The neuromuscular junction (NMJ) is a synapse formed between a motor neuron and a muscle fiber. NMJ formation requires the secreted proteoglycan neural agrin, which interacts with LRP4 to signal the receptor kinase MuSK. Zong et al. now provide the first crystal structure of the agrin–LRP4 complex, comprising two heterodimers. The study reveals how the agrin neuron-specific splice insert confers the basis for agrin–LRP4 recognition and provides novel insight into receptor tyrosine kinase activation and early neuromuscular synaptogenesis.
Keywords: crystal structure, motoneurons, neuromuscular junction, receptor tyrosine kinase, synapse, synaptogenesis
Abstract
Synapses are the fundamental units of neural circuits that enable complex behaviors. The neuromuscular junction (NMJ), a synapse formed between a motoneuron and a muscle fiber, has contributed greatly to understanding of the general principles of synaptogenesis as well as of neuromuscular disorders. NMJ formation requires neural agrin, a motoneuron-derived protein, which interacts with LRP4 (low-density lipoprotein receptor-related protein 4) to activate the receptor tyrosine kinase MuSK (muscle-specific kinase). However, little is known of how signals are transduced from agrin to MuSK. Here, we present the first crystal structure of an agrin–LRP4 complex, consisting of two agrin–LRP4 heterodimers. Formation of the initial binary complex requires the z8 loop that is specifically present in neuronal, but not muscle, agrin and that promotes the synergistic formation of the tetramer through two additional interfaces. We show that the tetrameric complex is essential for neuronal agrin-induced acetylcholine receptor (AChR) clustering. Collectively, these results provide new insight into the agrin–LRP4–MuSK signaling cascade and NMJ formation and represent a novel mechanism for activation of receptor tyrosine kinases.
The brain contains billions of nerve cells, or neurons, that integrate signals from the environment and govern the body's responses to external stimuli. The activity of the nervous system is made possible by synapses, which are the fundamental units of neural circuitry that enable complex behaviors (Sanes and Lichtman 1999; Kim and Sheng 2004; Waites et al. 2005; Sudhof 2008). Many proteins critical for synaptic structure have been identified in recent years. However, the molecular mechanisms that govern their assembly to form functional synapses remain poorly understood. The neuromuscular junction (NMJ) is a peripheral, cholinergic synapse that rapidly conveys signals from motoneurons to muscle cells (Froehner 1993; Hall and Sanes 1993; Sanes and Lichtman 1999, 2001). The NMJ exhibits a high degree of subcellular specialization characteristic of chemical synapses. For example, acetylcholine receptors (AChRs) are heavily concentrated in the post-junctional membrane. Being large and easily accessible, this peripheral synapse has contributed greatly to our understanding of the general principles of synaptogenesis as well as of neuromuscular disorders. Interactions between presynaptic motoneurons and post-synaptic muscle fibers are essential to NMJ formation (Sanes and Lichtman 2001; Wu et al. 2010).
Agrin, a basal lamina proteoglycan, is a critical factor secreted by motoneurons to direct NMJ formation (McMahan 1990; Gautam et al. 1996). It is composed of an N-terminal basal membrane-binding domain, nine follistatin-like repeats, and two laminin-B-like modules, followed by three laminin globular (LG) domains at the C terminus (Fig. 1A). Intriguingly, muscles also secrete agrin; however, the muscle agrin isoform lacks a specific alternative splicing sequence at the B/z site within the LG3 domain and is incapable of inducing AChR clustering (Hoch et al. 1993; Gesemann et al. 1996; Burgess et al. 1999; Bezakova et al. 2001). Neural agrin activates muscle-specific kinase (MuSK), a type I receptor tyrosine kinase (RTK), which is a key organizer of NMJ formation (DeChiara et al. 1996; Glass et al. 1996). In MuSK−/− mice, muscle fibers do not form neural clusters or “prepattern” prior to innervation and do not form the NMJ (DeChiara et al. 1996; Lin et al. 2001; Yang et al. 2001). Similarly, neural agrin is unable to induce AChR clusters in MuSK−/− muscle cells (Glass et al. 1996), but agrin sensitivity can be restored by introduction of wild-type MuSK (Zhou et al. 1999; Herbst and Burden 2000). Unlike most conventional RTKs that are activated by a bound ligand, MuSK does not bind directly to neural agrin (Glass et al. 1996), suggesting the existence of a coreceptor and a unique mechanism of activation of MuSK.
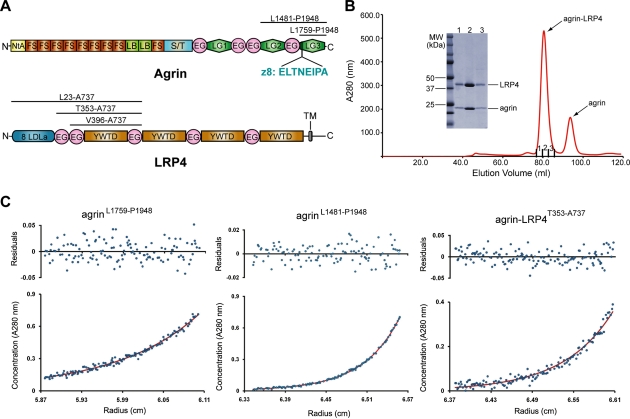
Figure 1.
Characterization of the agrin–LRP4 interaction. (A) Schematic domain organizations of rat agrin and LRP4. The boundaries of protein fragments used in this study are indicated. (NtA) N-terminal agrin domain; (FS) follistatin-like repeat; (LB) laminin-B-like domain; (S/T) serine/threonine glycosylation sites; (EG) EGF-like domain; (LG) laminin-G-like domain; (LDLa) LDL class A repeats; (YWTD) YWTD repeat-containing β propeller; (TM) transmembrane region. (B) Agrin LG3 (residues Leu 1759–Pro 1948) forms a stable complex with LRP4V396–A737 in solution, as demonstrated by size exclusion chromatography. (C) Analytic ultracentrifugation (AUC) studies of agrin LG3, agrin LG2/LG3 (residues Leu 1481–Pro 1948), and the agrin LG3–LRP4T353–A737 complex. Agrin LG3 and LG2/LG3 are both monomeric in solution, and the agrin–LRP4 complex dimerizes with an average Kd of ∼39 μM. (Bottom panel) Absorbance data (blue dots) fit to a single-species model (LG2/LG3 and LG3) or a monomer–dimer model (agrin LG3–LRP4 complex) (red line). (Top panel) Residuals from the fit.
