Poly(ADP-ribosyl)ation (PARylation) of axin was recently found to be required for its ubiquitination by the RNF146 ubiquitin ligase. Wang et al. now dissect the interplay between protein PARylation and ubiquitination and find that PARylation appears to be a general signaling mechanism targeting proteins for ubiquitination. The RNF146 WWE domain perceives PARylating through recognition of the iso-ADP-ribose moiety. Crystal structures depict the structural basis for this recognition and reveal that iso-ADP-ribose binding is a common function of WWE domains, which are contained in many ubiquitin ligases.
Keywords: poly(ADP-ribosyl)ation, ubiquitination, WWE domain, PARP-1, axin, crystal structure
Abstract
Protein poly(ADP-ribosyl)ation and ubiquitination are two key post-translational modifications regulating many biological processes. Through crystallographic and biochemical analysis, we show that the RNF146 WWE domain recognizes poly(ADP-ribose) (PAR) by interacting with iso-ADP-ribose (iso-ADPR), the smallest internal PAR structural unit containing the characteristic ribose–ribose glycosidic bond formed during poly(ADP-ribosyl)ation. The key iso-ADPR-binding residues we identified are highly conserved among WWE domains. Binding assays further demonstrate that PAR binding is a common function for the WWE domain family. Since many WWE domain-containing proteins are known E3 ubiquitin ligases, our results suggest that protein poly(ADP-ribosyl)ation may be a general mechanism to target proteins for ubiquitination.
Protein ubiquitination regulates diverse biological processes; however, the mechanism by which proteins are earmarked for ubiquitination remains incompletely understood. Other than phosphorylation, which is a general mechanism for many cases, hydroxylation of a substrate (i.e., HIF1-α) and the binding of small molecules (e.g., the plant hormone auxin) to E3 ligases have been shown to control protein ubiquitination in sporadic cases (Willems et al. 1999; Min et al. 2002; Bergink and Jentsch 2009; Tan and Zheng 2009). Protein poly(ADP-ribosyl)ation (PARylation), catalyzed by PAR polymerases (PARPs), also regulates a myriad of biological processes, including DNA damage responses, transcriptional regulation, intracellular trafficking, energy metabolism, circadian rhythm, and cell survival and cell death programs, among others (Curtin 2005; Jagtap and Szabo 2005; Kim et al. 2005; Schreiber et al. 2006; Krishnakumar and Kraus 2010). How PARylation affects so many biological functions remains largely mysterious. In many cases, such as PARylation of histones in transcriptional regulation, PARylation is considered to control activities of the substrate proteins via the negative charges in the PAR polymer (Schreiber et al. 2006; Krishnakumar and Kraus 2010). In other cases, PAR polymers have been implicated as signaling molecules that can induce cell death, especially in the brain (Andrabi et al. 2006, 2011).
Most recently, PARylation has been shown to control the polyubiquitination and degradation of axin, a key regulator of the Wnt signaling pathway (Huang et al. 2009; Callow et al. 2011; Kang et al. 2011; Zhang et al. 2011). In all of these reports, RNF146 (aka Iduna), which contains a WWE domain and a RING domain (Supplemental Fig. 1), is the only known E3 ubiquitin ligase to date that requires PARylation of the substrate for subsequent polyubiquitination (Callow et al. 2011; Kang et al. 2011; Zhang et al. 2011). The RNF146 WWE domain has been shown to bind PAR (Callow et al. 2011; Zhang et al. 2011), and it was reported that a short PAR-binding motif (PBM) within the domain retains this binding activity (Andrabi et al. 2011). The PBM was originally found in histones and several other proteins (Gagne et al. 2008). However, the PBM identified in RNF146 is not conserved in other WWE domains, so it remains unclear whether the WWE domain represents a novel PAR-binding domain. Here we reveal the structural basis of the RNF146 WWE domain/iso-ADPR interaction and, for the first time, define the PAR/iso-ADPR binding as a bona fide function of the WWE domain family. Importantly, the structural coupling of WWE domains and E3 ligase domains in many WWE domain-containing proteins suggests a functional coupling of protein PARylation and ubiquitination.
