Abstract
Serine/arginine-rich splicing factor 1 (SRSF1), previously designated SF2/ASF, belongs to a family of SR proteins that regulate constitutive and alternative splicing. SRSF1 expression is increased in tumors from several tissues and elicits changes in key target genes involved in tumor genesis. Several protein kinases phosphorylate SRSF1, which regulates its localization and function. It is previously reported that protein kinase A (PKA) phosphorylates SRSF1, but the importance of this modification is not well characterized. Here, we show that PKA phosphorylates SRSF1 on serine 119 in vitro. Phosphorylation of SRSF1 on this site enhanced the RNA binding capacity of SRSF1 in vivo and reduced the protein’s capacity to activate splicing of the Minx transcript in vitro. We also confirm an interaction between SRSF1 and PKA Cα1 and demonstrate that this interaction is not dependent on serine 119 phosphorylation but requires active PKA Cα1. We conclude that PKA phosphorylation of SRSF1 at serine 119 regulates SFRS1-dependent RNA binding and processing but not its interaction with PKA.
Keywords: pre-mRNA splicing regulation, SRSF1, PKA, phosphorylation
Introduction
Gene expression in eukaryotic cells is regulated at several levels and by numerous mechanisms. Transcription of protein encoding genes results in the synthesis of a precursor messenger RNA (pre-mRNA), which contains a sequence of coding and noncoding RNA in addition to a 5′ cap and a 3′ polyadenine tail. The coding exons are joined together by a process called pre-mRNA splicing to produce the mature RNA that is exported to the cytoplasm and translated into protein. This process is catalyzed by the spliceosome, which consists of large RNA protein complexes that assemble in a stepwise manner onto the pre-mRNA.1,2 Alternative splicing is an important mechanism to increase the proteome complexity by allowing production of structurally and functionally distinct protein isoforms from a single transcript. Estimates predict that as much as 92% to 94% of the human genes can produce alternatively spliced mRNAs, and this can be achieved by exon skipping, intron retention, mutual exclusion of exons, alternative first or last exon, or use of alternative 3′ and 5′ splice sites.3 Alternatively spliced isoforms are commonly found during development and in tissue-specific expression.4 In tumor biology, constitutively expressed proteins often possess control functions in the cell, such as growth and programmed cell death, while alternatively spliced isoforms tend to promote uncontrolled cell growth, invasive properties, and resistance towards apoptosis. These tumor-promoting behaviors are confirmed by the fact that these isoforms tend to be overexpressed in many cancer cells and tissues.5,6 Splice-site selection under normal physiological conditions is highly regulated, and errors resulting in aberrant transcripts encoding truncated proteins rarely occur. In contrast to this, errors in splicing events seem to be an intrinsic property of cancer cells.7,8
The pre-mRNA splicing process is regulated by both internal and external factors. Exonic and intronic splicing enhancers and silencers are RNA sequences that interact with external splicing factors, which again can promote or suppress the spliceosome assembly.9 Human serine/ arginine-rich proteins (SR proteins) are a family of 12 splicing factors characterized by 1 or 2 N-terminal RNA binding domains (RBDs) followed by a domain rich in arginine and serine dipeptide repeats (RS domain) of a minimum 50 amino acids.10 In general, SR proteins interact with exonic splicing enhancer elements (ESEs) and promote binding of the spliceosomal elements U2AF and/or U1 snRNP to splice sites.2 In addition to being essential in pre-mRNA splicing, SR proteins are implicated in mRNA nuclear export, mRNA stability, nonsense-mediated mRNA decay (NMD), translation, and oncogenic transformation.11-15 SR proteins are extensively phosphorylated in vivo, mainly in the RS domain, a modification that plays an important role in regulating their subcellular localization and activity.13 During recent years, it has become increasingly evident that molecular changes mediated by signal transduction pathways control splice-site selection in vivo.16 Reversible phosphorylation cascades are able to conduct rapid signals throughout the cell and are probably important in mediating extracellular signals to the spliceosome.17