The low-density lipoprotein receptor (LDLR) family is a large family of surface receptors that have been implicated in diverse biological functions (Nykjaer and Willnow 2002). One member of this family, LRP4 (LDLR-related protein 4; also known as MEGF7 [multiple epidermal growth factor-like domains 7]), has a large extracellular N-terminal region, a transmembrane domain, and a short C-terminal region (Fig. 1A; May et al. 2007). The ligand of LRP4 remains unknown although its closest relatives, LRP5 and LRP6, are receptors for Wnt (He et al. 2004). Several recent studies indicate that LRP4 is the obligate receptor of neural agrin. First, mice lacking LRP4 die at birth, with NMJ deficits that resemble those observed in MuSK mutant mice (Weatherbee et al. 2006). Second, biochemical studies indicate that LRP4 interacts with neural, but not muscle, agrin (Kim et al. 2008; Zhang et al. 2008). Third, LRP4 is required for neural agrin-induced AChR clustering (Weatherbee et al. 2006; Zhang et al. 2008). Conversely, coexpression of LRP4 reconstitutes agrin-binding activity, activation of MuSK, and phosphorylation of Abl in nonmuscle cells, which otherwise would not respond to agrin (Zhang et al. 2008). Finally, LRP4 also interacts with MuSK in a manner that is enhanced by neural agrin. These observations demonstrate that LRP4 is a coreceptor of neural agrin that is necessary and sufficient to activate MuSK and initiate downstream signaling cascades for AChR clustering. Despite these observations, little is known of the mechanism by which LRP4 transduces signals from neural agrin to MuSK or how it confers selectivity for the neural isoform of agrin. Understanding how agrin and LPR4 function in these capacities will require a detailed knowledge of the atomic structure of the agrin–LRP4 complex.
Here, we present the first crystal structure of a 2:2 tetrameric signaling complex formed by dimerization of two agrin–LRP4 binary complexes. In particular, formation of the binary complex requires a neuron-specific alternative splicing insertion in agrin, which projects into a pocket on the concave surface of the first β-propeller domain of LRP4. Subsequent tetramerization is synergistically mediated by both agrin and LRP4 through three additional interdimer interfaces. We show by mechanistic studies that agrin-induced dimerization of the agrin–LRP4 binary complex is essential for post-synaptic MuSK activation and AChR clustering. Collectively, our results provide novel insights into the agrin–LRP4–MuSK signaling cascade and NMJ formation. Furthermore, the demonstration that monomeric agrin indirectly activates MuSK through a novel tetrameric ligand–coreceptor complex represents a new paradigm in mechanisms for activation of RTKs.
Results
Characterization of the agrin–LRP4 interaction
Recombinant neural agrin containing the C-terminal LG2 and LG3 domains is as potent as the full-length agrin in terms of AChR clustering activity (Cornish et al. 1999). Furthermore, the LG3 domain containing a neuron-specific eight-amino-acid insert (ELTNEIPA, termed z8) is sufficient to trigger MuSK activation and AChR clustering, albeit with a lower potency (Gesemann et al. 1995; Cornish et al. 1999). These data suggest that the LG3 domain is likely the minimum LRP4-interacting domain in agrin. We thus focused on the two recombinant neural agrin fragments, agrin LG2/LG3 (residues Leu 1481–Pro 1948) and agrin LG3 (residues Leu 1759–Pro 1948), to study their interactions with LRP4 (Fig. 1A).
Until now, the region in LRP4 responsible for agrin association was not known. LRP4 has a large N-terminal extracellular segment (∼1700 residues) that begins with a LDLa (LDL class A) repeat region and is followed by two consecutive EGF modules and four YWTD motif-containing β-propeller domains, each of which is separated by an EGF domain (Fig. 1A). To map the agrin-binding domain in LRP4, we systematically expressed a large number of LRP4 ectodomain truncations using an insect cell expression system. Most of the LRP4 fragments failed to be expressed, except for LRP4L23–A737, which retained the robust binding capacity for agrin LG3 but was expressed at a level too low for structural studies. Using the recombinant LRP4L23–A737 as the starting point, we performed in vitro binding and limited proteolysis and identified the first β-propeller domain (β1) of LRP4 as the minimally functional domain for binding neural agrin LG3 (Supplemental Fig. 1). A second round of expression screening focusing on LRP4 β1 identified two fragments, LRP4T353–A737 and LRP4V396–A737, which had much improved expression yields compared with LRP4L23–A737 when coexpressed and copurified with the agrin LG3 domain. LRP4T353–A737 and LRP4V396–A737 were thus selected for further structural studies (Fig. 1A). Interestingly, LRP4T353–A737 and LRP4V396–A737 failed to be expressed in the absence of agrin.
Agrin LG3 forms a tight complex with LRP4T353–A737 or LRP4V396–A737 in solution, as demonstrated by size exclusion chromatography (Fig. 1B). Further quantification by analytic ultracentrifugation (AUC) suggested that the agrin–LRP4 complex dimerized with an average Kd (dissociation constant) of ∼39 μM (Fig. 1C). However, AUC analysis of the agrin LG3 domain and the agrin LG2/LG3 showed that they were all monomeric in solution (Fig. 1C), which is consistent with an electron microscopic study showing that full-length agrin purified from native sources is a monomer (Denzer et al. 1998). To our knowledge, agrin is the only known RTK ligand that functions as a monomer in concert with an obligate coreceptor (Lemmon and Schlessinger 2010).
Overall structure of the agrin–LRP4 binary complex
To better understand the molecular mechanism of the agrin–LRP4 interaction, we determined the crystal structure of neural agrin LG3 containing the z8 insert (hereafter referred to as agrin) in complex with LRP4V396–A737 (referred to as LRP4) at 2.85 Å resolution. The structure reveals two agrin–LRP4 binary complexes, each with 1:1 stoichiometry, packed in one asymmetric unit (ASU) (Fig. 2A). The agrin LG3 domain adopts a slightly curved β-sandwich structure with a total of 14 β strands, which is homologous to the LNS (laminin, neurexin, and sex hormone-binding globulin-like) domain (Timpl et al. 2000; Rudenko et al. 2001). The C terminus of agrin is anchored close to the N-terminal end by a disulfide bond that is conserved in many LNS domains. The central part of LRP4V396–A737 is a six-bladed β-propeller domain, which is conserved in the LDLR family in that the second strand of each blade contains the YWTD motif (Springer 1998; Takagi et al. 2003). There are two slightly concave surfaces perpendicular to the β propeller's central pseudo-sixfold axis. While one surface of β1 is covered by the two EGF modules flanking the β-propeller domain, the opposite surface is involved in agrin binding. The crystal structures of the β-propeller domains of LRP6, a close relative of LRP4 in the LDLR family, have been reported recently (Ahn et al. 2011; Bourhis et al. 2011; Chen et al. 2011; Cheng et al. 2011). These homologous β-propeller domains in the LDLR family and nidogen all use the same surface for ligand binding (Rudenko et al. 2002; Takagi et al. 2003; Ahn et al. 2011; Bourhis et al. 2011; Chen et al. 2011; Cheng et al. 2011).
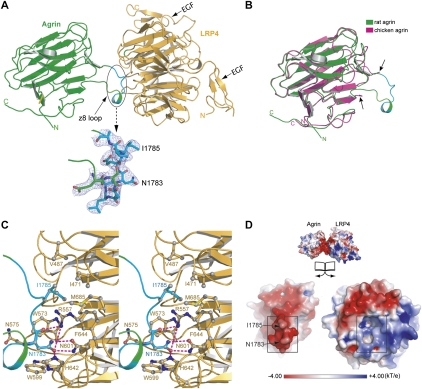
Figure 2.