Results and Discussion
The RNF146 WWE domain recognizes iso-ADPR, but not ADPR
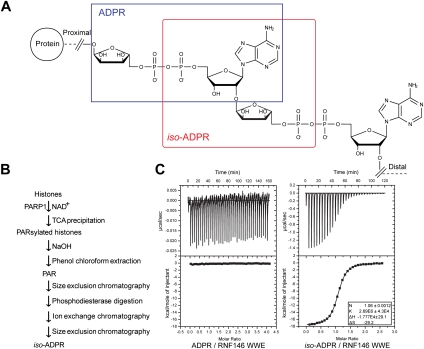
We sought to clarify the requirement of the entire RNF146 WWE domain structure for PAR binding through structural analysis. PAR polymers display high chemical heterogeneity (in both lengths and branching patterns) and are not suitable for quantitative and structural analysis. Thus, we first examined its interaction with ADP-ribose (ADPR), the building unit added to PAR during PAR synthesis (Fig. 1A). Isothermal titration calorimetry (ITC) analysis demonstrated that the RNF146 WWE domain does not interact with ADPR, even at high concentrations (>0.1 mM) (Fig. 1C). We then turned to iso-ADPR, which is the smallest PAR structural unit containing the ribose–ribose glycosidic bond unique to PAR, formed during PAR synthesis by PARPs (Fig. 1A). It remains a major challenge to obtain PAR of a specific length in sufficient quantities for biochemical analysis, and iso-ADPR is not commercially available, nor has it been used previously in structural and biochemical studies. We therefore developed a protocol to biosynthetically generate iso-ADPR (Fig. 1B). We synthesized PAR using histone PARylation by PARP1, following previously published protocols (Kiehlbauch et al. 1993; Fahrer et al. 2007). After removing small molecules by size exclusion chromatography (SEC), we digested PAR polymers with a phosphodiesterase to generate iso-ADPR. Finally, we purified iso-ADPR by ion exchange and a second SEC step to remove all remaining large molecules. The purity and identity of purified iso-ADPR were confirmed by reverse-phase high-performance liquid chromatography (RP-HPLC) and mass spectrometry. ITC analysis demonstrated that, in contrast to the poor interaction with ADPR, the RNF146 WWE domain interacted with iso-ADPR avidly, with a dissociation constant of 0.37 μM (Fig. 1C). Selectivity of the RNF146 WWE domain for iso-ADPR was also demonstrated by its comigration with iso-ADPR, but not ADPR, in SEC (Supplemental Fig. 2). Thus, the RNF146 WWE domain interacts with the PAR polymer, but not mono-ADPR. This is likely to be important for RNF146 function, since ADP-ribosylation and PARylation are performed by different enzymes and have distinct biological functions (Okazaki and Moss 1996; Corda and Di Girolamo 2002; Curtin 2005; Jagtap and Szabo 2005; Schreiber et al. 2006; Krishnakumar and Kraus 2010). We note that the ability to generate purified iso-ADPR provides a unique reagent for biochemical and structural analysis of PAR-binding proteins and mechanistic analysis of PAR-metabolizing enzymes, such as PARG that cleaves the glycosidic bond.
Figure 1.
The interaction between the RNF146 WWE domain and PAR, through the recognition of iso-ADPR. (A) Structures of PAR and its structural units. ADPR is the unit within the blue frame, and iso-ADPR is within the red frame. (B) Procedural outline of iso-ADPR in vitro biosynthesis and purification. (C) ITC analysis showed the binding of the RNF146 WWE domain with iso-ADPR (Kd ∼ 370 nM), but not with ADPR.