Serine/arginine-rich splicing factor 1 (SRSF1), previously designated alternative splicing factor/splicing factor 2 (SF2/ASF),10 is a prototypical SR protein. It is involved in both constitutive and alternative splicing and contains 2 RBDs and a C-terminal RS domain.18 Moreover, SRSF1 has been reported to be a proto-oncogene whose increased expression in tumors from several tissues elicits changes in key target genes involved in tumor genesis.19 In addition, SRSF1 is located to a genomic region, which is amplified in several cancers and is correlated with poor prognosis.20 SRSF1 has also been shown to promote tumor transformation and growth by several mechanisms, for example, by stabilizing mRNA of antiapoptotic factors21 and by generating inactive tumor suppressor proteins by alternative splicing.19,22 SRSF1’s activity and function are regulated by phosphorylation by several protein kinases, such as the SR-specific protein kinase 1 (SRPK1),23 Cdc2-like kinases (Clk/Sty),24 topoisomerase I,25 and dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A).26 Phosphorylation by SRPK1/2 and Clk/Sty modulates shuttling of SRSF1s between the cytoplasm, nucleus, nuclear speckles, and active transcription sites.17 Cyclic AMP (cAMP)–dependent protein kinase (PKA) also phosphorylates SRSF1, but the effect of this phosphorylation is not well characterized.24,27
The cAMP/PKA signaling pathway is involved in the regulation of numerous cellular processes such as cell growth and differentiation, metabolism, and gene expression. In the absence of cAMP, PKA is an inactive tetramer composed of a regulatory (R) subunit dimer and 2 catalytic (C) subunits. PKA subunits are encoded by a family of R and C subunit genes (RIα, RIIα, RIβ, RIIβ, Cα, Cβ, Cγ, and PRKX).28 Specificity in this signal transduction pathway is obtained by compartmentalization by A-kinase anchoring proteins (AKAPs) and by expression of multiple forms of the R and C subunits, which are expressed in a tissue- specific manner and harbor different physiochemical properties.29,30 Several C subunit–binding proteins have also been identified during recent years.29,31-34 PKA is activated upon binding of 4 molecules of cAMP to the R dimer, releasing the C subunits to phosphorylate relevant substrates on serine and threonine residues in its vicinity.35 However, a proportion of the C subunit translocates to the nucleus after activation, where it is involved in the regulation of transcription and splicing.36-38 We have previously shown that PKA Cα1 is involved in the regulation of pre-mRNA splicing in vivo, possibly through phosphorylation of splicing factors, because the C subunit was shown to phosphorylate SR proteins in vitro.38 We observed that the band representing SR proteins with molecular weight around 30 kDa, such as SRSF1, SRSF2, and SRSF9, had the highest amount of 32P incorporated.
Here, we have analyzed SRSF1 for potential sites for PKA phosphorylation and identified a site at serine 119. We also demonstrated an interaction between SRSF1 and PKA Cα1 that was not dependent on serine 119 phosphorylation but that required active PKA Cα1. Furthermore, mutation of serine 119 altered SRSF1’s ability to bind RNA in vivo and enhanced its ability to promote pre-mRNA splicing of the Minx transcript in vitro, implying that PKA may inhibit the ability of SRSF1 to activate splicing.