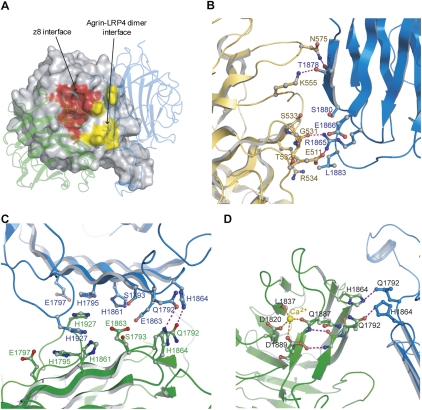
Binary complex of agrin LG3 (agrin) and LRP4V396–A737 (LRP4). (A) The association of agrin (green) and LRP4 (orange) is mostly mediated by the neuron-specific z8 alternative splicing sequence in agrin. This loop is unambiguously defined by excellent electron densities (2Fo − Fc map contoured at 1σ) (in cyan). (B) The structure of rat agrin (green) in the context of an agrin–LRP4 complex is superimposed on the structure of an apo form of chicken agrin that also has an eight-amino-acid insert at the B/z site (magenta; PDB code 1PZ8). The arrows indicate the boundaries of the disordered z8 loop of the apo chicken agrin. (C) Stereoview of the detailed interactions between agrin (green/cyan) and LRP4 (yellow). Key residues that are directly involved in complex interactions are shown as the ball-and-stick model. Hydrogen bonds are indicated by dotted lines. (D) Open-book view of the electrostatic potential of the z8-interacting surfaces (highlighted with squares). The negatively charged z8 loop is complementary to the positively charged surface in LRP4.
Agrin–LRP4 binary complex formation is mediated by the neuron-specific z8 loop
Agrin and LRP4 in the binary complex are tethered primarily by a long loop of agrin, mostly composed of the z8 alternative splicing sequence (we refer to this as the z8 loop). The entire z8 loop shows well-defined electron density (Fig. 2A). In contrast, the homologous loop was completely disordered in the apo form of chicken neural agrin (Protein Data Bank [PDB] code 1PZ8), where the binding partner LRP4 was not present (Stetefeld et al. 2004), indicating an induced-fit recognition between agrin and LRP4 (Fig. 2B). It appears unlikely that the muscle isoform of agrin that lacks the z8 loop would interact with LRP4, consistent with the finding that the B/z insert is indispensable for the function of neural agrin in NMJ synaptogenesis (McMahan et al. 1992; Gesemann et al. 1995).
Close inspection of the binary complex showed that Asn 1783 and Ile 1785 on the tip of the agrin z8 loop project into two deep pockets on the concave surface of LRP4 (Fig. 2C). Specifically, agrin Asn 1783 forms multiple hydrogen bonds with Arg 557, Asn 601, and His 642 of LRP4. In addition, agrin Ile 1785 is bound in a deep hydrophobic pocket composed of Phe 644, Ile 471, Val 487, Trp 658, and Met 685 on the surface of LRP4. The total buried solvent-accessible area of the z8–LRP4 interaction is ∼525 Å2. The critical role of agrin Asn 1783 and Ile 1785 in agrin–LRP4 complex formation was verified by our mutagenesis studies (discussed below). In addition to the direct intracomplex interactions, complementary electrostatic interactions observed between the z8 loop of agrin and its binding surface in LRP4 may further stabilize the binary complex (Fig. 2D).
Agrin–LRP4 forms a 2:2 tetrameric complex
The two agrin–LRP4 binary complexes in the ASU further assemble into a dimer with a twofold noncrystallographic symmetry (Fig. 3A). In addition to the z8 loop-mediated binary complex interface (termed the z8 interface), the dimeric status of the binary complexes is maintained by three new interfaces that bury a large solvent-accessible surface of ∼900 Å2 per tetramer. Two interfaces are between agrin and LRP4 (termed the agrin–LRP4 dimer interface) that are symmetrically equivalent. The third is between two agrin molecules (termed the agrin–agrin dimer interface) (Fig. 3A). No direct interaction is observed between the two LRP4 molecules. The dimerization of the binary complex in the crystal is consistent with its dimerization in solution, as shown by AUC (Fig. 1C).
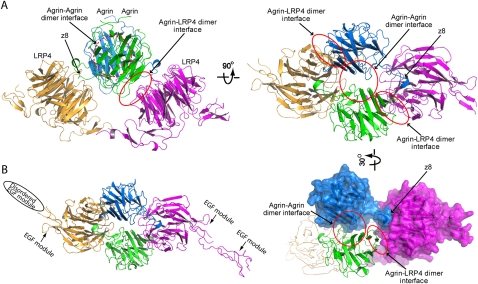
Figure 3.
The tetrameric architecture of the agrin–LRP4 complex. (A) Two agrin–LRP4V396–A737 binary complexes associate with each other with a noncrystallographic twofold symmetry. Besides the z8 interface, the tetrameric complex is stabilized by a second interface between agrin and LRP4 as well as an agrin–agrin interface, which are highlighted by red cycles. (B) The crystal structure of agrin–LRP4T353–A737 has a crystal packing different from that of agrin–LRP4V396–A737, but adopts the identical 2:2 tetrameric assembly of agrin and LRP4 in an asymmetry unit.
To further verify the unique assembly of the agrin–LRP4 complex, we determined a second crystal structure of a complex composed of agrin LG3 and LRP4T353–A737 at 3.30 Å resolution (Table 1). LRP4T353–A737 contains one extra N-terminal EGF module in comparison with LRP4V396–A737, which leads to a different crystal packing of the agrin–LRP4T353–A737 complex. Nevertheless, the two structures show the identical 2:2 tetrameric arrangement of agrin and LRP4, suggesting that the dimerization of the agrin–LRP4 binary complex is likely a faithful representation of the physiological conformation of the agrin–LRP4 association (Fig. 3B). Only one of the two LRP4T353–A737 in the tetrameric complex shows clear electron densities for the first N-terminal EGF module, indicating the flexibility of this domain.
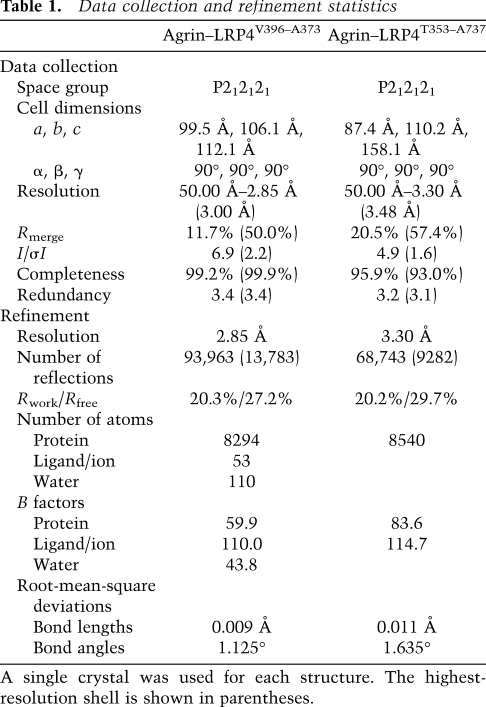
Table 1.