Crystal structure of the RNF146 WWE domain in complex with iso-ADPR
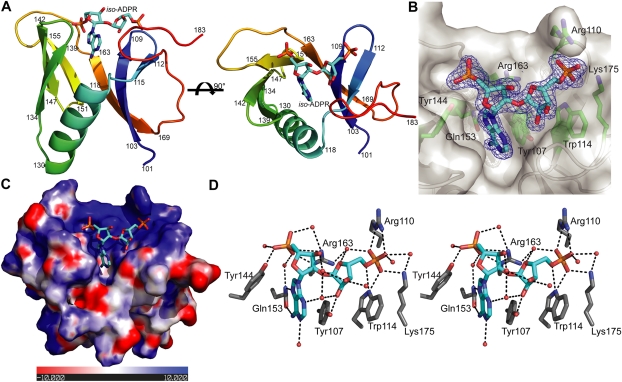
To understand how the RNF146 WWE domain interacts with iso-ADPR, we determined the crystal structure of the RNF146 WWE domain in complex with iso-ADPR at 1.63 Å resolution (Fig. 2A,B; Supplemental Table 1). The WWE domain contains six β strands, forming half of a β barrel, with the other side of this half β barrel covered by an α helix. The high resolution of our structure allowed us to define unambiguously that the ribose–ribose linkage in iso-ADPR is an α(1 → 2) glycosidic bond (Fig. 2B). This confirmed that PAR synthesis catalyzed by PARP1 is α(1 → 2)-specific. The adenine ring of iso-ADPR inserts into the pocket formed by the half β barrel and the α helix (Fig. 2A). The two ribose–phosphate moieties on both sides of iso-ADPR sit on the edge of the half β barrel, which is highly positively charged (Fig. 2C). Both sides of iso-ADPR, especially the two separated phosphate groups, are involved in extensive interactions with WWE domain residues (Fig. 2D), providing an explanation for why the RNF146 WWE domain specifically binds to iso-ADPR and thus to PAR, but not ADPR, which has the two phosphate groups on the same side (Fig. 1A). Compared with a previously determined nuclear magnetic resonance (NMR) structure of an unliganded RNF146 WWE domain (Protein Data Bank [PDB] code 1UJR), it appears that iso-ADPR binding induces significant conformational changes in the WWE domain, particularly in the C-terminal tail region (residues 169–183), which folds back to support the distal ribose–phosphate groups of iso-ADPR (Supplemental Fig. 3). The mode of binding displayed in our crystal structure, in which the two phosphate groups lie on an open surface of the RNF146 WWE domain, should allow internal iso-ADPR units in PAR polymers to bind the WWE domain in the same manner.
Figure 2.
Crystal structure of the RNF146 WWE domain in complex with iso-ADPR. (A) Overall structure of the complex. The protein is shown in rainbow, and iso-ADPR is shown in cyan. (B) Fo − Fc difference density (blue mesh) calculated when iso-ADPR is omitted (contoured at 2.5σ). Protein is shown in gray, and key binding residues are highlighted in sticks. (C) The electrostatic surface potentials of the iso-ADPR-binding region, calculated using Pymol. Negative potential patches are shown in red, and positive potential patches are shown in blue. (D) Stereoview of key interactions (in black dash lines) involved in iso-ADPR binding with the RNF146 WWE domain. Key binding residues are highlighted in sticks, and water molecules are shown as pink spheres.
Mutagenesis analysis of the RNF146 WWE domain
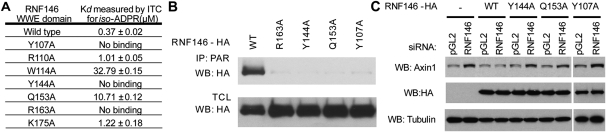
Seven RNF146 WWE domain surface residues are involved in iso-ADPR binding (Fig. 2D). Among them, the Tyr 107 phenol group stacks on the side of the iso-ADPR adenine ring within the pocket, and Gln 153 near the bottom of the adenine-binding pocket forms two hydrogen bonds with the adenine ring and appears to confer binding specificity as well as binding affinity. The Arg 163 and Tyr 144 side chain groups interact with the proximal phosphate group, and Trp 114, Arg 110, and Lys 175 interact with the ribose–phosphate groups in the distal side of iso-ADPR (Fig. 2D). To validate our structure and define key interactions between the RNF146 WWE domain and iso-ADPR, we analyzed the iso-ADPR binding of seven RNF146 WWE domain mutants by ITC analysis. Mutants Y107A, Y144A, and R163A lack a detectable interaction, and mutants W114A and Q153A have much lower affinity than the wild-type RNF146 WWE domain. The mutations R110A and K175A had only a minor effect on the interaction (summarized in Fig. 3A; Supplemental Fig. 4). Then we tested the binding of RNF146 mutants to PAR in the context of a full-length protein. Consistent with the ITC analysis using the WWE domain, the RNF146 mutants Y107A, Y144A, R163A, and Q153A all lost the ability to interact with PAR in a coimmunoprecipitation assay (Fig. 3B).
