2003; Yang et al. 2003; O'Mahony et al. 2004). In cell lines, an S536A mutation prevents CBP recruitment (Chen et al. 2005), although this result appears to be inconsistent (Chen et al. 2005). Ser 529 within the TAD is also inducibly phosphorylated, with unclear consequences (Bird et al. 1997; Wang and Baldwin 1998; Wang et al. 2000). Phosphorylation has also been reported at S311, within the dimerization domain of p65 (Duran et al. 2003). PKCζ can phosphorylate, and reduced binding of CBP to p65 occurs upon stimulation of PKCζ-deficient cells (Duran et al. 2003). The role of the S276 phosphorylation is discussed in the text.
Acetylation. Although well known to bind histone acetyltransferases (HATs) and recruit these coactivators to DNA, it was some time before it was found that p65 could also be a HAT substrate (Chen et al. 2001). While multiple lysines in p65 can be acetylated, most investigations have focused on K310 (Chen et al. 2002). Acetylation of K310 is blocked in the absence of S276 phosphorylation (Chen et al. 2005), consistent with the requirement for S276 phosphorylation for HAT recruitment (Zhong et al. 1998; Dong et al. 2008). S536 phosphorylation also promotes K310 acetylation (Chen et al. 2005), perhaps by decreasing the association of histone deacetylases (HDACs) with p65 (Hoberg et al. 2006).
Others. Ubiquitination of p65 was initialy reported to regulate DNA binding and stability of nuclear p65 (Ryo et al. 2003; Saccani et al. 2004). Subsequently, it was also reported that p65 ubiquitination may regulate the subnuclear localization of p65 (Maine et al. 2007; Tanaka et al. 2007). Poly(ADP-ribosyl)ation of p65 has been reported and is thought to affect nuclear localization of p65 by reducing the association of p65 with the nuclear export machinery (Kameoka et al. 2000; Zerfaoui et al. 2010). However, it is not clear that targeting of p65 or p50 for direct poly(ADP-ribosyl)ation, as opposed to autopoly(ADP-ribosyl)ation, of PARP1 or targeting of other substrates is responsible for the lack of NF-κB transcriptional responses in PARP1-deficient cells (Chang and Alvarez-Gonzalez 2001; Hassa et al. 2005). Monomethylation of p65 has been reported to augment promoter binding and target gene transcription (Ea and Baltimore 2009; Lu et al. 2009). S-Nitrosylation of cysteine residues in p50 (DelaTorre et al. 1997; Marshall and Stamler 2001) and tyrosine and cysteine residues in p65 (Park et al. 2005; Kelleher et al. 2007; Nishida et al. 2011) has been reported to inhibit transcriptional responses.
Inducible phosphorylation of p65 was described >15 years ago (Naumann and Scheidereit 1994; Neumann et al. 1995). Phosphorylation of p65 on Ser 276 is catalyzed by PKA, which is constitutively associated with cytosolic p65-containing complexes (Zhong et al. 1997). Following degradation of IκBα, PKA phosphorylates p65 on Ser 276, leading to the interaction of p65 with the transcriptional coactivator CBP/p300 (Zhong et al. 1998, 2002). In subsequent studies, multiple additional kinases have been shown to phosphorylate Ser 276 , including MSK1 and MSK2 (Hoffmann et al. 2006). Loss of both MSK1 and MSK2 reduces TNF-induced transcriptional responses, consistent with the proposed role of S276 phosphorylation (Vermeulen et al. 2003). The physiological importance of S276 phosphorylation was established by knocking in a serine-to-alanine mutation at position 276. These experiments demonstrated the inability of p65 that cannot be phosphorylated at Ser 276 to recruit CBP and p300 to the κB sites and led to a lack of inducible expression of a subset of NF-κB target genes (Dong et al. 2008). Conversely, introduction of a phosphomimetic mutation at this site resulted in constitutive recruitment of CBP by p65 homodimers, resulting in systemic inflammation via aberrant expression of TNF (Dong et al. 2010). These and other findings highlight two fundamental aspects of NF-κB biology: First, NF-κB family members, like other transcription factors, lack enzymatic activity and therefore can effectively be considered specialized adapter proteins linking DNA sequences to enzymatic transcriptional coregulatory proteins. Second, regulation of the capacity of NF-κB to bind DNA and coregulators confers inducibility to the NF-κB system.
Because NF-κB lacks enzymatic activity and cannot recruit CBP in the absence of S276 phosphorylation, one might predict that the knock-in mouse expressing S276A would lack all p65-dependent transcription. Instead, expression of only a subset of NF-κB-regulated genes is affected (Dong et al. 2008). Because phosphorylation of S276 is critical for CBP binding and displacement of HDAC, NF-κB-regulated genes must differ in their requirement for induced histone acetylation, although the basis of this difference remains unclear. These data are consistent with earlier studies that showed that NF-κB-regulated genes can be categorized by their requirement for chromatin modification (Natoli et al. 2005). Initially, NF-κB-dependent genes were subdivided into those with constitutively and immediately accessible (CIA) promoters, which do not require chromatin modification, and promoters with regulated and late accessibility (RLA), which are dependent on stimulus-induced chromatin modification (Saccani et al. 2001). This basic classification has been extended to demonstrate that chromatin remodeling complexes are used differentially for inflammatory gene expression (Sanjabi et al. 2005).
Can NF-κB be a pioneer transcription factor? Several recent reports, including from our own laboratory, have suggested a role for NF-κB as a so-called pioneer transcription factor. It is quite clear that NF-κB can promote transcriptional responses that rely on activation and/or recruitment of additional transcription factors. However, a pioneer transcription factor should be one that is capable of binding to nucleosomal DNA. It could be argued that this might be an important property of effective inducible transcription factors as well, as many inducibly activated genes have enriched nucleosomal content (Cairns 2009). It is generally believed that NF-κB dimers could not bind to nucleosomal DNA in part because the structural relationship between NF-κB and DNA leaves little space for the nucleosome. This would suggest, therefore, that NF-κB could not act as a pioneer transcription factor. However, at least for p50, it has been shown that NF-κB can readily bind nucleosomal DNA in vitro (Angelov et al. 2004). Consistent with these data, both p50 and p52 were found constitutively associated with nucleosomes in a proteomics approach (Bartke et al. 2010). Therefore, it seems plausible that NF-κB is a true pioneer transcription factor. Recruitment of HATs by NF-κB may result in nucleosome remodeling, providing DNA access to other transcription factors. Alternatively, it is also possible, as has been suggested (Natoli et al. 2005), that NF-κB binding to a nucleosomal κB site could fix the nucleosome in a given position. Indeed, it was found that p50 binding to nucleosomal κB sites had little effect on nucleosome structure (Angelov et al. 2004), suggesting that in the absence of coregulatory proteins, NF-κB binding could inhibit nucleosome remodeling. Further studies are needed to assess the binding of NF-κB to nucleosomal κB sites in vivo and the consequences of such binding for specific transcriptional responses.
Nucleosome remodeling complexes (e.g., SWI/SNF) relocate or remove histones, providing access to DNA for the PIC or other transctiption factors. The precise mechanism by which nucleosomal remodeling complexes are regulated by transctiption factors remains unclear. The primary mechanism is thought to be histone acetylation via HAT recruitment. Acetylation marks histones for remodeling, allowing promoter access for inducible gene expression. The interplay between NF-κB and nucleosome remodeling is complex. It is thought that for many late genes, particularly those that require de novo protein synthesis for transcription, access of NF-κB to binding sites requires Swi/Snf-mediated nucleosomal remodeling (Natoli 2009). Brg1/Brm-dependent chromatin remodeling is required for most late-activated NF-κB-dependent genes (Ramirez-Carrozzi et al. 2006). It is thought that nucleosome remodeling precedes NF-κB binding in these cases, although this has not been definitively established for large numbers of genes (Natoli 2009) and may depend on the structure of the nucleosome. As discussed recently (Natoli 2009), nucleosomes in transcription start site-proximal promoter elements may be particularly resistant to NF-κB binding. As a result, Swi/Snf-mediated nucleosome remodeling, independent of NF-κB, may be required for NF-κB binding to some nucleosomal κB DNA sites (Weinmann et al. 1999, 2001; El Gazzar et al. 2010)
Concluding remarks
There has been incredible progress made over the past 25 years in understanding the regulation and biology of the NF-κB family of transcription factors. This work has also shed considerable light on general mechanisms of inducible transcription, cell signaling, and immune and inflammatory responses. Based on these many areas of progress, there is a sense that many of the fundamental questions about the biology of this transcription factor family have been answered, and what remains to be understood are details. However, as is apparent from our condensed overview of the field, there are numerous major questions that remain to be answered. Indeed, the quantity of research in this field and the detailed understanding of NF-κB signaling pathways and transcription responses that has, in some cases, resulted from these efforts provide the tools needed to tackle these big picture questions. Technical, methodological, and computational advances now offer the opportunity to address some of these fundamental questions that have either eluded clear answers or been set aside in recent years. We organized this review around three such areas in which significant progress over the past quarter-century has equipped the field to make even more significant advances in coming years: the regulation of IKK activity by receptor/adapter complexes, the regulation of NF-κB transcriptional activity by the IκB family, and the mechanism through which NF-κB induces transcription of target genes. Just as the NF-κB pathway has been the vanguard of knowledge in immune signaling and inducible transcription, so, too, will research into these outstanding issues increase our understanding of signaling and inducible transcription more broadly.