Data collection and refinement statistics
Notably, a structural similarity search in the PDB revealed that the architecture of LRP4T353–A737, including the β1 domain and flanking EGF modules, is very similar to the ectodomain of LDLR (PDB code 1N7D) (Supplemental Fig. 2A; Rudenko et al. 2002). Intriguingly, both LRP4 and LDLR have multiple LDLa repeats at the N terminus, and in LDLR, the LDLa repeats form an extended arc-like structure that folds back to interact with the ligand-binding surface of its β-propeller domain. Structural modeling suggested that, if the LDLa repeats of LRP4 adopt a conformation similar to that of LDLR, then the z8 interface and the agrin–LRP4 dimer interface in LRP4 will be partially occupied by LDLa repeats. It is thus tempting to propose that the LDLa repeats of LRP4 could protect the ligand-binding surface in the β1 propeller when the ligand is absent. Subsequently, agrin binding would displace the associated LDLa and initiate formation of the signaling complex (Supplemental Fig. 2B). If true, this model could explain our observation that only the LDLa-containing fragment of LRP4 (i.e., LRP4L23–A737) could be expressed in the absence of agrin.
Agrin–LRP4 dimer interface
The agrin–LRP4 dimer interface in the LRP4 β1 propeller is formed mainly by the loops connecting blades 3 and 4 (yellow in Fig. 4A), which is in close vicinity to the central z8 interface (red in Fig. 4A). Thus, the ligand-binding surface of LRP4 β1 splits to bind two molecules of agrin. Interestingly, the equivalent surface in the homologous nidogen binds one molecule of laminin EGF-like (LE) domain through a loop in LE4 that resembles the z8 loop of agrin (Supplemental Fig. 3; Takagi et al. 2003). This supports the importance and flexibility of this β-propeller surface for ligand binding.
Figure 4.
Agrin–LRP4 tetramer interfaces and the Ca2+-binding site in agrin. (A) In the LRP4 β1 propeller, the two agrin-binding interfaces (highlighted in red and yellow) are close to each other. The two bound agrin molecules are shown in the transparent cartoon. (B) A close-up view of the agrin–LRP4 dimer interface, where agrin and LRP4 are colored in blue and orange, respectively. (C) A close-up view of the agrin–agrin dimer interface, where the two agrin molecules are colored in blue and green. (D) A Ca2+ atom is coordinated by side chains of D1820 and D1889 and carbonyl oxygen of Q1887 and L1837. Q1887 is only 5 Å away from Q1792, which mediates the agrin–agrin dimer interface.
The agrin–LRP4 dimer interface in agrin locates on the rim of its β sandwich, on the opposite side of the z8 loop and its N/C termini (Fig. 3A). Specifically, the LRP4-binding rim is composed mainly of two β strands (Gln 1840–Thr 1847 and Ala 1875–Leu 1883) that are in van der Waals interaction distance with LRP4. Complementing this, Arg 1865 and Thr 1878 of agrin form hydrogen bonds with Glu 511/Gly 531 and Lys 555/Asn 575 of LRP4, respectively (Fig. 4B). Notably, a similar ligand-binding motif is observed in several LNS domain-containing proteins—e.g., laminin, neurexin, perlecan, and SHBG—where the same rim of the β-sandwich structure is used for interactions with a protein and/or a carbohydrate, suggesting that this rim of the β sandwich is a conserved ligand-binding site (Rudenko et al. 2001).
Agrin–agrin dimer interface and the Ca2+-binding site in agrin
Although agrin LG3 is monomeric in solution, it forms a dimer in the tetrameric agrin–LRP4 complex. In the center of the agrin–agrin dimer interface, His 1927 in one agrin is sandwiched between His 1927 and His 1975 in the other agrin. The aromatic rings of these four histidine residues—two from each agrin—thus make three pairs of interlaced stacking contacts that likely provide the major force to stabilize this interface (Fig. 4C). Additional interactions bridging the two agrin molecules involve residues His 1864 and Gln 1792, making two pairs of interagrin hydrogen bonds.
Notably, the agrin–agrin dimer interface is in close proximity to a Ca2+-binding site in agrin, where a Ca2+ is chelated by the side chains of Asp 1820/Asp 1889 and the carbonyl oxygen of Leu 1837/Gln 1887 (Fig. 4D). Illustrating this proximity, Gln 1792 is only 5 Å away from Gln 1887. The bound Ca2+ may thus stabilize the local conformation of agrin and facilitate agrin–agrin dimerization. This might explain how Ca2+ enhances the agrin–LRP4 interaction and facilitates agrin function (Tseng et al. 2003; Stetefeld et al. 2004; Kim et al. 2008).
Assembly of the tetrameric supercomplex is a stepwise process
The crystal structure has provided detailed information that allows us to interrogate the role of the three distinct interaction interfaces within the tetrameric agrin–LRP4 complex: the z8 interface, the agrin–LRP4 dimer interface, and the agrin–agrin dimer interface. To do this, we designed three groups of single-point mutants of agrin LG3, with each group selectively disrupting one intratetramer interface, while having minimal effects on the other interfaces. The group 1 mutations include N1783A, which eliminates the hydrogen bonding mediated by Asn 1783, and I1785S, which attenuates the hydrophobicity of Ile 1785. These two mutations are designed to disrupt the z8 interfaces of the binary complexes. The group 2 mutation is R1865E, which abolishes the hydrogen bonding between Arg 1865 and LRP4 Glu 511/Gly 531 and subsequently disrupts the agrin–LRP4 dimer interfaces. The group 3 mutations are H1795L and H1927L, which interfere with the aromatic stacking interactions that are mainly responsible for stabilizing the agrin–agrin dimer interface.
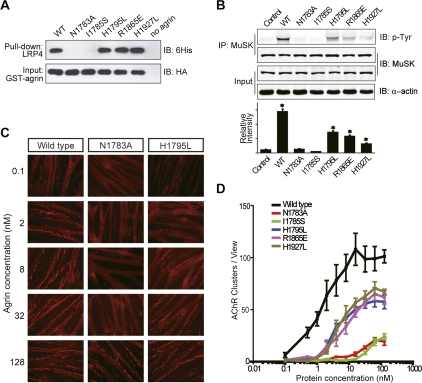
All agrin mutants had the same protein-melting temperature as wild-type agrin, indicating their proper folding (Supplemental Fig. 4). We first performed in vitro GST pull-down assays to examine the binding between the agrin variants and LRP4. We used a longer fragment of LRP4, LRP4L23–A737, for this assay, since the two shorter fragments used in the structural studies need to be coexpressed with agrin. In the GST pull-down assays, agrin-R1865E, H1795L, and H1927L each bound well to LRP4L23–A737, indicating that the agrin–LRP4 and agrin–agrin dimer interfaces are not major forces linking agrin and LRP4. However, agrin-N1783A and I1785S mutants bearing a defective z8 loop did not pull down LRP4 (Fig. 5A).
Figure 5.