Figure 3.
Mutagenic analysis of crucial WWE domain residues involved in iso-ADPR binding. (A) Dissociation constants of RNF146 WWE domain mutants with iso-ADPR as measured by ITC analysis. (B) Coimmunoprecipitation of wild-type RNF146—but not RNF146 mutants Y107A, Y144A, Q153A, and R163A—by anti-PAR antibody in HEK293 cells. (C) Expression of siRNA-resistant wild-type RNF146—but not RNF146 mutants Y107A, Y144A, and Q153A—rescues RNF146 siRNA-induced stabilization of Axin1 in HEK293 cells.
To demonstrate the importance of the RNF146 WWE domain residues involved in PAR binding, we examined the effect of their mutation in full-length RNF146 protein on axin turnover in a cellular context. Expression of siRNA-resistant RNF146 completely rescued the effect of RNF146 siRNA on Axin1, whereas expression of RNF146 mutants failed to do so (Fig. 3C). These experiments confirm that the specific interactions observed in the crystal structure are important for the in vivo functions of RNF146.
The previously proposed PBM in RNF146 (residues 144–167) (Andrabi et al. 2011) only accounts for three β strands in the WWE domain structure (Fig. 2A). Residues outside the PBM region interact extensively with iso-ADPR (Fig. 2C,D), and missense mutations of the RNF146 WWE domain outside the PBM, such as Y107A, abolish the PAR-binding activity (Fig. 3). Therefore, we conclude that the RNF146 WWE domain is a bona fide PAR-binding domain, which specifically recognizes the iso-ADPR moiety of PAR.
PAR/iso-ADPR binding is a common function of WWE domains
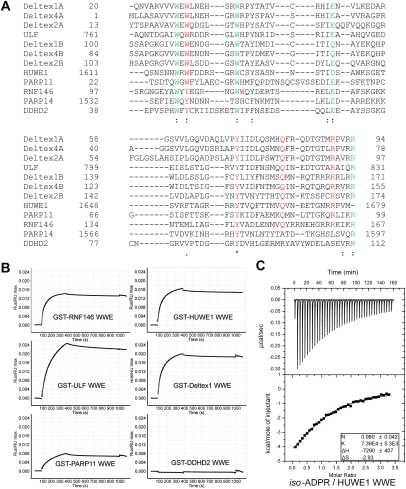
The WWE domain family exhibits a low degree of sequence homology (Fig. 4A). The most conserved residues include the two Trp and one Glu residues that give rise to the name “WWE” domain. These residues are involved in stabilizing the WWE domain fold (Supplemental Fig. 5). Based on the crystal structure of the RNF146 WWE domain/iso-ADPR complex and our mutagenesis analysis, we note that critical residues for iso-ADPR and PAR binding are conserved in most WWE domains, suggesting that PAR binding may be a common function of the WWE domain family (Fig. 4A). Surface plasma resonance (SPR) analysis demonstrated that GST-tagged WWE domains from HUWE1, ULF, Deltex1, and PARP11 bind to the PAR polymers, whereas the DDHD2 WWE domain did not interact with PAR (Fig. 4B). This is in perfect concordance with our structure and sequence alignment. While the four crucial residues involved in iso-ADPR binding in the RNF146 WWE domain are conserved among most WWE domains in the human genome, including these from RNF146, HUWE1, ULF, Deltex1, Deltex2, Deltex4, and PARP11, residues corresponding to RNF146 Q153 and R163 are not conserved in DDHD2 and PARP14 (Fig. 4A).