Results
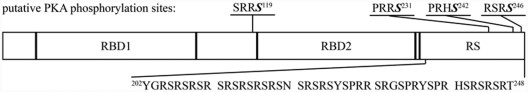
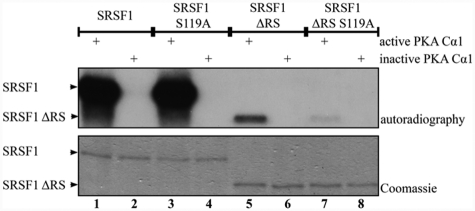
SRSF1 is phosphorylated by PKA in vitro and in vivo, but the actual phosphorylation site(s) are not identified.24,27 We analyzed the SRSF1 sequence (Uniprot:Q07955) for potential PKA phosphorylation sites, and the NetPHosK Server39 identified 4 putative PKA phosphorylation motifs at serines 119, 231, 242, and 246 (scoring between 0.51-0.77) (Fig. 1). It should be noted that serine 119 is the only site located outside of the RS domain. Based on this, we initially investigated which serine residues in SRSF1 become phosphorylated by PKA. Several plasmids of SRSF1 for prokaryotic expression were made. In order to test serine 119 as a potential phosphorylation site, one of the constructs made was devoid of the RS domain (SRSF1 ΔRS) (Fig. 1, lower sequence). The RS domain was removed because we suspected that the high number of arginine/serine repeats would lead to unspecific phosphorylation under our experimental conditions that could conceal phosphorylation outside of the RS domain. We also consider serine 119 as the most likely PKA phosphorylation site for phosphorylation in vivo because the serine residues in the RS domain are extensively phosphorylated by the SR protein–specific kinases SRPK1 and Clk/Sty via processive mechanisms.40-42 This residue was therefore mutated to alanine both in the full-length (SRSF1 S119A) and the RS-deleted (SRSF1 ΔRS S119A) constructs. After recombinant expression of the 4 SRSF1 constructs, the protein products were subjected to in vitro phosphorylation. The full-length SRSF1 wild-type (Fig. 2, lanes 1 and 2) and S119A-mutated (Fig. 2, lanes 3 and 4) proteins were both heavily phosphorylated by PKA as determined by the incorporation of 32P in the presence of active but not inactive PKA Cα1 and γ-[32P]-ATP. No apparent reduction in the phosphorylation levels due to the mutation at serine 119 was observed. However, when the RS domain was removed (Fig. 2, lanes 5 and 6), the protein was still phosphorylated but to a lower extent than the full-length proteins. This suggested that serine 119 may be a site in SRSF1 for PKA phosphorylation, a suggestion that was supported by markedly reduced phosphorylation when SRSF1 ΔRS S119A was incubated with PKA Cα1 in the presence of γ-[32P]-ATP (Fig. 2, lanes 7 and 8).
Figure 1.
Domains and putative PKA phosphorylation sites in SRSF1. Schematic representation of SRSF1 showing the sequence and location of the 4 putative PKA phosphorylation sites predicted by the NetPHosK server. The deleted sequence in SRSF1 ΔRS (aa 202-248) is expanded.
Figure 2.
PKA phosphorylates SRSF1 at serine 119 in vitro. Purified SRSF1 (lanes 1 and 2), SRSF1 S119A (lanes 3 and 4), SRSF1 ΔRS (lanes 5 and 6), and SRSF1 ΔRS S119A (lanes 7 and 8) were incubated with active or heat-inactivated PKA Cα1 and γ-[32P]-ATP in a reaction buffer. The samples were analyzed by SDS-PAGE followed by Coomassie staining (lower panel) and autoradiography (upper panel).
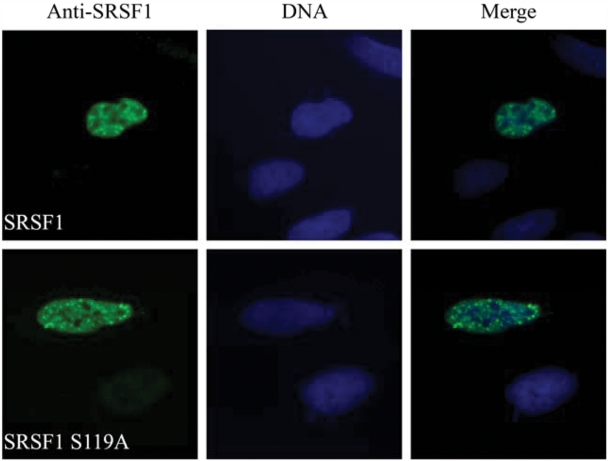
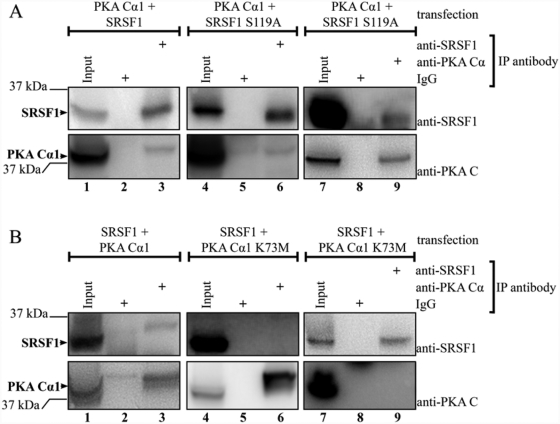