Agrin-induced tetramerization of agrin–LRP4 is critical for AChR clustering and MuSK phosphorylation. (A) The z8 interface plays a predominant role in assembly of the agrin–LRP4 complex. GST pull-down assays were performed between GST-agrin variants (containing a HA tag) and LRP4L23–A737 (containing a His tag). Agrin and LRP4 were detected by an anti-HA antibody and an anti-His antibody, respectively. (B) MuSK phosphorylation induced by agrin variants. Fully differentiated myotubes were treated with wild-type or mutant agrin at 100 nM. MuSK was immunoprecipitated with anti-MuSK polyclonal antibody, and phosphorylation was detected by immunoblotting with anti-phosphotyrosine antibody 4G10. The bar graph shows mean ± SD, n = 3; (*) P < 0.05 in comparison with the wild-type agrin. (C) Representative images of AChR clustering (red dots) on C2C12 myotubes induced by wild-type LG3, LG3-N1783A, or LG3-H1795L. (D) Quantitative analyses of AChR clustering assay for agrin variants. Data are mean ± SD based on three independent experiments.
We further analyzed the effects of agrin mutants on the dimerization of the agrin–LRP4 binary complex. We successfully copurified LRP4T353–A737 in complex with agrin-H1795L, R1865E, or H1927L. The AUC studies showed that all three agrin mutants formed stable binary complexes with LRP4. However, the dimerization Kd of the binary agrin–LRP4 complex increased by ∼5.0-fold, ∼3.0-fold, and ∼4.4-fold for agrin-H1795L, R1865E, and H1927L, respectively, in comparison with that of wild-type agrin (Supplemental Fig. 5). Attempts to copurify LRP4T353–A737 in complex with agrin-N1783A or I1785S failed, likely due to the defective z8 interface that disrupted the binary complex. Collectively, these data support our hypothesis that the formation of the binary agrin–LRP4 complex is primarily mediated by the neuron-specific z8 interface and is likely an independent and prerequisite event for subsequent formation of the 2:2 tetrameric complex.
Agrin–LRP4 tetramerization is required for MuSK activation and AChR clustering
To investigate the physiological function of the agrin–LRP4 binary complex and the 2:2 tetrameric complex, we next examined the effects of the agrin mutants in ex vivo cell-based experiments. Muscle cells were stimulated with wild-type or mutant agrin LG3 and then analyzed for MuSK phosphorylation and AChR clustering. As shown in Figure 5B, wild-type agrin LG3 induced robust MuSK phosphorylation; however, the agrin mutants N1783A and I1785S, which were unable to interact with LRP4 via the z8 interface (Fig. 5A), lost the ability to stimulate MuSK. Similarly, the ability of these mutants to induce AChR clusters was severely impaired (Fig. 5C,D). These results indicate that the z8 interface is essential for agrin function, which is consistent with a recent report that mutating Asn 1783 eliminates the AChR clustering activity of neural agrin (Tseng et al. 2010). Intriguingly, the agrin mutants R1865E, H1795L, or H1927L, which primarily disrupt tetramerization of the complex but have no effect on the z8 interface, significantly reduced but did not eliminate MuSK phosphorylation and AChR clustering (Fig. 5B–D). Specifically, the maximal response elicited by saturating concentrations of mutant agrin was only 50%–60% of the response elicited by wild-type agrin. Collectively, these data show that the binary agrin–LRP4 complex is necessary but not sufficient to fulfill agrin's physiological function. On the other hand, the maximal physiological response of agrin requires agrin-induced dimerization of the agrin–LRP4 binary complex.
Discussion
Neuron-specific trans-synaptic regulation
The B/z alternative splicing insert of agrin plays a critical role in triggering AChR clustering; this role has been appreciated ever since the different isoforms of agrin were discovered (Campanelli et al. 1992; McMahan et al. 1992). Interestingly, synthetic peptides containing the z8 sequence did not replicate the function of neural agrin or interfere with the function of endogenous agrin (Stetefeld et al. 2004; Tseng et al. 2010), suggesting that the insert itself is not separately functional, but instead functions in the context of the three-dimensional structure of the agrin LG3 domain. Previous attempts to visualize the structure of this loop have been unsuccessful, as it is completely disordered in the structure of the apo form of neural agrin (Stetefeld et al. 2004). We show here that the z8 loop has a well-defined structure when the agrin LG3 is bound with its coreceptor, LRP4, suggesting an induced-fit molecular recognition between the ligand and receptor. Consistent with this, disrupting the z8 interface by mutagenesis almost completely abolished agrin's neuronal function.
Surprisingly, only two residues on the tip of the z8 loop (Asn 1783 and Ile 1785) directly interact with LRP4 β1, and as a result, this loop buries only ∼525 Å2 of the solvent-accessible area. To our knowledge, this is one of the smallest protein–protein interaction interfaces known that supports high binding affinity (Takagi et al. 2003; Janin et al. 2007). Interestingly, a similar loop-to-β-propeller-binding pattern was observed in the interactions between laminin and nidogen and between DKK1 and the LRP6 β1-propeller domain (Supplemental Fig. 3; Takagi et al. 2003; Ahn et al. 2011; Bourhis et al. 2011; Cheng et al. 2011). Despite no similarity in the amino acid sequences, laminin, agrin, and the Wnt signaling inhibitors such as DKK1 and SOST mediate receptor recognition through an almost identical N-X-I/V (where X is any residue) motif located in an extended loop. Thus, it could represent a universal structural motif by which a signaling protein recognizes a YWTD β-propeller-containing receptor.
Dimerization of the agrin–LRP4 binary complex
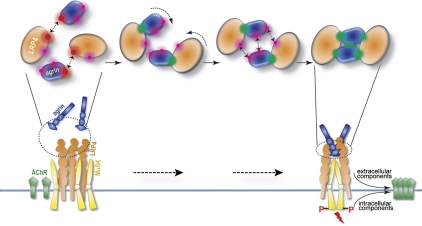
Unlike the neuron-specific z8 interface, the composition of the agrin–LRP4 and agrin–agrin dimer interfaces do not provide strong driving forces for the intermolecular interactions. Therefore, the z8 interface appears crucial for initial binary complex formation and sets the stage for the subsequent multivalent weak contacts. We thus propose a two-step model for agrin function (Fig. 6), in which monomeric agrin first assembles with LRP4 into a binary complex, predominantly mediated by the neuron-specific z8 loop. The binary agrin–LRP4 complex then reconfigures the relatively weak agrin–LRP4 dimer interface and agrin–agrin dimer interface to promote synergistic dimerization of the binary complex. The affinity for dimerization of the agrin–LRP4 binary complex appears to be moderate (Kd ∼39 μM) based on the in vitro binding assays using the minimum interacting domains, but the overall avidity should be strengthened in vivo due to high local concentrations of the full-length proteins in the membrane at the NMJ. In addition to facilitating MuSK-mediated signaling, the agrin–LRP4–MuSK supercomplex may provide a structural scaffold to recruit additional molecules for synapse formation. For example, agrin is known to increase rapsyn targeting to the NMJ post-synaptic membrane to enhance AChR clustering (Brockhausen et al. 2008).
Figure 6.