Figure 4.
PAR-binding properties of the WWE domain family. (A) Multiple sequence alignment by ClustalW of known human WWE domain sequences. Most conserved residues important for WWE domain folding (see Supplemental Fig. 5) are shown in green. Highly conserved residues potentially involved in iso-ADPR binding (corresponding to Tyr 107, Tyr 144, Gln 153, and Arg 163 in RNF146) are highlighted in red, and two other nonconserved iso-ADPR-binding residues in RNF146 (Arg 110 and Trp 114) are in purple. The sequence surrounding RNF146 Lys 175 in the C-terminal tail is not conserved and is not shown here. In accordance with the mutagenesis results shown in Figure 3, conserved residues are important for iso-ADPR binding, whereas nonconserved residues are not. Deltex proteins contain tandem WWE domains (A and B represent the N-terminal and C-terminal WWE domains, respectively). (B) SPR analysis of the interactions between various GST-tagged WWE domains and PAR. Except for DDHD2, all other WWE domains tested interact with PAR. (C) ITC analysis of the interaction between the HUWE1 WWE domain and iso-ADPR.
Given the sequence conservation, we predict that other PAR-binding WWE domains also recognize iso-ADPR in the same way as the RNF146 WWE domain. While many of the WWE domains we tested were not suitable for ITC analysis due to aggregation, our ITC analysis with the HUWE1 WWE domain showed that this domain interacts with iso-ADPR with a Kd of 13 μM (Fig. 4C). The interactions between WWE domains and PAR are further enhanced by the high local concentrations of iso-ADPR around PAR, as there can be as many as ∼200 units in each PAR chain. Deltex1/2/4 proteins contain two tandem WWE domains. Structural modeling based on the unliganded Drosophila Deltex WWE domain structure (Zweifel et al. 2005) also suggests that these two WWE domains may recognize two neighboring iso-ADPR units, with each interacting in a manner similar to the RNF146 WWE domain (Supplemental Fig. 6). Therefore, with the exception of DDHD2 and PARP14, PAR binding through the recognition of iso-ADPR is a common function for most WWE domains.
Structural and functional coupling of protein PARylation and ubiquitination
It has been previously noticed that the majority of WWE domains are structurally coupled with E3 ubiquitin ligases (Supplemental Fig. 1; Aravind 2001). Here we show that the WWE domains from all four E3 domain-containing proteins tested (RNF146, HUWE1, ULF, and Deltex1) interact specifically with PAR. HUWE1 (aka Mule, ARF-BP1, LASU1, and HectH9) is a HECT-type E3 ligase critical for cell regulation; it ubiquitinates the tumor suppressor p53, core histones, and the Bcl-2 family member Mcl-1 (Chen et al. 2006; Bernassola et al. 2008). ULF/TRIP12 is an E3 ubiquitin ligase of ARF, a key activator of p53 (Chen et al. 2010). The Deltex family, which plays an important role in Notch signaling, contains a RING domain and two tandem WWE domains (Zweifel et al. 2005; Katoh 2007). The definition of WWE domains as a novel PAR-binding domain and the structural coupling of the WWE domain with E3 domains suggest that PAR may be a signal for protein ubiquitination—either polyubiquitination that may lead to protein degradation by proteasome, or divergent monoubiquitination that controls protein activities in the cell. The WWE domains may therefore be a key link between protein PARylation and ubiquitination. Furthermore, since the small WWE domains recognize only one (or two) internal iso-ADPR unit, numerous WWE domain-containing proteins may potentially cluster around a PAR polymer, a property that may have functional importance. The role of PARPs and PARylation is well established in many biological processes, including DNA repair. The insights provided by the studies reported here will facilitate design of specific mutations in WWE domain-containing E3s that can be used to unravel the role and the molecular mechanisms of PARylation in biological systems. In summary, protein PARylation may be another general mechanism to label proteins for ubiquitination other than protein phosphorylation, and many of the protein PARylation events may function through ubiquitination. Our studies have not only identified the iso-ADPR as the critical signaling unit in PAR, but also suggested a role for the WWE domains in a superfamily of ubiquitin ligases in decoding the protein PARylation signal.