While progress in understanding many facets of NF-κB biology has been undeniably remarkable, in one key respect, research on NF-κB has failed to live up to its potential. Although implicated in a host of human ailments, no pharmaceutical approach designed to selectively inhibit the NF-κB pathway has yet been shown to have efficacy in human disease. This failure is, perhaps, the clearest evidence of what yet remains to be accomplished in the future. The utility of NF-κB blockade in laboratory models of inflammatory, autoimmune, and oncological diseases has been proven time and again using both pharmacological inhibitors and genetically modified animals. Yet concerns about untoward consequences of strong NF-κB inhibition, as revealed in studies on genetic and pharmacologic models, have dissuaded rapid translation to the clinic. We argue that as research begins to focus more on the selective function of individual NF-κB and IκB family members, and as we progress toward a better understanding of the mechanisms governing IKK activation in pathological settings, past optimism and current ambivalence about the therapeutic utility of NF-κB blockade will be replaced with real-world therapeutic successes.
Acknowledgments
Work in our laboratories is supported by grants from the NIH (including AI033443, AI068977, and AI093985 to S.G., and P30AR058886 to M.S.H.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.183434.111.
References
- Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC 2007. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol 27: 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agou F, Ye F, Goffinont S, Courtois G, Yamaoka S, Israel A, Veron M 2002. NEMO trimerizes through its coiled-coil C-terminal domain. J Biol Chem 277: 17464–17475 [DOI] [PubMed] [Google Scholar]
- Agou F, Traincard F, Vinolo E, Courtois G, Yamaoka S, Israel A, Veron M 2004. The trimerization domain of NEMO is comprised of the interacting C-terminal CC2 and LZ coiled-coil subdomains. J Biol Chem. 279: 27861–27869 [DOI] [PubMed] [Google Scholar]
- Ahmed N, Zeng M, Sinha I, Polin L, Wei WZ, Rathinam C, Flavell R, Massoumi R, Venuprasad K 2011. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 12: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertella MR, Campbell RD 1994. Characterization of a novel gene in the human major histocompatibility complex that encodes a potential new member of the IκB family of proteins. Hum Mol Genet 3: 793–799 [DOI] [PubMed] [Google Scholar]
- Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y 1995. Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci 92: 10599–10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Haecker H, Karin M, Ciechanover A 2004. Mechanism of processing of the NF-κB2 p100 precursor: Identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(β-TrCP) ubiquitin ligase. Oncogene 23: 2540–2547 [DOI] [PubMed] [Google Scholar]
- Angelov D, Lenouvel F, Hans F, Muller CW, Bouvet P, Bednar J, Moudrianakis EN, Cadet J, Dimitrov S 2004. The histone octamer is invisible when NF-κB binds to the nucleosome. J Biol Chem 279: 42374–42382 [DOI] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ 2000. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol 1: 113–118 [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Henkel T 1994. Function and activation of NF-κB in the immune system. Annu Rev Immunol 12: 141–179 [DOI] [PubMed] [Google Scholar]
- Baker RG, Hayden MS, Ghosh S 2011. NF-κB, inflammation, and metabolic disease. Cell Metab 13: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ, Karin M 1997. Nuclear factor-κB: A pivotal transcription factor in chronic inflammatory diseases. N Engl J Med 336: 1066–1071 [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, et al. 2007. A fourth IκB protein within the NF-κB signaling module. Cell 128: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Villunger A, Labi V, Fischer SF, Strasser A, Wagner H, Schmid RM, Hacker G 2006. The NF-κB regulator Bcl-3 and the BH3-only proteins Bim and Puma control the death of activated T cells. Proc Natl Acad Sci 103: 10979–10984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AA, Baldwin AS Jr 1993. The IκB proteins: Multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev 7: 2064–2070 [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Baltimore D 1995. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev 9: 2736–2746 [DOI] [PubMed] [Google Scholar]
- Beinke S, Robinson MJ, Hugunin M, Ley SC 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol Cell Biol 24: 9658–9667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M 2011. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol 12: 715–723 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ 2009. Ubiquitylation in innate and adaptive immunity. Nature 458: 430–437 [DOI] [PubMed] [Google Scholar]
- Bird TA, Schooley K, Dower SK, Hagen H, Virca GD 1997. Activation of nuclear transcription factor NF-κB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J Biol Chem 272: 32606–32612 [DOI] [PubMed] [Google Scholar]
- Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U 1993. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell 72: 729–739 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. 2004. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Brasier AR, Lu M, Hai T, Lu Y, Boldogh I 2001. NF-κB-inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-κB1 residence. J Biol Chem 276: 32080–32093 [DOI] [PubMed] [Google Scholar]
- Brocke-Heidrich K, Ge B, Cvijic H, Pfeifer G, Loffler D, Henze C, McKeithan TW, Horn F 2006. BCL3 is induced by IL-6 via Stat3 binding to intronic enhancer HS4 and represses its own transcription. Oncogene 25: 7297–7304 [DOI] [PubMed] [Google Scholar]
- Brown K, Park S, Kanno T, Franzoso G, Siebenlist U 1993. Mutual regulation of the transcriptional activator NF-κB and its inhibitor, IκB-α. Proc Natl Acad Sci 90: 2532–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulek K, Liu C, Swaidani S, Wang L, Page RC, Gulen MF, Herjan T, Abbadi A, Qian W, Sun D, et al. 2011. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat Immunol 12: 844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy DL, McKeithan TW 1997. Diverse effects of BCL3 phosphorylation on its modulation of NF-κB p52 homodimer binding to DNA. J Biol Chem 272: 33132–33139 [DOI] [PubMed] [Google Scholar]
- Caamano JH, Perez P, Lira SA, Bravo R 1996. Constitutive expression of Bc1-3 in thymocytes increases the DNA binding of NF-κB1 (p50) homodimers in vivo. Mol Cell Biol 16: 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383: 443–446 [DOI] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH 2007. Negative regulation of toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science 317: 675–678 [DOI] [PubMed] [Google Scholar]
- Chang WJ, Alvarez-Gonzalez R 2001. The sequence-specific DNA binding of NF-κB is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. J Biol Chem 276: 47664–47670 [DOI] [PubMed] [Google Scholar]
- Chang SH, Park H, Dong C 2006. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem 281: 35603–35607 [DOI] [PubMed] [Google Scholar]
- Chen ZJ 2005. Ubiquitin signalling in the NF-κB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T 1995. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev 9: 1586–1597 [DOI] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293: 1653–1657 [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J 21: 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC 2005. NF-κB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol 25: 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, Huang HL, Pike KA, Hao Z, Su YW, et al. 2008. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci 105: 12429–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JD, Ryseck RP, Attar RM, Dambach D, Bravo R 1998. Functional redundancy of the nuclear factor κB inhibitors IκBα and IκBβ. J Exp Med 188: 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Matsuzaka Y, Warita T, Sugoh T, Miyashita K, Tajima A, Nakamura M, Inoko H, Sato T, Kimura M 2011. NFKBIL1 confers resistance to experimental autoimmune arthritis through the regulation of dendritic cell functions. Scand J Immunol 73: 478–485 [DOI] [PubMed] [Google Scholar]
- Clark JM, Aleksiyadis K, Martin A, McNamee K, Tharmalingam T, Williams RO, Memet S, Cope AP 2011. Inhibitor of κBɛ (IκBɛ) is a non-redundant regulator of c-Rel-dependent gene expression in murine T and B cells. PLoS ONE 6: e24504 doi: 10.1371/journal.pone.0024504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Orian A, Ciechanover A 2001. Processing of p105 is inhibited by docking of p50 active subunits to the ankyrin repeat domain, and inhibition is alleviated by signaling via the carboxyl-terminal phosphorylation/ ubiquitin-ligase binding domain. J Biol Chem 276: 26769–26776 [DOI] [PubMed] [Google Scholar]
- Cohen S, Achbert-Weiner H, Ciechanover A 2004. Dual effects of IκB kinase β-mediated phosphorylation on p105 fate: SCF(β-TrCP)-dependent degradation and SCF(β-TrCP)-independent processing. Mol Cell Biol 24: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G, Gilmore TD 2006. Mutations in the NF-κB signaling pathway: Implications for human disease. Oncogene 25: 6831–6843 [DOI] [PubMed] [Google Scholar]