PKA Cα1 and SRSF1 are both located in splicing factor compartments.38,43 To explore whether the S119A mutation altered subcellular localization, we conducted immunofluorescence experiments of U2OS cells transfected with full-length wild-type and S119A-mutated SRSF1. Cells were fixated and permeabilized 16 hours after transfection and stained with anti-SRSF1 and Hoechst. Both SRSF1 wild-type (Fig. 3, upper panel) and SRSF1 S119A (Fig. 3, lower panel) were localized in the nucleus, and we did not observe any apparent changes in subnuclear localization. In a recent paper, it was shown by GST pull-down and co-immunoprecipitation (co-IP) that Cα1 targets SRSF1.27 Based on the results depicted in Figure 2, we tested whether PKA phosphorylation of SRSF1 at serine 119 regulates this interaction. We first verified the interaction by cross-IP of co-transfected PKA Cα1 and SRSF1 in 293T cells using anti-SRSF1 (Fig. 4A, lane 3) and anti-PKA Cα (Fig. 4B, lane 3). PKA Cα1 was further co-transfected with SRSF1 S119A and subjected to the same cross-IP using anti-SRSF1 (Fig. 4A, lane 6) and anti-PKA Cα (Fig. 4A, lane 9). This demonstrated that the S119A mutant of SRSF1 still interacted with active Cα1, indicating that serine 119 may not be important for the interaction. To further investigate the role of PKA in these interactions, we introduced an inactive form of Cα1 mutated at lysine 73 (lysine 72 when the first methionine is not included) (PKA Cα1 K73M).44 In these experiments, the interaction was abolished, as anti-PKA Cα was only able to immunoprecipitate PKA Cα1 (Fig. 4B, lower panel, lane 6) and not SRSF1 (Fig. 4B, upper panel, lane 6). This result was confirmed by applying anti-SRSF1, as this antibody was able to immunoprecipitate SRSF1 but not PKA Cα1 K73M (Fig. 4B, lane 9). Taken together, this firstly demonstrated that wild-type SRSF1 and PKA Cα1 interact in 293T cells. Secondly, mutation of PKA Cα1 at lysine 73 abolished not only Cα1 activity as previously reported44 but also the interaction of Cα1 with SRSF1.
Figure 3.
SRSF1 and SRSF1 S119A are localized in the nucleus. Immunofluorescence of PF-fixated U2OS cells transfected with SRSF1 wild-type (upper panel) or SRSF1 S119A (lower panel) and stained with anti-SRSF1 (left column, blue). DNA was visualized by Hoechst staining (middle column, blue). Photograph overlays are shown to the right (Merge).
Figure 4.
SRSF1 interacts with catalytically active PKA Cα1. (A) 293T cells were co-transfected with either PKA Cα1 and SRSF1 (lanes 1-3) or PKA Cα1 and SRSF1 S119A (lanes 4-9). The cell lysates were adjusted to equal protein concentration for each experiment and precleared with magnetic beads before input samples were collected (lanes 1, 4, and 7). The lysates were subjected to IPs using anti-SRSF1 (lanes 3 and 6), anti-PKA Cα (lane 9), mouse IgG (lanes 2 and 5), or rabbit IgG (lane 8) and magnetic beads. All samples were analyzed by SDS-PAGE and immunoblotting using anti-SRSF1 (upper panel) and anti-PKA C (lower panel). (B) 293T cells were co-transfected with either SRSF1 and PKA Cα1 (lanes 1-3) or SRSF1 and PKA Cα1 K73M (lanes 4-9). The cell lysates were adjusted to equal protein concentration for each experiment and precleared with magnetic beads before input samples were collected (lanes 1, 4, and 7). The lysates were subjected to IPs using anti-PKA Cα (lanes 3 and 6), anti-SRSF1 (lane 9), mouse IgG (lane 8), or rabbit IgG (lanes 2 and 5) and magnetic beads. All samples were analyzed by SDS-PAGE and immunoblotting using anti-SRSF1 (upper panel) and anti-PKA C (lower panel).