Model of the agrin–LRP4–MuSK signaling pathway. MuSK (yellow) interacts with self-associated LRP4 (orange) in a nerve-independent manner. Monomeric agrin (blue) secreted by motor neurons first binds to the LRP4 β1 domain through a neuron-specific z8 insert (interface shown as a red glowing cycle). The binary complex then reorganizes the agrin–LRP4 dimer interface and the agrin–agrin dimer interface (magenta glowing cycles) into a unique configuration that is competent to further dimerize. Finally, the tetrameric complex of agrin–LRP4 is stabilized by five separate interfaces in a cooperative manner. Such interaction is necessary for MuSK activation, which leads to AChR clustering and synaptic differentiation
A working model of MuSK activation
Considering that LRP4 could self-associate and bind MuSK independently of agrin, we propose a working model for MuSK activation (Fig. 6). In the absence of neural agrin, MuSK forms a heterodimer with LRP4 to maintain basal activity (Kim et al. 2008; Zhang et al. 2008). At the early stage of NMJ differentiation, agrin secreted by motoneurons binds with LRP4 initially through the z8 interface to form the binary complex and subsequently promotes the assembly of a tetrameric agrin–LRP4 complex. This supercomplex will likely reorganize the preformed MuSK–LRP4 complex as well as LRP4 self-association at immature NMJ and subsequently promote activation and trans-phosphorylation of MuSK, leading to AChR clustering and synaptic differentiation. This scenario is consistent with the enhanced LRP4–MuSK interaction observed in the presence of agrin (Kim et al. 2008; Zhang et al. 2008). However, it is not yet clear whether activation of MuSK—presumably by MuSK dimerization (Hopf and Hoch 1998)—involves a new composite surface formed by the tetrameric agrin–LRP4 or conformational changes in LRP4 and/or MuSK.
MuSK is a member of a unique family of RTKs for which activation requires not only a soluble ligand, but also the involvement of additional coreceptors (Lemmon and Schlessinger 2010). Another prominent member of this family is the RET (rearranged during transfection) receptor that is indirectly activated by GDNF (glial-derived neurotrophic factor) family ligands (GFLs) through the coreceptor GFRα (GDNF family receptor-α) (Runeberg-Roos and Saarma 2007). However, key differences exist between the agrin–LRP4 and GFL–GFRα interactions. First, agrin is a monomeric ligand, while the RET ligand GDNF functions as a homodimer (Wang et al. 2006; Parkash et al. 2008). Second, the formation of the agrin–LRP4 complex is a two-step synergistic process, while GFLs dimerize with GFRα in a 1:1 stoichiometry (Schlee et al. 2006; Wang et al. 2006; Parkash et al. 2008). Finally, MuSK prebinds to its coreceptor, LRP4, before activation, while RET does not (Kim et al. 2008; Zhang et al. 2008). Therefore, our current model of MuSK activation may represent a new paradigm in mechanisms of RTK activation.
Materials and methods
Protein expression and purification
Rat neural agrin LG3 (residues Leu 1759–Pro 1948), containing the z8 (ELTNEIPA) sequence, was subcloned into a modified pAcGP67 vector (BD Biosciences) that harbors an N-terminal six-histidine (6His) tag and a HRV 3C protease cleavage site. The same agrin fragment with an additional C-terminal HA tag was subcloned into a pGEX-6P vector (GE Healthcare). Point mutations of agrin were introduced by QuikChange site-directed mutagenesis (Stratagene). Fragments of rat LRP4, LRP4V396–A737 (residues Val 396–Ala 737), LRP4T353–A737 (residues Thr 353–Ala 737), and LRP4L23–A737 (residues Leu 23–Ala 737) were cloned into the modified pAcGP67 vector.
For insect cell expression, high-titer baculoviruses made in Sf9 cells were used to infect Hi5 cells for secreted protein expression. Specifically, LRP4V396–A737 and LRP4T353–A737 were coexpressed with agrin LG3 in Hi5 cells, and LRP4L23–A737 was expressed alone. The agrin–LRP4 complexes were first purified with a Ni-NTA (nitrilotriacetic acid) affinity column, followed by 3C cleavage to remove the 6His tag, and were further purified by a Superdex-200 size exclusion column (GE Healthcare). The pGEX-agrin constructs were transformed into the Escherichia coli strain BL21Star (Novagen). Bacteria were grown in LB medium at 37°C to OD600 of 0.6–0.7 and induced with IPTG (isopropyl-β-D-thiogalactopyranoside) for 4–5 h at 30°C. The GST-agrin fusion proteins were purified with glutathione resins in batch mode. They were eluted and later used for pull-down experiments or were digested with 3C on-column to prepare GST-free agrin for Tm measurement, AUC, AChR clustering, and MuSK phosphorylation experiments.
Protein characterization
The thermal stability of agrin variants was measured using a fluorescence-based thermal shift assay on Roche Lightcycler 480 II. Ten microliters of 10 μM each agrin variant was mixed with 5× fluorescent dye SYPRO Orange (Sigma-Aldrich) immediately before heating. The samples were then heated from 20°C to 95°C in 20 min. The midpoint (Tm value) of the protein-melting curve was determined using the melting curve analysis software provided by the instrument manufacturer. Three independent experiments were performed, and the data were averaged.
Sedimentation equilibrium experiments were performed at 20°C in a ProteomeLab XL-I (BeckmanCoulter) analytical ultracentrifuge with an An-50 Ti eight-place rotor. Purified agrin LG2/LG3 and LG3 domains and various agrin–LRP4 complexes were dialyzed extensively against a buffer containing 15 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM CaCl2. Samples were loaded into six-channel equilibrium cells at 0.26, 0.13, and 0.065 mg/mL for agrin and run at speeds of 20,000 and 23,000 rpm until equilibrium was reached; for the wild-type agrin–LRP4 complexes, concentrations were 0.19, 0.10, and 0.05 mg/mL, and speeds were 14,000 and 16,000 rpm. For the mutant agrin–LRP4 complexes, protein concentrations were all adjusted to 0.10 mg/mL, and speeds were 14,000 and 16,000 rpm. Three independent experiments were repeated for each sample. Data analysis using HeteroAnalysis software (by J.L. Cole and J.W. Lary, University of Connecticut) showed that agrin LG2/LG3 and LG3 are monomeric in solution. Best fits for the wild-type and the mutated agrin–LRP4 complexes were achieved using a monomer–dimer model.
Crystallization and structure determination
The agrin–LRP4 complexes were concentrated to 7–8 mg/mL before crystallization. Agrin–LRP4V396–A737 was crystallized in 20% PEG 3350, 0.2 M KI, and 20 mM Tris (pH 8.0). Agrin–LRP4T353–A737 was crystallized in 20% PEG 3350 and 0.2 M phosphate buffer (pH 8.0). In both cases, single crystals were obtained at 18°C by seeding from initial low-quality small crystals. All crystals were protected in the original mother liquor complemented with 15% glycerol, flash-frozen in liquid nitrogen, and analyzed at the NE-CAT beamline 24-ID-C/E of Advanced Photon Source (APS). The best X-ray diffraction data for agrin–LRP4V396–A737 and agrin–LRP4T353–A737 were collected up to 2.85 Å and 3.30 Å resolution, respectively. The data were processed with iMOSFLM and scaled with SCALA of CCP4 (Collaborative Computational Project, Number 4 1994; Battye et al. 2011). Crystals of agrin–LRP4V396–A737 belong to space group P212121, with unit cell dimensions a = 99.5 Å, b = 106.1 Å, and c = 112.1 Å; α = β = γ = 90°. Crystals of agrin–LRP4T353–A737 belong to space group P212121, with unit cell dimensions a = 87.4 Å, b = 110.2 Å, and c = 158.1 Å; α = β = γ = 90°.