- Courtois G, Israel A 2011. IKK regulation and human genetics. Curr Top Microbiol Immunol 349: 73–95 [DOI] [PubMed] [Google Scholar]
- Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA 2005. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NFκB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem 280: 36560–36566 [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. 2008. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelaTorre A, Schroeder RA, Kuo PC 1997. Alteration of NF-κB p50 DNA binding kinetics by S-nitrosylation. Biochem Biophys Res Commun 238: 703–706 [DOI] [PubMed] [Google Scholar]
- Delhase M, Hayakawa M, Chen Y, Karin M 1999. Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science 284: 309–313 [DOI] [PubMed] [Google Scholar]
- de Martin R, Vanhove B, Cheng Q, Hofer E, Csizmadia V, Winkler H, Bach FH 1993. Cytokine-inducible expression in endothelial cells of an IκBα-like gene is regulated by NFκB. EMBO J 12: 2773–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey PW, Doyle SE, He JQ, Cheng G 2003. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev 14: 193–209 [DOI] [PubMed] [Google Scholar]
- Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z 2000. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity 12: 419–429 [DOI] [PubMed] [Google Scholar]
- Dhamija S, Doerrie A, Winzen R, Dittrich-Breiholz O, Taghipour A, Kuehne N, Kracht M, Holtmann H 2010. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IκBζ mRNA. J Biol Chem 285: 29165–29178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388: 548–554 [DOI] [PubMed] [Google Scholar]
- Doerre S, Corley RB 1999. Constitutive nuclear translocation of NF-κB in B cells in the absence of IκB degradation. J Immunol 163: 269–277 [PubMed] [Google Scholar]
- Doerre S, Mesires KP, Daley KM, McCarty T, Knoetig S, Corley RB 2005. Reductions in IκBɛ and changes in NF-κB activity during B lymphocyte differentiation. J Immunol 174: 983–991 [DOI] [PubMed] [Google Scholar]
- Donath MY, Shoelson SE 2011. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol 11: 98–107 [DOI] [PubMed] [Google Scholar]
- Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S 2008. Repression of gene expression by unphosphorylated NF-κB p65 through epigenetic mechanisms. Genes Dev 22: 1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Jimi E, Zeiss C, Hayden MS, Ghosh S 2010. Constitutively active NF-κB triggers systemic TNFα-dependent inflammation and localized TNFα-independent inflammatory disease. Genes Dev 24: 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D, Shimada E, Huynh K, Bergqvist S, Talwar R, Karin M, Ghosh G 2007. Inhibitor κB kinase β binding by inhibitor κB kinase γ. Biochemistry 46: 12482–12490 [DOI] [PubMed] [Google Scholar]
- Duran A, Diaz-Meco MT, Moscat J 2003. Essential role of RelA Ser311 phosphorylation by ζPKC in NF-κB transcriptional activation. EMBO J 22: 3910–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J 2010. Identification of the MMS22L–TONSL complex that promotes homologous recombination. Mol Cell 40: 632–644 [DOI] [PubMed] [Google Scholar]
- Ea CK, Baltimore D 2009. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci 106: 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ 2006. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 245–257 [DOI] [PubMed] [Google Scholar]
- El Gazzar M, Yoza BK, Hu JY, Cousart SL, McCall CE 2007. Epigenetic silencing of tumor necrosis factor α during endotoxin tolerance. J Biol Chem 282: 26857–26864 [DOI] [PubMed] [Google Scholar]
- El Gazzar M, Liu T, Yoza BK, McCall CE 2010. Dynamic and selective nucleosome repositioning during endotoxin tolerance. J Biol Chem 285: 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, Tschopp J, Pasparakis M 2008. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol 9: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Eto A, Muta T, Yamazaki S, Takeshige K 2003. Essential roles for NF-κB and a Toll/IL-1 receptor domain-specific signal(s) in the induction of IκB-ζ. Biochem Biophys Res Commun 301: 495–501 [DOI] [PubMed] [Google Scholar]
- Fan CM, Maniatis T 1991. Generation of p50 subunit of NF-κB by processing of p105 through an ATP-dependent pathway. Nature 354: 395–398 [DOI] [PubMed] [Google Scholar]
- Ferrier R, Nougarede R, Doucet S, Kahn-Perles B, Imbert J, Mathieu-Mahul D 1999. Physical interaction of the bHLH LYL1 protein and NF-κB1 p105. Oncogene 18: 995–1005 [DOI] [PubMed] [Google Scholar]
- Fiorini E, Schmitz I, Marissen WE, Osborn SL, Touma M, Sasada T, Reche PA, Tibaldi EV, Hussey RE, Kruisbeek AM, et al. 2002. Peptide-induced negative selection of thymocytes activates transcription of an NF-κB inhibitor. Mol Cell 9: 637–648 [DOI] [PubMed] [Google Scholar]
- Fong A, Sun SC 2002. Genetic evidence for the essential role of β-transducin repeat-containing protein in the inducible processing of NF-κB2/p100. J Biol Chem 277: 22111–22114 [DOI] [PubMed] [Google Scholar]
- Fong A, Zhang M, Neely J, Sun SC 2002. S9, a 19 S proteasome subunit interacting with ubiquitinated NF-κB2/p100. J Biol Chem 277: 40697–40702 [DOI] [PubMed] [Google Scholar]
- Franzoso G, Bours V, Azarenko V, Park S, Tomita-Yamaguchi M, Kanno T, Brown K, Siebenlist U 1993. The oncoprotein Bcl-3 can facilitate NF-κB-mediated transactivation by removing inhibiting p50 homodimers from select κB sites. EMBO J 12: 3893–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Scharton-Kersten T, Shores EW, Epstein S, Grinberg A, Tran T, Shacter E, Leonardi A, Anver M, et al. 1997. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity 6: 479–490 [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Chen A, Xiong Y, Pan ZQ, Ronai Z 1999. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene 18: 2039–2046 [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Yasuda H, Sato Y, Yamamoto K 1995. A role for phosphorylation in the proteolytic processing of the human NF-κB1 precursor. Gene 165: 183–189 [DOI] [PubMed] [Google Scholar]
- Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D 1993. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev 7: 1354–1363 [DOI] [PubMed] [Google Scholar]
- Fujita S, Seino K, Sato K, Sato Y, Eizumi K, Yamashita N, Taniguchi M 2006. Regulatory dendritic cells act as regulators of acute lethal systemic inflammatory response. Blood 107: 3656–3664 [DOI] [PubMed] [Google Scholar]
- Fusco AJ, Savinova OV, Talwar R, Kearns JD, Hoffmann A, Ghosh G 2008. Stabilization of RelB requires multidomain interactions with p100/p52. J Biol Chem 283: 12324–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL 2009. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 9: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantke T, Sriskantharajah S, Ley SC 2011. Regulation and function of TPL-2, an IκB kinase-regulated MAP kinase kinase kinase. Cell Res 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Li O, Wilder P, Rizzino A, McKeithan TW 2003. NF-κB regulates BCL3 transcription in T lymphocytes through an intronic enhancer. J Immunol 171: 4210–4218 [DOI] [PubMed] [Google Scholar]
- Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR, Nachbur U, Rickard J, Anderton H, Moulin M, et al. 2011. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-κB and activation of caspase-8. J Biol Chem 286: 13282–13291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596 [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A 2006. Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 25: 6781–6799 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Baltimore D 1990. Activation in vitro of NF-κB by phosphorylation of its inhibitor IκB. Nature 344: 678–682 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M 2002. Missing pieces in the NF-κB puzzle. Cell 109: S81–S96 doi: 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB 1998. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16: 225–260 [DOI] [PubMed] [Google Scholar]
- Gilmore TD 2006. Introduction to NF-κB: Players, pathways, perspectives. Oncogene 25: 6680–6684 [DOI] [PubMed] [Google Scholar]
- Goudeau B, Huetz F, Samson S, Di Santo JP, Cumano A, Beg A, Israel A, Memet S 2003. IκBα/IκBɛ deficiency reveals that a critical NF-κB dosage is required for lymphocyte survival. Proc Natl Acad Sci 100: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greetham D, Ellis CD, Mewar D, Fearon U, an Ultaigh SN, Veale DJ, Guesdon F, Wilson AG 2007. Functional characterization of NF-κB inhibitor-like protein 1 (NFκBIL1), a candidate susceptibility gene for rheumatoid arthritis. Hum Mol Genet 16: 3027–3036 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P 2010. IAPs: From caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Cancer 10: 561–574 [DOI] [PubMed] [Google Scholar]
- Haas AL 2009. Linear polyubiquitylation: The missing link in NF-κB signalling. Nat Cell Biol 11: 116–118 [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844 [DOI] [PubMed] [Google Scholar]
- Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z 2004. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-κB. EMBO J 23: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M 2006. Regulation and function of IKK and IKK-related kinases. Sci STKE 2006: re13 doi: 10.1126/stke.3572006re13 [DOI] [PubMed] [Google Scholar]
- Hacker H, Tseng PH, Karin M 2011. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol 11: 457–468 [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM 2011. Deubiquitinases in the regulation of NF-κB signaling. Cell Res 21: 22–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Maggirwar SB, Sun SC 1996. Inhibition of p105 processing by NF-κB proteins in transiently transfected cells. Oncogene 12: 2385–2392 [PubMed] [Google Scholar]
- Hartupee J, Liu C, Novotny M, Li X, Hamilton T 2007. IL-17 enhances chemokine gene expression through mRNA stabilization. J Immunol 179: 4135–4141 [DOI] [PubMed] [Google Scholar]
- Hartupee J, Liu C, Novotny M, Sun D, Li X, Hamilton TA 2009. IL-17 signaling for mRNA stabilization does not require TNF receptor-associated factor 6. J Immunol 182: 1660–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta H, Kato A, Todokoro K 2001. Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J Biol Chem 276: 12485–12488 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N 2008. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 27: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskill S, Beg AA, Tompkins SM, Morris JS, Yurochko AD, Sampson-Johannes A, Mondal K, Ralph P, Baldwin AS Jr 1991. Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell 65: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO 2005. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-κB-dependent transcription. J Biol Chem 280: 40450–40464 [DOI] [PubMed] [Google Scholar]
- Hatada EN, Nieters A, Wulczyn FG, Naumann M, Meyer R, Nucifora G, McKeithan TW, Scheidereit C 1992. The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc Natl Acad Sci 89: 2489–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2004. Signaling to NF-κB. Genes Dev 18: 2195–2224 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2008. Shared principles in NF-κB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2011. NF-κB in immunobiology. Cell Res 21: 223–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G 2006. Rescue of TRAF3-null mice by p100 NF-κB deficiency. J Exp Med 203: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V, Krappmann D, Wulczyn FG, Scheidereit C 1999. NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3-p50 complexes. EMBO J 18: 4766–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C 2001. Shared pathways of IκB kinase-induced SCF(βTrCP)-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBα. Mol Cell Biol 21: 1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle PA 1993. Rapid proteolysis of IκB-α is necessary for activation of transcription factor NF-κB. Nature 365: 182–185 [DOI] [PubMed] [Google Scholar]
- Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C 2010. A cytoplasmic ATM–TRAF6–cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol Cell 40: 63–74 [DOI] [PubMed] [Google Scholar]
- Hirotani T, Lee PY, Kuwata H, Yamamoto M, Matsumoto M, Kawase I, Akira S, Takeda K 2005. The nuclear IκB protein IκBNS selectively inhibits lipopolysaccharide-induced IL-6 production in macrophages of the colonic lamina propria. J Immunol 174: 3650–3657 [DOI] [PubMed] [Google Scholar]
- Hishiki T, Ohshima T, Ego T, Shimotohno K 2007. BCL3 acts as a negative regulator of transcription from the human T-cell leukemia virus type 1 long terminal repeat through interactions with TORC3. J Biol Chem 282: 28335–28343 [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Popko AE, Ramsey CS, Mayo MW 2006. IκB kinase α-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol Cell Biol 26: 457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D 2002. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science 298: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G 2006. Transcriptional regulation via the NF-κB signaling module. Oncogene 25: 6706–6716 [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H, Shu HB, Pan MG, Goeddel DV 1996. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84: 299–308 [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science 284: 316–320 [DOI] [PubMed] [Google Scholar]
- Huang GJ, Zhang ZQ, Jin DY 2002. Stimulation of IKK-γ oligomerization by the human T-cell leukemia virus oncoprotein Tax. FEBS Letters 531: 494–498 [DOI] [PubMed] [Google Scholar]
- Hur GM, Lewis J, Yang Q, Lin Y, Nakano H, Nedospasov S, Liu ZG 2003. The death domain kinase RIP has an essential role in DNA damage-induced NF-κB activation. Genes Dev 17: 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, Ogura Y, Nunez G 2000. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem 275: 27823–27831 [DOI] [PubMed] [Google Scholar]
- Inoue J, Kerr LD, Kakizuka A, Verma IM 1992. IκBγ, a 70 kd protein identical to the C-terminal half of p110 NF-κB: A new member of the IκB family. Cell 68: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Ishimaru N, Kishimoto H, Hayashi Y, Sprent J 2006. Regulation of naive T cell function by the NF-κB2 pathway. Nat Immunol 7: 763–772 [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J 2005. PIDD mediates NF-κB activation in response to DNA damage. Cell 123: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K 2000. Evidence for regulation of NF-κB by poly(ADP-ribose) polymerase. Biochem J 346: 641–649 [PMC free article] [PubMed] [Google Scholar]
- Karin M 2006. Nuclear factor-κB in cancer development and progression. Nature 441: 431–436 [DOI] [PubMed] [Google Scholar]
- Karin M 2009. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol 1: a000141 doi: 10.1101/cshperspect.a000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Delhase M 2000. The IκB kinase (IKK) and NF-κB: Key elements of proinflammatory signalling. Semin Immunol 12: 85–98 [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A 2006. IκBɛ provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol 173: 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ZT, Matsumoto A, Stamler JS, Marshall HE 2007. NOS2 regulation of NF-κB by S-nitrosylation of p65. J Biol Chem 282: 30667–30672 [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P 1998. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity 8: 297–303 [DOI] [PubMed] [Google Scholar]
- Kerr LD, Inoue J, Davis N, Link E, Baeuerle PA, Bose HR Jr, Verma IM 1991. The rel-associated pp40 protein prevents DNA binding of Rel and NF-κB: Relationship with IκBβ and regulation by phosphorylation. Genes Dev 5: 1464–1476 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Omori E, Matsumoto K, Nunez G, Ninomiya-Tsuji J 2008. TAK1 is a central mediator of NOD2 signaling in epidermal cells. J Biol Chem 283: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Sharma N, Nyborg JK 2008. The proto-oncogene Bcl3, induced by Tax, represses Tax-mediated transcription via p300 displacement from the human T-cell leukemia virus type 1 promoter. J Virol 82: 11939–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel EA, Lemieux ME, Xia X, Davis TN, Rebel VI, Kung AL 2009. Systematic in vivo structure-function analysis of p300 in hematopoiesis. Blood 114: 4804–4812 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H, Kanehira K, Okita K, Morimatsu M, Saito M 2000. MAIL, a novel nuclear IκB protein that potentiates LPS-induced IL-6 production. FEBS Lett 485: 53–56 [DOI] [PubMed] [Google Scholar]
- Klement JF, Rice NR, Car BD, Abbondanzo SJ, Powers GD, Bhatt PH, Chen CH, Rosen CA, Stewart CL 1996. IκBα deficiency results in a sustained NF-κB response and severe widespread dermatitis in mice. Mol Cell Biol 16: 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Kadono Y, Naito A, Matsumoto K, Yamamoto T, Tanaka S, Inoue J 2001. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J 20: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 2007. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci 104: 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann D, Hatada EN, Tegethoff S, Li J, Klippel A, Giese K, Baeuerle PA, Scheidereit C 2000. The IκB kinase (IKK) complex is tripartite and contains IKK γ but not IKAP as a regular component. J Biol Chem 275: 29779–29787 [DOI] [PubMed] [Google Scholar]
- Kreisel D, Sugimoto S, Tietjens J, Zhu J, Yamamoto S, Krupnick AS, Carmody RJ, Gelman AE 2011. Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. J Clin Invest 121: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll M, Margottin F, Kohl A, Renard P, Durand H, Concordet JP, Bachelerie F, Arenzana-Seisdedos F, Benarous R 1999. Inducible degradation of IκBα by the proteasome requires interaction with the F-box protein h-βTrCP. J Biol Chem 274: 7941–7945 [DOI] [PubMed] [Google Scholar]
- Kuwata H, Watanabe Y, Miyoshi H, Yamamoto M, Kaisho T, Takeda K, Akira S 2003. IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-α production in macrophages. Blood 102: 4123–4129 [DOI] [PubMed] [Google Scholar]
- Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, Koga R, Takeda K 2006. IκBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity 24: 41–51 [DOI] [PubMed] [Google Scholar]
- Lang V, Janzen J, Fischer GZ, Soneji Y, Beinke S, Salmeron A, Allen H, Hay RT, Ben-Neriah Y, Ley SC 2003. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol Cell Biol 23: 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T 2009. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb Perspect Biol 1: a001651 doi: 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail O, Schmidt-Ullrich R, Israel A 1993. Promoter analysis of the gene encoding the IκB-α/MAD3 inhibitor of NF-κB: Positive regulation by members of the rel/NF-κB family. EMBO J 12: 5043–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Hannink M 2002. Characterization of the nuclear import and export functions of IκB(ɛ). J Biol Chem 277: 23358–23366 [DOI] [PubMed] [Google Scholar]
- Lee TH, Shank J, Cusson N, Kelliher MA 2004. The kinase activity of Rip1 is not required for tumor necrosis factor-α-induced IκB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem 279: 33185–33191 [DOI] [PubMed] [Google Scholar]
- Lee KY, D'Acquisto F, Hayden MS, Shim JH, Ghosh S 2005. PDK1 nucleates T cell receptor-induced signaling complex for NF-κB activation. Science 308: 114–118 [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D 1989. NF-κB: A pleiotropic mediator of inducible and tissue-specific gene control. Cell 58: 227–229 [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM 1999a. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev 13: 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM 1999b. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science 284: 321–325 [DOI] [PubMed] [Google Scholar]
- Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, Wallach D, Horwitz MS 1999. Identification of a cell protein (FIP-3) as a modulator of NF-κB activity and as a target of an adenovirus inhibitor of tumor necrosis factor α-induced apoptosis. Proc Natl Acad Sci 96: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M 1999. The IKKβ subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J Exp Med 189: 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Estepa G, Memet S, Israel A, Verma IM 2000. Complete lack of NF-κB activity in IKK1 and IKK2 double-deficient mice: Additional defect in neurulation. Genes Dev 14: 1729–1733 [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang J, Chen D, Shu HB 2003. Casper/c-FLIP is physically and functionally associated with NF-κB1 p105. Biochem Biophys Res Commun 309: 980–985 [DOI] [PubMed] [Google Scholar]
- Li H, Kobayashi M, Blonska M, You Y, Lin X 2006. Ubiquitination of RIP is required for tumor necrosis factor α-induced NF-κB activation. J Biol Chem 281: 13636–13643 [DOI] [PubMed] [Google Scholar]
- Liang C, Zhang M, Sun SC 2006. β-TrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal 18: 1309–1317 [DOI] [PubMed] [Google Scholar]
- Liang Y, Seymour RE, Sundberg JP 2011. Inhibition of NF-κB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J Invest Dermatol 131: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, Sun SC 2004. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem 279: 26243–26250 [DOI] [PubMed] [Google Scholar]
- Lin L, Ghosh S 1996. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol 16: 2248–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Brown K, Siebenlist U 1995. Activation of NF-κB requires proteolysis of the inhibitor IκB-α: Signal-induced phosphorylation of IκB-α alone does not release active NF-κB. Proc Natl Acad Sci 92: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, DeMartino GN, Greene WC 1998. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell 92: 819–828 [DOI] [PubMed] [Google Scholar]
- Lin X, Mu Y, Cunningham ET Jr, Marcu KB, Geleziunas R, Greene WC 1998. Molecular determinants of NF-κB-inducing kinase action. Mol Cell Biol 18: 5899–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Cao Z, Goeddel DV 1998. NF-κB-inducing kinase activates IKK-α by phosphorylation of Ser- 176. Proc Natl Acad Sci 95: 3792–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZG 2005. Molecular mechanism of TNF signaling and beyond. Cell Res 15: 24–27 [DOI] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev 13: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, Myszka D, Han J, Wu H 2007. XIAP induces NF-κB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell 26: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Zougman A, Pudelko M, Bebenek M, Ziolkowski P, Mann M, Wisniewski JR 2009. Mapping of lysine monomethylation of linker histones in human breast and its cancer. J Proteome Res 8: 4207–4215 [DOI] [PubMed] [Google Scholar]
- MacKichan ML, Logeat F, Israel A 1996. Phosphorylation of p105 PEST sequence via a redox-insensitive pathway up-regulates processing of p50 NF-κB. J Biol Chem 271: 6084–6091 [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, Arora V, Mak TW, Lacasse EC, Waring J, et al. 2008. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc Natl Acad Sci 105: 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E 2007. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J 26: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek S, Huang DB, Huxford T, Ghosh S, Ghosh G 2003. X-ray crystal structure of an IκBβ x NF-κB p65 homodimer complex. J Biol Chem 278: 23094–23100 [DOI] [PubMed] [Google Scholar]
- Marienfeld RB, Palkowitsch L, Ghosh S 2006. Dimerization of the IκB kinase-binding domain of NEMO is required for tumor necrosis factor α-induced NF-κB activity. Mol Cell Biol 26: 9209–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall HE, Stamler JS 2001. Inhibition of NF-κB by S-nitrosylation. Biochemistry 40: 1688–1693 [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X 2005. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity 23: 575–585 [DOI] [PubMed] [Google Scholar]
- May MJ, Larsen SE, Shim JH, Madge LA, Ghosh S 2004. A novel ubiquitin-like domain in IκB kinase β is required for functional activity of the kinase. J Biol Chem 279: 45528–45539 [DOI] [PubMed] [Google Scholar]
- McKenzie FR, Connelly MA, Balzarano D, Muller JR, Geleziunas R, Marcu KB 2000. Functional isoforms of IκB kinase α(IKKα) lacking leucine zipper and helix–loop–helix domains reveal that IKKα and IKKβ have different activation requirements. Mol Cell Biol 20: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellits KH, Hay RT, Goodbourn S 1993. Proteolytic degradation of MAD3 (IκBα) and enhanced processing of the NF-κB precursor p105 are obligatory steps in the activation of NF-κB. Nucleic Acids Res 21: 5059–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, DiDonato JA, Rosette C, Karin M 1993. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev 7: 705–718 [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, et al. 1997. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science 278: 860–866 [DOI] [PubMed] [Google Scholar]
- Mercurio F, Murray BW, Shevchenko A, Bennett BL, Young DB, Li JW, Pascual G, Motiwala A, Zhu H, Mann M, et al. 1999. IκB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Mol Cell Biol 19: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Tschopp J 2005. The RIP kinases: Crucial integrators of cellular stress. Trends Biochem Sci 30: 151–159 [DOI] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat Immunol 5: 503–507 [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J 2001. NF-κB signals induce the expression of c-FLIP. Mol Cell Biol 21: 5299–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BS, Zandi E 2001. Complete reconstitution of human IκB kinase (IKK) complex in yeast. Assessment of its stoichiometry and the role of IKKγ on the complex activity in the absence of stimulation. J Biol Chem 276: 36320–36326 [DOI] [PubMed] [Google Scholar]
- Mitchell TC, Hildeman D, Kedl RM, Teague TK, Schaefer BC, White J, Zhu Y, Kappler J, Marrack P 2001. Immunological adjuvants promote activated T cell survival via induction of Bcl-3. Nat Immunol 2: 397–402 [DOI] [PubMed] [Google Scholar]
- Moorthy AK, Savinova OV, Ho JQ, Wang VY, Vu D, Ghosh G 2006. The 20S proteasome processes NF-κB1 p105 into p50 in a translation-independent manner. EMBO J 25: 1945–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama M, Yamazaki S, Eto-Kimura A, Takeshige K, Muta T 2005. Positive and negative regulation of nuclear factor-κB-mediated transcription by IκB-ζ, an inducible nuclear protein. J Biol Chem 280: 7444–7451 [DOI] [PubMed] [Google Scholar]
- Muhlbauer M, Chilton PM, Mitchell TC, Jobin C 2008. Impaired Bcl3 up-regulation leads to enhanced lipopolysaccharide-induced interleukin (IL)-23P19 gene expression in IL-10−/− mice. J Biol Chem 283: 14182–14189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muta T, Yamazaki S, Eto A, Motoyama M, Takeshige K 2003. IκB-ζ, a new anti-inflammatory nuclear protein induced by lipopolysaccharide, is a negative regulator for nuclear factor-κB. J Endotoxin Res 9: 187–191 [DOI] [PubMed] [Google Scholar]
- Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, et al. 1999. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4: 353–362 [DOI] [PubMed] [Google Scholar]
- Nakano H, Sakon S, Koseki H, Takemori T, Tada K, Matsumoto M, Munechika E, Sakai T, Shirasawa T, Akiba H, et al. 1999. Targeted disruption of Traf5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc Natl Acad Sci 96: 9803–9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G 2009. When sirtuins and NF-κB collide. Cell 136: 19–21 [DOI] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I 2005. Interactions of NF-κB with chromatin: The art of being at the right place at the right time. Nat Immunol 6: 439–445 [DOI] [PubMed] [Google Scholar]
- Naumann M, Scheidereit C 1994. Activation of NF-κB in vivo is regulated by multiple phosphorylations. EMBO J 13: 4597–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Wulczyn FG, Scheidereit C 1993. The NF-κB precursor p105 and the proto-oncogene product Bcl-3 are IκB molecules and control nuclear translocation of NF-κB. EMBO J 12: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Grieshammer T, Chuvpilo S, Kneitz B, Lohoff M, Schimpl A, Franza BR Jr, Serfling E 1995. RelA/p65 is a molecular target for the immunosuppressive action of protein kinase A. EMBO J 14: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M, Ogushi M, Suda R, Toyotaka M, Saiki S, Kitajima N, Nakaya M, Kim KM, Ide T, Sato Y, et al. 2011. Heterologous down-regulation of angiotensin type 1 receptors by purinergic P2Y2 receptor stimulation through S-nitrosylation of NF-κB. Proc Natl Acad Sci 108: 6662–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan GP, Ghosh S, Liou HC, Tempst P, Baltimore D 1991. DNA binding and IκB inhibition of the cloned p65 subunit of NF-κB, a rel-related polypeptide. Cell 64: 961–969 [DOI] [PubMed] [Google Scholar]
- Nolan GP, Fujita T, Bhatia K, Huppi C, Liou HC, Scott ML, Baltimore D 1993. The bcl-3 proto-oncogene encodes a nuclear IκB-like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation-dependent manner. Mol Cell Biol 13: 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman DA, Barton PJ 2000. Isolation, sequence, and chromosomal localisation of the human IκBR gene (NFKBIL2). Ann Hum Genet 64: 15–23 [DOI] [PubMed] [Google Scholar]
- Novack DV 2011. Role of NF-κB in the skeleton. Cell Res 21: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW 2010. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L–NFKBIL2 complex required for genomic stability. Mol Cell 40: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. 2010. The MMS22L–TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 40: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D 2007. Malt1 ubiquitination triggers NF-κB signaling upon T-cell activation. EMBO J 26: 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Hayden MS, Ghosh S 2011. Crosstalk in NF-κB signaling pathways. Nat Immunol 12: 695–708 [DOI] [PubMed] [Google Scholar]
- Ohno H, Takimoto G, McKeithan TW 1990. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell 60: 991–997 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, Ohya K, Jetten AM, Akira S, Muta T, et al. 2010. IκBζ regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 464: 1381–1385 [DOI] [PubMed] [Google Scholar]
- O'Mahony AM, Montano M, Van Beneden K, Chen LF, Greene WC 2004. Human T-cell lymphotropic virus type 1 tax induction of biologically active NF-κB requires IκB kinase-1-mediated phosphorylation of RelA/p65. J Biol Chem 279: 18137–18145 [DOI] [PubMed] [Google Scholar]
- Orian A, Schwartz AL, Israel A, Whiteside S, Kahana C, Ciechanover A 1999. Structural motifs involved in ubiquitin-mediated processing of the NF-κB precursor p105: Roles of the glycine-rich region and a downstream ubiquitination domain. Mol Cell Biol 19: 3664–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Gonen H, Bercovich B, Fajerman I, Eytan E, Israel A, Mercurio F, Iwai K, Schwartz AL, Ciechanover A 2000. SCF(β)(-TrCP) ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J 19: 2580–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18: 6853–6866 [DOI] [PubMed] [Google Scholar]
- Palkowitsch L, Leidner J, Ghosh S, Marienfeld RB 2008. Phosphorylation of serine 68 in the IκB kinase (IKK)-binding domain of NEMO interferes with the structure of the IKK complex and tumor necrosis factor-α-induced NF-κB activity. J Biol Chem 283: 76–86 [DOI] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78: 773–785 [DOI] [PubMed] [Google Scholar]
- Park SW, Huq MD, Hu X, Wei LN 2005. Tyrosine nitration on p65: A novel mechanism to rapidly inactivate nuclear factor-κB. Mol Cell Proteomics 4: 300–309 [DOI] [PubMed] [Google Scholar]
- Park SG, Schulze-Luehrman J, Hayden MS, Hashimoto N, Ogawa W, Kasuga M, Ghosh S 2009. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-κB and activate T cells. Nat Immunol 10: 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RJ, Ghosh S 1997. Regulation of IκBβ in WEHI 231 mature B cells. Mol Cell Biol 17: 4390–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, Pasero P, Gerlich DW, Peter M 2010. RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J 29: 4210–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C, Liu Z 2008. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol 9: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Lin JH, Fernandes-Alnemri T, Yamaoka S, Tsichlis PN, Alnemri ES 2000. Activation of the IκB kinases by RIP via IKKγ/NEMO-mediated oligomerization. J Biol Chem 275: 37966–37977 [DOI] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Alnemri ES 2001. vCLAP, a caspase-recruitment domain-containing protein of equine Herpesvirus-2, persistently activates the IκB kinases through oligomerization of IKKγ. J Biol Chem 276: 3183–3187 [DOI] [PubMed] [Google Scholar]
- Qing G, Xiao G 2005. Essential role of IκB kinase α in the constitutive processing of NF-κB2 p100. J Biol Chem 280: 9765–9768 [DOI] [PubMed] [Google Scholar]
- Quann EJ, Liu X, Altan-Bonnet G, Huse M 2011. A cascade of protein kinase C isozymes promotes cytoskeletal polarization in T cells. Nat Immunol 12: 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangelova S, Kirschnek S, Strasser A, Hacker G 2008. FADD and the NF-κB family member Bcl-3 regulate complementary pathways to control T-cell survival and proliferation. Immunology 125: 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala JK, Pouwels J, Pellinen T, Veltel S, Laasola P, Mattila E, Potter CS, Duffy T, Sundberg JP, Kallioniemi O, et al. 2011. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat Cell Biol 13: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P, Hayden MS, Long M, Scott ML, West AP, Zhang D, Oeckinghaus A, Lynch C, Hoffmann A, Baltimore D, et al. 2010. IκBβ acts to inhibit and activate gene expression during the inflammatory response. Nature 466: 1115–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Zhang DH, Elias JA, Ray A 1995. Cloning of a differentially expressed IκB-related protein. J Biol Chem 270: 10680–10685 [DOI] [PubMed] [Google Scholar]
- Rebollo A, Dumoutier L, Renauld JC, Zaballos A, Ayllon V, Martinez AC 2000. Bcl-3 expression promotes cell survival following interleukin-4 deprivation and is controlled by AP1 and AP1-like transcription factors. Mol Cell Biol 20: 3407–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M 1997. Identification and characterization of an IκB kinase. Cell 90: 373–383 [DOI] [PubMed] [Google Scholar]
- Rice NR, MacKichan ML, Israel A 1992. The precursor of NF-κB p50 has IκB-like functions. Cell 71: 243–253 [DOI] [PubMed] [Google Scholar]
- Rocha S, Martin AM, Meek DW, Perkins ND 2003. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-κB subunit with histone deacetylase 1. Mol Cell Biol 23: 4713–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwarf DM, Karin M 1999. The NF-κB activation pathway: A paradigm in information transfer from membrane to nucleus. Sci STKE 1999: re1 doi: 10.1126/stke.1999.5.re1 [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M 1998. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature 395: 297–300 [DOI] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW 2000. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes Dev 14: 854–862 [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM 2003. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science 302: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19: 749–758 [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP 2003. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12: 1413–1426 [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2001. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2003. Modulation of NF-κB activity by exchange of dimers. Mol Cell 11: 1563–1574 [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G 2004. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med 200: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Suzuki S, Kawasaki N, Nakano H, Okazaki T, Chino A, Doi T, Saiki I 2003. Tumor necrosis factor-α-induced IKK phosphorylation of NF-κB p65 on serine 536 is mediated through the TRAF2, TRAF5, and TAK1 signaling pathway. J Biol Chem 278: 36916–36923 [DOI] [PubMed] [Google Scholar]