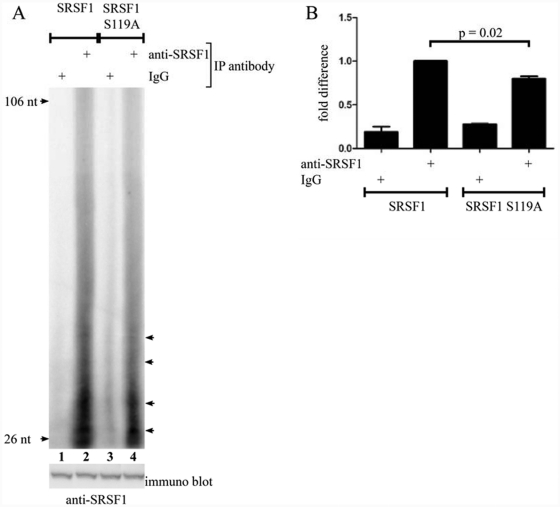
This made us want to investigate other roles of serine 119 in SRSF1 function. It has previously been shown that phosphorylation of RNA binding proteins can change their RNA binding properties.45-48 In fact, it was recently demonstrated that PKA phosphorylation of SRSF1 enhances binding to tau pre-mRNA.27 The serine 119 PKA phosphorylation site in SRSF1 is located close to the N-terminal end of RBD2 (Fig. 1). Furthermore, a structural analysis of RNA binding to SRSF1 showed that when the 2 arginines preceding serine 119 are mutated, RNA binding is severely reduced.49 Hence, serine 119 may potentially be involved in regulating RNA binding. To test this, we conducted RNA-IP with transfected SRSF1 and SRSF1 S119A. This confirmed that wild-type SRSF1 binds RNA (Fig. 5A, lane 2) whereas mutation of serine 119 reduced RNA binding significantly (P = 0.02; n = 3) (Fig. 5A [lane 4] and 5B).
Figure 5.
Mutation of serine 119 decreases the ability of SRSF1 to interact with RNA. (A) 293T cells were transfected with SRSF1 (lanes 1 and 2) or SRSF1 S119A (lanes 3 and 4). Twenty hours posttransfection, the cells were UV cross-linked and lysed. The cell lysates were adjusted to equal protein concentration and treated with T1 RNase before IPs with magnetic beads conjugated with either mouse IgG (lanes 1 and 3) or anti-SRSF1 (lanes 2 and 4). Immunoprecipitated samples were dephosphorylated, labeled with γ-[32P]-ATP by PNK kinase, and run on a denaturating polyacrylamide gel before analysis by autoradiography. The immunoblot (lower panel) shows the amount of SRSF1 in the cell lysate. The arrows indicate accumulation of specific RNA species. (B) Lanes in unsaturated images were manually detected in Adobe Photoshop using identical frames. The obtained intensities were adjusted for background and analyzed in GraphPad Prism by a paired t test (n = 3).
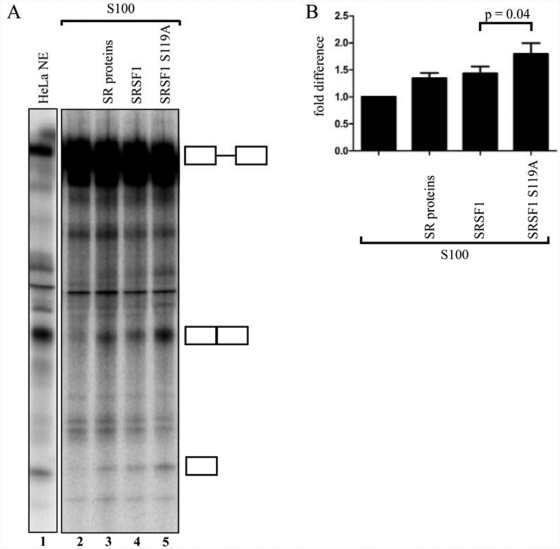
Knowing that mutation of serine 119 affects SRSF1’s RNA-binding capacity, we next investigated if this mutation also affects SRSF1-dependent pre-mRNA splicing. Due to the high endogenous levels of SRSF1, it is difficult to analyze the effect of the S119A mutant by an in vivo minigene splicing assay or in vitro splicing assays using nuclear extracts. We therefore took advantage of an in vitro S100 complementation assay using adenoviral Minx pre-mRNA as a transcript. This model pre-mRNA derived from the major late transcription unit of adenovirus is known to be spliced in cytoplasmic S100 extracts complemented with SRSF1.50 We therefore considered the Minx pre-mRNA as well suited for the analysis of differences in splicing capacity between SRSF1 and SRSF1 S119A. As controls for these experiments, we performed splicing of the Minx transcript in HeLa nuclear extracts (Fig. 6A, lane 1), uncomplemented S100 extract (Fig. 6A, lane 2), and SR protein–complemented S100 extract (Fig. 6A, lane 3). This showed a minimal amount of splicing activity in uncomplemented compared to the complemented S100 extract (Fig. 6A, lanes 2 and 3). These controls provided a threshold for monitoring the relative contribution of wild-type and mutated SRSF1 to pre-mRNA splicing (Fig. 6A, lanes 4 and 5). The results revealed that the SRSF1 S119A mutant protein had a significantly higher capacity to splice the Minx pre-mRNA in the S100 extract compared to the wild-type protein (P = 0.04; n = 9) (Fig. 6B).