The agrin–LRP4V396–A737 structure was determined using Phaser (McCoy et al. 2007) by molecular replacement using the structures of chicken agrin (PDB code 1PZ8) (Stetefeld et al. 2004) and nidogen β-propeller domain (PDB code 1NPE) (Takagi et al. 2003) as models. The structural modeling and refinement were carried out iteratively using COOT and Refmac5.5 of the CCP4 suite (Collaborative Computational Project, Number 4 1994; Murshudov et al. 1997; Emsley and Cowtan 2004). Several N-terminal residues in LRP4, including Val 396–Gly 403 and Glu 413–Ala 415 in one chain and Val 396–Gly 403 and Asn 411–Ala 415 in the other chain, do not have visible electron densities. The final refinement of the structure was performed with PHENIX (Adams et al. 2010), resulting in Rwork/Rfree of 20.3%/27.2%. The agrin–LRP4T353–A737 structure was determined by molecular replacement using the agrin–LRP4V396–A737 structure as the search model and refined to Rwork/Rfree of 20.2%/29.7%. Due to the low resolution, residues Thr 353–Asn 357 and Ser 412–Ala 415 in one chain of LRP4 and residues Thr 353–Gly 403 and Asn 411–Ala 415 in the other are not visible. The final structures were validated with MolProbity (Chen et al. 2010) and showed excellent stereochemistry. In the Ramachandran plots, 92% of residues are in the most favored regions, 8% are in the additionally allowed region, and no residues are in the disallowed regions for the agrin–LRP4V396–A737 complex. The corresponding measurements for agrin–LRP4T353–A737 are 81.5%, 15.0%, and 3.5%, respectively. All structure figures were prepared with PyMol (http://www.pymol.org).
GST pull-down assay
For GST pull-down assays, purified GST-agrin variants (with HA tag) were prebound to glutathione resins for 1 h. After washing, the resins were incubated with insect culture medium containing the secreted 6His-LRP4L23–A737. After 5 h binding at 4°C, the resins were washed twice with PBS, boiled in SDS loading buffer, and resolved by SDS-PAGE. LRP4L23–A737 and agrin were visualized by Western blot using anti-6His (GE Healthcare) and anti-HA (Sigma-Aldrich) antibodies, respectively.
AChR clustering assay
AChR clusters in C2C12 myotubes were measured as described previously with modification (Luo et al. 2008; Zhang et al. 2008). C2C12 cells were maintained in 35-mm dishes as undifferentiated myoblasts in DMEM with high glucose supplemented with 20% fetal bovine serum, 0.5% chicken embryo extract, and 2 mM L-glutamine. Fusion of myoblasts into myotubes was induced by culturing cells in differentiation medium consisting of DMEM supplemented with 5% horse serum and 2 mM L-glutamine. Two days later, fully differentiated myotubes were treated with wild-type or mutant agrin proteins at the indicated concentrations for 16 h. The cells were then washed with PBS and fixed in 4% paraformaldehyde (in PBS) for 30 min. After washing with PBS, AChR clusters were labeled by incubating cells with 50 nM rhodamine-conjugated bungarotoxin (R-BTX) (Invitrogen) for 60 min at room temperature. After three washes with PBS, the dishes were mounted in VectaSheild (Vector Laboratories), covered with coverslips, and viewed under a Zeiss epifluorescence microscope. Images were collected with Axiovision 3.1 software. AChR clusters with diameters or a longer axis ≥4 μm were scored. At least 10 views per dish and at least two dishes were scored in each of three independent experiments.
MuSK phosphorylation assay
The assay was performed as previously described (Zhang et al. 2008; Zhu et al. 2008). Fully differentiated myotubes were treated with wild-type or mutant agrin proteins (100 nM) for 1 h, and cells were then lysed with RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate at pH 8.0). Cell lysates were cleared by centrifugation and subjected to immunoprecipitation with anti-MuSK polyclonal antibody (G3652) and protein A or protein G beads (Roche) overnight at 4°C. After a brief centrifuge, the supernatants were removed, and pellets were washed three times in lysis buffer. Bound proteins were denatured by SDS loading buffer, resolved by SDS-PAGE, and analyzed by immunoblotting with anti-phosphotyrosine antibody 4G10 (Millipore). The blots were scanned using an Odyssey infrared imager (LI-COR). Immunoblots were quantified with NIH image software. Data are presented as mean ± SD, n = 3; (*) P < 0.05 in comparison with the wild-type agrin.
Acknowledgments
We thank the staff at NE-CAT of the APS, particularly Dr. Kanagalaghatta Rajashankar, for assistance in data collection. This work is based on research conducted at the APS on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under contract number DE-AC02-06CH11357. This work was partly supported by the Alfred P. Sloan Research Fellowship (to R.J.) and by grants from the NIH (to L.M.). Atomic coordinates and structure factors for the agrin–LRP4V396–A737 and agrin– LRP4T353–A737 complexes have been deposited with the PDB under accession codes 3V64 and 3V65, respectively.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.180885.111.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, Weis WI 2011. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell 21: 862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG 2011. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezakova G, Helm JP, Francolini M, Lomo T 2001. Effects of purified recombinant neural and muscle agrin on skeletal muscle fibers in vivo. J Cell Biol 153: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Wang W, Tam C, Hwang J, Zhang Y, Spittler D, Huang OW, Gong Y, Estevez A, Zilberleyb I, et al. 2011. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure 19: 1433–1442 [DOI] [PubMed] [Google Scholar]
- Brockhausen J, Cole RN, Gervasio OL, Ngo ST, Noakes PG, Phillips WD 2008. Neural agrin increases postsynaptic ACh receptor packing by elevating rapsyn protein at the mouse neuromuscular synapse. Dev Neurobiol 68: 1153–1169 [DOI] [PubMed] [Google Scholar]
- Burgess RW, Nguyen QT, Son YJ, Lichtman JW, Sanes JR 1999. Alternatively spliced isoforms of nerve- and muscle-derived agrin: Their roles at the neuromuscular junction. Neuron 23: 33–44 [DOI] [PubMed] [Google Scholar]
- Campanelli JT, Ferns M, Hoch W, Rupp F, von Zastrow M, Hall Z, Scheller RH 1992. Agrin: A synaptic basal lamina protein that regulates development of the neuromuscular junction. Cold Spring Harb Symp Quant Biol 57: 461–472 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bubeck D, Macdonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, et al. 2011. Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell 21: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, Xu W 2011. Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol 18: 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cornish T, Chi J, Johnson S, Lu Y, Campanelli JT 1999. Globular domains of agrin are functional units that collaborate to induce acetylcholine receptor clustering. J Cell Sci 112: 1213–1223 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Bowen DC, Valenzuela DM, Simmons MV, Poueymirou WT, Thomas S, Kinetz E, Compton DL, Rojas E, Park JS, et al. 1996. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell 85: 501–512 [DOI] [PubMed] [Google Scholar]