- Salmeron A, Janzen J, Soneji Y, Bump N, Kamens J, Allen H, Ley SC 2001. Direct phosphorylation of NF-κB1 p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J Biol Chem 276: 22215–22222 [DOI] [PubMed] [Google Scholar]
- Samson SI, Memet S, Vosshenrich CA, Colucci F, Richard O, Ndiaye D, Israel A, Di Santo JP 2004. Combined deficiency in IκBα and IκBɛ reveals a critical window of NF-κB activity in natural killer cell differentiation. Blood 103: 4573–4580 [DOI] [PubMed] [Google Scholar]
- Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST 2005. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev 19: 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjo H, Zajonc DM, Braden R, Norris PS, Ware CF 2010. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-β receptor. J Biol Chem 285: 17148–17155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel M, Klein B, Merkle H, Schulz M, Fritsch R, Greten FR, Arkan MC, Schneider G, Schmid RM 2010. IκBβ is an essential co-activator for LPS-induced IL-1β transcription in vivo. J Exp Med 207: 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinman RI, Beg AA, Baldwin AS Jr 1993. NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol Cell Biol 13: 6089–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M 2000. NEMO/IKK γ-deficient mice model incontinentia pigmenti. Mol Cell 5: 981–992 [DOI] [PubMed] [Google Scholar]
- Schutze S, Schneider-Brachert W 2009. Impact of TNF-R1 and CD95 internalization on apoptotic and antiapoptotic signaling. Results Probl Cell Differ 49: 63–85 [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Krimpenfort P, Berns A, Verma IM 1997. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev 11: 187–197 [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, et al. 2001a. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293: 1495–1499 [DOI] [PubMed] [Google Scholar]
- Senftleben U, Li ZW, Baud V, Karin M 2001b. IKKβ is essential for protecting T cells from TNFα-induced apoptosis. Immunity 14: 217–230 [DOI] [PubMed] [Google Scholar]
- Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A 2009. Kinetic control of negative feedback regulators of NF-κB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci 106: 9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Konno A, Oonuma T, Kitamura H, Imaoka K, Takeda N, Todokoro K, Morimatsu M 2004. Targeted disruption of MAIL, a nuclear IκB protein, leads to severe atopic dermatitis-like disease. J Biol Chem 279: 55493–55498 [DOI] [PubMed] [Google Scholar]
- Shinkura R, Kitada K, Matsuda F, Tashiro K, Ikuta K, Suzuki M, Kogishi K, Serikawa T, Honjo T 1999. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-κb-inducing kinase. Nat Genet 22: 74–77 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Maeda S, Watarai H, Kurosaki T 2007. IκB kinase β-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med 204: 3285–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane M, Hatakeyama S, Hattori K, Nakayama K 1999. Common pathway for the ubiquitination of IκBα, IκBβ, and IκBɛ mediated by the F-box protein FWD1. J Biol Chem 274: 28169–28174 [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Franzoso G, Brown K 1994. Structure, regulation and function of NF-κB. Annu Rev Cell Biol 10: 405–455 [DOI] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML 2012. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat Immunol. 13: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonidis S, Liang S, Chen G, Thanos D 1997. Cloning and functional characterization of mouse IκBɛ. Proc Natl Acad Sci 94: 14372–14377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore N, Lerner N, Dombrowski N, Sakurai H, Stark GR 2002. Distinct roles of the IκB kinase α and β subunits in liberating nuclear factor κB (NF-κB) from IκB and in phosphorylating the p65 subunit of NF-κB. J Biol Chem 277: 3863–3869 [DOI] [PubMed] [Google Scholar]
- Smale ST 2011. Hierarchies of NF-κB target-gene regulation. Nat Immunol 12: 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV 2002. RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem 277: 1405–1418 [DOI] [PubMed] [Google Scholar]
- Solt LA, Madge LA, Orange JS, May MJ 2007. Interleukin-1-induced NF-κB activation is NEMO-dependent but does not require IKKβ. J Biol Chem 282: 8724–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ 2005. Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 23: 561–574 [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ 1999. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev 13: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskantharajah S, Belich MP, Papoutsopoulou S, Janzen J, Tybulewicz V, Seddon B, Ley SC 2009. Proteolysis of NF-κB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat Immunol 10: 38–47 [DOI] [PubMed] [Google Scholar]
- Stefansson B, Brautigan DL 2006. Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IκBɛ. J Biol Chem 281: 22624–22634 [DOI] [PubMed] [Google Scholar]
- Sun SC 2011. Non-canonical NF-κB signaling pathway. Cell Res 21: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Liu ZG 2011. A special issue on NF-κB signaling and function. Cell Res 21: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC 1993. NF-κB controls expression of inhibitor IκBα: Evidence for an inducible autoregulatory pathway. Science 259: 1912–1915 [DOI] [PubMed] [Google Scholar]
- Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg PL, et al. 2000. PKC-theta is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature 404: 402–407 [DOI] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell 14: 289–301 [DOI] [PubMed] [Google Scholar]
- Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T 2011. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat Immunol 12: 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyang H, Phillips R, Douglas I, Ghosh S 1996. Role of unphosphorylated, newly synthesized IκBβ in persistent activation of NF-κB. Mol Cell Biol 16: 5444–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Kobayashi M, Takeuchi M, Suzuki T, Ichiyama A, Ikenoue T, Omata M, Furuichi K, Tanaka K 1999. IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem Biophys Res Commun 256: 127–132 [DOI] [PubMed] [Google Scholar]
- Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K, et al. 2001. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J Biol Chem 276: 36530–36534 [DOI] [PubMed] [Google Scholar]
- Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB 2003. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J Mol Biol 326: 105–115 [DOI] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S 1999. Limb and skin abnormalities in mice lacking IKKα. Science 284: 313–316 [DOI] [PubMed] [Google Scholar]
- Tam WF, Lee LH, Davis L, Sen R 2000. Cytoplasmic sequestration of rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol 20: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV 1999. Embryonic lethality, liver degeneration, and impaired NF-κB activation in IKK-β-deficient mice. Immunity 10: 421–429 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T 2007. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol 8: 584–591 [DOI] [PubMed] [Google Scholar]
- Tang ED, Inohara N, Wang CY, Nunez G, Guan KL 2003. Roles for homotypic interactions and transautophosphorylation in IκB kinase (IKKβ) activation. J Biol Chem 278: 38566–38570 [DOI] [PubMed] [Google Scholar]
- Tegethoff S, Behlke J, Scheidereit C 2003. Tetrameric oligomerization of IκB kinase γ (IKKγ) is obligatory for IKK complex activity and NF-κB activation. Mol Cell Biol 23: 2029–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tergaonkar V, Correa RG, Ikawa M, Verma IM 2005. Distinct roles of IκB proteins in regulating constitutive NF-κB activity. Nat Cell Biol 7: 921–923 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Phillips RJ, Erdjument-Bromage H, Tempst P, Ghosh S 1995. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell 80: 573–582 [DOI] [PubMed] [Google Scholar]
- Ting AT, Pimentel-Muinos FX, Seed B 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J 15: 6189–6196 [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. 2009. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K 2011. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471: 633–636 [DOI] [PubMed] [Google Scholar]
- Touma M, Antonini V, Kumar M, Osborn SL, Bobenchik AM, Keskin DB, Connolly JE, Grusby MJ, Reinherz EL, Clayton LK 2007. Functional role for IκBNS in T cell cytokine regulation as revealed by targeted gene disruption. J Immunol 179: 1681–1692 [DOI] [PubMed] [Google Scholar]
- Tran K, Merika M, Thanos D 1997. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol 17: 5386–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik M, Good L, Xiao G, Harhaj EW, Zandi E, Karin M, Sun SC 1998. NF-κB-inducing kinase and IκB kinase participate in human T-cell leukemia virus I Tax-mediated NF-κB activation. J Biol Chem 273: 21132–21136 [DOI] [PubMed] [Google Scholar]
- Urban MB, Baeuerle PA 1990. The 65-kD subunit of NF-κB is a receptor for IκB and a modulator of DNA-binding specificity. Genes Dev 4: 1975–1984 [DOI] [PubMed] [Google Scholar]