Figure 6.
Mutation of serine 119 enhances the ability of SRSF1 to activate splicing of the Minx transcript in vitro. (A) In vitro splicing of Minx pre-mRNA in HeLa-NE (lane 1) or S100 extracts (lanes 2-5). The splicing-deficient S100 extracts were supplemented with either no addition (lane 2), SR mix (lane 3), SRSF1 (lane 4), or SRSF1 S119A (lane 5). The identities of the RNAs are shown schematically at the right side of the panel. (B) Unsaturated images were quantified in Scion Image, adjusted for background, and analyzed by a paired t test in GraphPad Prism (n = 9).
Discussion
Here, we present evidence that PKA phosphorylates the splicing factor SRSF1 at serine 119 in vitro. Prevention of phosphorylation at this site led to a reduction of SRSF1’s capacity to bind RNA in vivo and increased its capability to splice the Minx transcript in vitro. We also present evidence that PKA Cα1 and SRSF1 interact by a mechanism that is independent of phosphorylation at serine 119 but that requires active PKA. PKA phosphorylation of full-length SRSF1 is documented in 3 independent reports.24,27 The NetPHosK server depicted 4 possible PKA phosphorylation sites, of which the consensus sequence for the first 3 is RXS (arginine-random-serine), while the last one is RXXS (arginine-random-random-serine). These consensus sites have been postulated to have a relatively low probability of being phosphorylated.51 However, there are several examples of physiological PKA substrates with these consensus sites, such as elongation factor-2 kinase (PRRS499),52 hormone-sensitive lipase (MRRS563 and PRRS659),53-55 heat shock factor 1 (RPSS320),33 and hSLO BKCa α subunit of large conductance Ca2+-dependent K+ channel (RQPS869).56 Based on this, we cannot rule out that SRSF1 is phosphorylated at any of these sites in the RS domain, especially because S119A-mutated full-length SRSF1 was strongly phosphorylated by active PKA Cα1 in vitro. Furthermore, Shi et al. also showed that the RS domain is extensively phosphorylated by PKA in vitro, and they observed reduced phosphorylation with increasing deletions of the RS domain.27 It is, however, difficult to determine the importance of phosphorylation of the RS domain due to the high probability of unspecific phosphorylation under in vitro conditions, where PKA may act in a promiscuous way. To address this further, one would have to mutate every putative PKA phosphorylation site individually and accordingly analyze alterations in SRSF1 function in vivo systematically.