Materials and methods
Generation and purification of iso-ADPR
Based on the methods reported previously (Kiehlbauch et al. 1993; Fahrer et al. 2007), PAR polymer was synthesized in vitro with some modifications. The histone PARylation reaction was set up in a 15-mL incubation mixture comprising 50 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 50 mM NaCl, 10 mM DTT, 1.5 mM β-NAD+, 0.1 mg/mL histone H1 (Millipore), 0.1 mg/mL histone type IIA (Sigma-Aldrich), 67 mg/mL activator oligonucleotide GGAATTCC, and 5000 U of human PARP-1 (Trevigen). The reaction was stopped after 1.5 h at room temperature by ice-cold trichloroacetic acid. The white pellet was washed with ice-cold ethanol and was further dissolved in buffer containing 0.2 mg/mL Protease K (Invitrogen) and 0.1% SDS. PAR polymer was detached using NaOH and was purified by phenol/chloroform extraction and isopropanol precipitation. After removing small molecules by Superdex 75 on fast protein liquid chromatography (FPLC) (GE healthcare), we digested purified PAR polymer by 50 U of snake venom phosphodiesterase (Worthington) with 15 mM MgCl2 overnight at room temperature. The product of the phosphodiesterase digestion, iso-ADPR, was further purified by anion exchange chromatography and Superdex 75 on FPLC (GE Healthcare). Purified iso-ADPR was air-dried and dissolved by ddH2O to 40 mM final concentration and stored at −20°C. LC-MS detected m/e 558.1 (M-H)− with high purity (the calculated mass of iso-ADPR in acidic form is 559.1).
Protein purification, crystallization, and structure determination
WWE domains were purified as GST fusion proteins, and the GST tag was removed for crystallographic analysis. Purified Se-Met RNF146 WWE domain (residues 99–183) at 10 mg/mL was mixed at 1:1.5 molar ratio with purified iso-ADPR and incubated for 30 min on ice prior to cocrystallization. The hanging-drop method was used to prepare crystals of the Se-Met RNF146 WWE domain in complex with iso-ADPR. One microliter of protein–ligand mixture solution was mixed with 1 μL of well solution containing 45% PEG 400, 100 mM Tris-HCl (pH 8.0), and 10 mM DTT. Ship-shaped crystals usually appeared in 1 d at 22°C and grew to their full sizes in 3 d. The crystals were directly flash-frozen in liquid nitrogen. Screening and data collection were performed at the Advanced Light Source (ALS), beamline 8.2.1. All diffraction data were processed by HKL2000 (Otwinowski and Minor 1997). The structure was determined by single-wavelength anomalous dispersion (SAD) using one data set collected at wavelength 0.9793 Å, which was also used for refinement (Supplemental Table 1). The selenium sites and the initial phases were determined by PHENIX (Adams et al. 2010). Four selenium sites were found in one asymmetric unit, and the experimental electron density map clearly showed the presence of two WWE molecules with two ligands in one asymmetric unit. The complex model was improved using iterative cycles of manual rebuilding with the program COOT (Emsley et al. 2010) and refinement with Refmac5 of the CCP4 6.1.2 program suite (Collaborative Computational Project, Number 4 1994). There is no Ramachandran outlier (98.1% most favored, 1.9% allowed). The electrostatic potential surfaces shown were generated by the APBS tool in Pymol (DeLano and Brunger 1994).
ITC
ITC analyses were carried out using a VP-ITC Microcal calorimeter (MicroCal) at 30°C for the RNF146 WWE domain and its mutants and at 16°C for the HUWE1 WWE domain. All proteins underwent buffer exchange to 20 mM HEPES (pH 7.5), 150 mM NaCl, and 1 mM DTT by a Superdex 75 column, with a final concentration of ∼20 μM. Ligands (ADPR and iso-ADPR) were also diluted by the same buffer to ∼500 μM. A typical titration consisted of injecting 30–40 5-μL aliquots of the ligand into the protein sample (1.4218-mL chamber) at time intervals of 4 min, to ensure that the titration peak reached the baseline. The ITC data were analyzed using the software Origin 7.0 provided by the manufacturer. Data were fit by a one-site model.