- Valenzuela JO, Hammerbeck CD, Mescher MF 2005. Cutting edge: Bcl-3 up-regulation by signal 3 cytokine (IL-12) prolongs survival of antigen-activated CD8 T cells. J Immunol 174: 600–604 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat Immunol 9: 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, et al. 2007. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell 131: 669–681 [DOI] [PubMed] [Google Scholar]
- Verhelst K, Verstrepen L, Carpentier I, Beyaert R 2011. Linear ubiquitination in NF-κB signaling and inflammation: What we do understand and what we do not. Biochemical pharmacology. 82: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S 1995. Rel/NF-κB/IκB family: Intimate tales of association and dissociation. Genes Dev 9: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G 2003. Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J 22: 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, et al. 2007. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell 131: 682–693 [DOI] [PubMed] [Google Scholar]
- Vivarelli MS, McDonald D, Miller M, Cusson N, Kelliher M, Geha RS 2004. RIP links TLR4 to Akt and is essential for cell survival in response to LPS stimulation. J Exp Med 200: 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Scheurich P 2011. TNFR1-induced activation of the classical NF-κB pathway. FEBS J 278: 862–876 [DOI] [PubMed] [Google Scholar]
- Wang D, Baldwin AS Jr 1998. Activation of nuclear factor-κB-dependent transcription by tumor necrosis factor-α is mediated through phosphorylation of RelA/p65 on serine 529. J Biol Chem 273: 29411–29416 [DOI] [PubMed] [Google Scholar]
- Wang D, Westerheide SD, Hanson JL, Baldwin AS Jr 2000. Tumor necrosis factor α-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J Biol Chem 275: 32592–32597 [DOI] [PubMed] [Google Scholar]
- Waterfield M, Jin W, Reiley W, Zhang M, Sun SC 2004. IκB kinase is an essential component of the Tpl2 signaling pathway. Mol Cell Biol 24: 6040–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R 1995. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell 80: 331–340 [DOI] [PubMed] [Google Scholar]
- Weil R, Laurent-Winter C, Israel A 1997. Regulation of IκBβ degradation. Similarities to and differences from IκBα. J Biol Chem 272: 9942–9949 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11: 665–675 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol 2: 51–57 [DOI] [PubMed] [Google Scholar]
- Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z 1997. MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7: 837–847 [DOI] [PubMed] [Google Scholar]
- Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, Johnson PF 2004. BCL-3 and NF-κB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem 279: 49995–50003 [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Mayo MW, Anest V, Hanson JL, Baldwin AS Jr 2001. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol Cell Biol 21: 8428–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside ST, Epinat JC, Rice NR, Israel A 1997. IκBɛ, a novel member of the IκB family, controls RelA and cRel NF-κB activity. EMBO J 16: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J 2010. RIPK1 is not essential for TNFR1-induced activation of NF-κB. Cell Death Differ 17: 482–487 [DOI] [PubMed] [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV 1997. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science 278: 866–869 [DOI] [PubMed] [Google Scholar]
- Wu C, Ghosh S 1999. β-TrCP mediates the signal-induced ubiquitination of IκBβ. J Biol Chem 274: 29591–29594 [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol 8: 398–406 [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Naumann M, Scheidereit C 1992. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. Nature 358: 597–599 [DOI] [PubMed] [Google Scholar]
- Wullaert A, Bonnet MC, Pasparakis M 2011. NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res 21: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell 7: 401–409 [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong A, Sun SC 2004. Induction of p100 processing by NF-κB-inducing kinase involves docking IKKα to p100 and IKKα-mediated phosphorylation. J Biol Chem. 279: 30099–30105 [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol Cell 36: 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Lo YC, Li Q, Napolitano G, Wu X, Jiang X, Dreano M, Karin M, Wu H 2011. Crystal structure of inhibitor of κB kinase β. Nature 472: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Takeda K, Akira S 2004a. TIR domain-containing adaptors define the specificity of TLR signaling. Mol Immunol 40: 861–868 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. 2004b. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430: 218–222 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, et al. 2006. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol 7: 962–970 [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israel A 1998. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell 93: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Yamauchi S, Ito H, Miyajima A 2010. Iκβ, a nuclear IκB protein, positively regulates the NF-κB-mediated expression of proinflammatory cytokines. Proc Natl Acad Sci 107: 11924–11929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Muta T, Takeshige K 2001. A novel IκB protein, IκB-ζ, induced by proinflammatory stimuli, negatively regulates nuclear factor-κB in the nuclei. J Biol Chem 276: 27657–27662 [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Muta T, Matsuo S, Takeshige K 2005. Stimulus-specific induction of a novel nuclear factor-κB regulator, IκB-ζ, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. J Biol Chem 280: 1678–1687 [DOI] [PubMed] [Google Scholar]
- Yang L, Ross K, Qwarnstrom EE 2003. RelA control of IκBα phosphorylation: A positive feedback loop for high affinity NF-κB complexes. J Biol Chem 278: 30881–30888 [DOI] [PubMed] [Google Scholar]
- Yang J, Williams RS, Kelly DP 2009. Bcl3 interacts cooperatively with peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α to coactivate nuclear receptors estrogen-related receptor α and PPARα. Mol Cell Biol 29: 4091–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y 1998. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396: 590–594 [DOI] [PubMed] [Google Scholar]
- Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, et al. 2002. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418: 443–447 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, et al. 1997. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 7: 715–725 [DOI] [PubMed] [Google Scholar]
- Yin Q, Lamothe B, Darnay BG, Wu H 2009. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry 48: 10558–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoza BK, Hu JY, Cousart SL, Forrest LM, McCall CE 2006. Induction of RelB participates in endotoxin tolerance. J Immunol 177: 4080–4085 [DOI] [PubMed] [Google Scholar]
- Zak DE, Schmitz F, Gold ES, Diercks AH, Peschon JJ, Valvo JS, Niemisto A, Podolsky I, Fallen SG, Suen R, et al. 2011. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc Natl Acad Sci 108: 11536–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M 1997. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell 91: 243–252 [DOI] [PubMed] [Google Scholar]
- Zandi E, Chen Y, Karin M 1998. Direct phosphorylation of IκB by IKKα and IKKβ: Discrimination between free and NF-κB-bound substrate. Science 281: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Zarnegar B, Yamazaki S, He JQ, Cheng G 2008. Control of canonical NF-κB activation through the NIK–IKK complex pathway. Proc Natl Acad Sci 105: 3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, et al. 2010. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-κB upon TLR4 stimulation. J Immunol 185: 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SQ, Kovalenko A, Cantarella G, Wallach D 2000. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity 12: 301–311 [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu LG, Han KJ, Shu HB 2004. Identification of a ZU5 and death domain-containing inhibitor of NF-κB. J Biol Chem 279: 17819–17825 [DOI] [PubMed] [Google Scholar]
- Zhang D, Lin J, Han J 2010. Receptor-interacting protein (RIP) kinase family. Cell Mol Immunol 7: 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Blackwell K, Shi Z, Habelhah H 2010. The RING domain of TRAF2 plays an essential role in the inhibition of TNFα-induced cell death but not in the activation of NF-κB. J Mol Biol 396: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J 2011. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471: 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S 1997. The transcriptional activity of NF-κB is regulated by the IκB- associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89: 413–424 [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell 1: 661–671 [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S 2002. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell 9: 625–636 [DOI] [PubMed] [Google Scholar]