- Denzer AJ, Schulthess T, Fauser C, Schumacher B, Kammerer RA, Engel J, Ruegg MA 1998. Electron microscopic structure of agrin and mapping of its binding site in laminin-1. EMBO J 17: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Froehner SC 1993. Regulation of ion channel distribution at synapses. Annu Rev Neurosci 16: 347–368 [DOI] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, Sanes JR 1996. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell 85: 525–535 [DOI] [PubMed] [Google Scholar]
- Gesemann M, Denzer AJ, Ruegg MA 1995. Acetylcholine receptor-aggregating activity of agrin isoforms and mapping of the active site. J Cell Biol 128: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesemann M, Cavalli V, Denzer AJ, Brancaccio A, Schumacher B, Ruegg MA 1996. Alternative splicing of agrin alters its binding to heparin, dystroglycan, and the putative agrin receptor. Neuron 16: 755–767 [DOI] [PubMed] [Google Scholar]
- Glass DJ, Bowen DC, Stitt TN, Radziejewski C, Bruno J, Ryan TE, Gies DR, Shah S, Mattsson K, Burden SJ, et al. 1996. Agrin acts via a MuSK receptor complex. Cell 85: 513–523 [DOI] [PubMed] [Google Scholar]
- Hall ZW, Sanes JR 1993. Synaptic structure and development: The neuromuscular junction. Cell 72: 99–121 [DOI] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X 2004. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: Arrows point the way. Development 131: 1663–1677 [DOI] [PubMed] [Google Scholar]
- Herbst R, Burden SJ 2000. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J 19: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch W, Ferns M, Campanelli JT, Hall ZW, Scheller RH 1993. Developmental regulation of highly active alternatively spliced forms of agrin. Neuron 11: 479–490 [DOI] [PubMed] [Google Scholar]
- Hopf C, Hoch W 1998. Dimerization of the muscle-specific kinase induces tyrosine phosphorylation of acetylcholine receptors and their aggregation on the surface of myotubes. J Biol Chem 273: 6467–6473 [DOI] [PubMed] [Google Scholar]
- Janin J, Rodier F, Chakrabarti P, Bahadur RP 2007. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr D Biol Crystallogr 63: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M 2004. PDZ domain proteins of synapses. Nat Rev Neurosci 5: 771–781 [DOI] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ 2008. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 135: 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF 2001. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Luo S, Zhang B, Dong XP, Tao Y, Ting A, Zhou Z, Meixiong J, Luo J, Chiu FC, Xiong WC, et al. 2008. HSP90 β regulates rapsyn turnover and subsequent AChR cluster formation and maintenance. Neuron 60: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Woldt E, Matz RL, Boucher P 2007. The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann Med 39: 219–228 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan UJ 1990. The agrin hypothesis. Cold Spring Harb Symp Quant Biol 55: 407–418 [DOI] [PubMed] [Google Scholar]
- McMahan UJ, Horton SE, Werle MJ, Honig LS, Kroger S, Ruegg MA, Escher G 1992. Agrin isoforms and their role in synaptogenesis. Curr Opin Cell Biol 4: 869–874 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE 2002. The low-density lipoprotein receptor gene family: A cellular Swiss army knife? Trends Cell Biol 12: 273–280 [DOI] [PubMed] [Google Scholar]
- Parkash V, Leppanen VM, Virtanen H, Jurvansuu JM, Bespalov MM, Sidorova YA, Runeberg-Roos P, Saarma M, Goldman A 2008. The structure of the glial cell line-derived neurotrophic factor-coreceptor complex: Insights into RET signaling and heparin binding. J Biol Chem 283: 35164–35172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, Hohenester E, Muller YA 2001. LG/LNS domains: Multiple functions—one business end? Trends Biochem Sci 26: 363–368 [DOI] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science 298: 2353–2358 [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos P, Saarma M 2007. Neurotrophic factor receptor RET: Structure, cell biology, and inherited diseases. Ann Med 39: 572–580 [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW 1999. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci 22: 389–442 [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW 2001. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci 2: 791–805 [DOI] [PubMed] [Google Scholar]
- Schlee S, Carmillo P, Whitty A 2006. Quantitative analysis of the activation mechanism of the multicomponent growth-factor receptor Ret. Nat Chem Biol 2: 636–644 [DOI] [PubMed] [Google Scholar]
- Springer TA 1998. An extracellular β-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol 283: 837–862 [DOI] [PubMed] [Google Scholar]
- Stetefeld J, Alexandrescu AT, Maciejewski MW, Jenny M, Rathgeb-Szabo K, Schulthess T, Landwehr R, Frank S, Ruegg MA, Kammerer RA 2004. Modulation of agrin function by alternative splicing and Ca2+ binding. Structure 12: 503–515 [DOI] [PubMed] [Google Scholar]
- Sudhof TC 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455: 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Yang Y, Liu JH, Wang JH, Springer TA 2003. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature 424: 969–974 [DOI] [PubMed] [Google Scholar]
- Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E 2000. Structure and function of laminin LG modules. Matrix Biol 19: 309–317 [DOI] [PubMed] [Google Scholar]
- Tseng CN, Zhang L, Cascio M, Wang ZZ 2003. Calcium plays a critical role in determining the acetylcholine receptor-clustering activities of alternatively spliced isoforms of Agrin. J Biol Chem 278: 17236–17245 [DOI] [PubMed] [Google Scholar]
- Tseng CN, Zhang L, Wu SL, Wang WF, Wang ZZ, Cascio M 2010. Asparagine of z8 insert is critical for the affinity, conformation, and acetylcholine receptor-clustering activity of neural agrin. J Biol Chem 285: 27641–27651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC 2005. Mechanisms of vertebrate synaptogenesis. Annu Rev Neurosci 28: 251–274 [DOI] [PubMed] [Google Scholar]
- Wang X, Baloh RH, Milbrandt J, Garcia KC 2006. Structure of artemin complexed with its receptor GFRα3: Convergent recognition of glial cell line-derived neurotrophic factors. Structure 14: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Anderson KV, Niswander LA 2006. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development 133: 4993–5000 [DOI] [PubMed] [Google Scholar]
- Wu H, Xiong WC, Mei L 2010. To build a synapse: Signaling pathways in neuromuscular junction assembly. Development 137: 1017–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Arber S, William C, Li L, Tanabe Y, Jessell TM, Birchmeier C, Burden SJ 2001. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30: 399–410 [DOI] [PubMed] [Google Scholar]
- Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L 2008. LRP4 serves as a coreceptor of agrin. Neuron 60: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Glass DJ, Yancopoulos GD, Sanes JR 1999. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J Cell Biol 146: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Yang Z, Luo Z, Luo S, Xiong WC, Mei L 2008. Muscle-specific receptor tyrosine kinase endocytosis in acetylcholine receptor clustering in response to agrin. J Neurosci 28: 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]