SPR
GST-tagged proteins were coupled to Biacore CM5 sensor chip coated with anti-GST antibody. PAR (625 nM; Trevigen) was then profiled at a flow rate of 30 mL/min for 300 sec, followed by a 600-sec flow of wash buffer. After analysis in BiaEvalution (Biacore), the normalized resonance units were plotted over time with the assumption of 1:1 binding.
Immunoblotting and immunoprecipitation
The RNF146 cDNA rescue experiment and the coimmunoprecipitation experiment were performed as described previously (Zhang et al. 2011). Details of experiments can be found in the Supplemental Material.
Accession number
Coordinates and structure factors have been deposited in the PDB (http://www.rcsb.org/pdb) under ID code 3V3L.
Acknowledgments
We are grateful to the staff at ALS beamlines BL 8.2.1 and 8.2.2 for assistance with synchrotron data collection. We thank Dr. Ning Zheng and Dr. Rachel Klevit for helpful discussions. This work was in part supported by a University of Washington RRF award to W.X.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.182618.111.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, et al. 2006. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci 103: 18308–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, et al. 2011. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med 17: 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L 2001. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci 26: 273–275 [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G 2008. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 14: 10–21 [DOI] [PubMed] [Google Scholar]
- Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lill JR, et al. 2011. Ubiquitin ligase RNF146 regulates tankyrase and axin to promote wnt signaling. PLoS ONE 6: e22595 doi: 10.1371/journal.pone.0022595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Brooks CL, Gu W 2006. ARF-BP1 as a potential therapeutic target. Br J Cancer 94: 1555–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shan J, Zhu WG, Qin J, Gu W 2010. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 464: 624–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Corda D, Di Girolamo M 2002. Mono-ADP-ribosylation: A tool for modulating immune response and cell signaling. Sci STKE pe53 doi: 10.1126/stke.2002.163.pe53 [DOI] [PubMed] [Google Scholar]
- Curtin NJ 2005. PARP inhibitors for cancer therapy. Expert Rev Mol Med 7: 1–20 [DOI] [PubMed] [Google Scholar]
- DeLano WL, Brunger AT 1994. Helix packing in proteins: Prediction and energetic analysis of dimeric, trimeric, and tetrameric GCN4 coiled coil structures. Proteins 20: 105–123 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A 2007. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res 35: e143 doi: 10.1093/nar/gkm944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG 2008. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36: 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620 [DOI] [PubMed] [Google Scholar]
- Jagtap P, Szabo C 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov 4: 421–440 [DOI] [PubMed] [Google Scholar]
- Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, et al. 2011. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci 108: 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M 2007. Notch signaling in gastrointestinal tract. Int J Oncol 30: 247–251 [PubMed] [Google Scholar]
- Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK 1993. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem 208: 26–34 [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19: 1951–1967 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Yang H, Ivan M, Gertler F, Kaelin WG Jr, Pavletich NP 2002. Structure of an HIF-1α pVHL complex: Hydroxyproline recognition in signaling. Science 296: 1886–1889 [DOI] [PubMed] [Google Scholar]
- Okazaki IJ, Moss J 1996. Mono-ADP-ribosylation: A reversible posttranslational modification of proteins. Adv Pharmacol 35: 247–280 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Academic Press, New York [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G 2006. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Tan X, Zheng N 2009. Hormone signaling through protein destruction: A lesson from plants. Am J Physiol Endocrinol Metab 296: E223–E227 doi: 10.1152/ajpendo.90807.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Goh T, Taylor L, Chernushevich I, Shevchenko A, Tyers M 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos Trans R Soc Lond B Biol Sci 354: 1533–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13: 623–629 [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Barrick D 2005. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure 13: 1599–1611 [DOI] [PubMed] [Google Scholar]