That PKA Cα1 and SRSF1 interact confirms results from a recent paper by Shi et al. where they show that GST-tagged SRSF1 pulls down PKA Cα1 and that anti-HA immunoprecipitated PKA Cα1 together with several HA-tagged SRSF1 variants.27 In the present study, we immunoprecipitated untagged PKA Cα1 with anti-SRSF1 and vice versa and were therefore able to verify the interaction by cross-IPs. In these experiments, we also observed that mutation of serine 119 to alanine, which prevents phosphorylation, did not abrogate the interaction. This indicates that this phosphorylation event is not essential for interaction. The amount of co-immunoprecipitated proteins was relatively low, however, not unexpected, taking into account the relatively low abundance of PKA C subunits in the nucleus compared to the whole cell38; hence, only a small fraction of the C subunit will be associated with SRSF1 at all times. When transfected into 293T cells, the C subunit is expressed at high levels in the cytoplasm (results not shown). Shi et al. documented that an SRSF1 deletion mutant lacking the most C-terminal end of the RS domain (aa 220-248) did not show any reduction in the amount of pulled-down PKA Cα1 compared to the full-length protein. They also showed that the RS domain alone was not able to immunoprecipitate detectable levels of PKA Cα1. All together, this implies that phosphorylation of neither serine 119 nor any of the putative PKA sites in the RS domain is required for the interaction between SRSF1 and PKA Cα1. More detailed mapping studies will be required to fully explain the basis for the interaction between the 2 proteins. Interestingly, we show that the interaction actually requires that PKA Cα1 is catalytically active because mutation of lysine 73 to methionine abrogated the interaction. Lysine 73 is conserved throughout the whole family of eukaryotic protein kinases, and replacement of this lysine, which is involved in ATP binding, has become a standard approach for establishing the importance of kinase activity.57 Lysine 73 is located in β strand 3 in the small lobe, and after mutation of this site, the protein is less stable than the wild-type, lacks catalytic activity, but is still able to bind ATP.58 The unphosphorylated lysine 73 mutant is unable to bind RI and RII subunits, but when it is activated by phosphorylation at threonine 197 in the activation loop, these interactions are restored. The PKA Cα1 K73M mutant is, however, not able to finish the activation cascade that is initiated by threonine 197 phosphorylation.59 The interaction between PKA Cα1 and SRSF1 is abolished along with mutation of lysine 73. This may implicate that binding of SRSF1 requires the conformal changes that an active PKA Cα1 displays, such as a stabile small lobe with high affinity–bound ATP and completed activation cascade. SRSF1 may interact with PKA Cα1 through the hydrophobic pocket, as is the case for RI, RII, and PKI.60 When the crystal structure of SRSF1 is solved, this interaction will be more accessible for investigation.
Based on the localization of serine 119 in proximity to RBD2, we assumed that phosphorylation of this site could be involved in the regulation of RNA binding. Both RBDs in SRSF1 are capable of binding RNA in the absence of other factors.18 Our RNA-IP results show that when serine 119 is mutated to alanine, the total amount of RNA bound to SRSF1 is reduced. The actual effect is probably higher because there are endogenous levels of SRSF1 that could mask the effect of the mutation. The measured reduction in bound RNA due to lack of phosphorylation was approximately 20%. This may imply that this phosphorylation event does not lead to a switch in activity but rather acts as a modulator of RNA binding. Interestingly, it has been shown that SRSF1 is bridged to other RNA binding proteins, such as U1-70K, through binding via the RBDs.61 Whether the RNA IPs presented here are influenced by or that S119 phosphorylation is involved in such a mechanism will need further investigation. The results documented here indirectly suggest that PKA phosphorylation at serine 119 enables SRSF1 to bind broader spectra of RNA. In fact, this correlates well with the report showing that PKA phosphorylation enhances the binding of SRSF1 to exon 10 of tau pre-mRNA.27 This result is, however, only valid if S119 really is phosphorylated by PKA in the intact cell.
For constitutive splicing regulated by SRSF1, both RBDs and the RS domain are required. In the regulation of alternative splicing, and more specifically for 5′ splice-site selection, RBD1 and RBD2 are sufficient.18 The in vitro splicing assay with the Minx transcript demonstrated an increased amount of splice product when serine 119 was mutated to alanine, indicating that PKA phosphorylation of this particular site inhibits the ability of SRSF1 to activate splicing. We cannot, however, rule out that lack of phosphorylation by other kinases influences the effect we observe; however, this will need further investigation. To our knowledge, there are no reports describing a direct interaction between PKA Cα1 and RNA or that PKA Cα1 (Uniprot P17612) contains any RBDs. We, therefore, assume that PKA Cα1’s involvement in pre-mRNA splicing occurs indirectly by interaction and phosphorylation of other proteins, such as SRSF1. We have previously described that PKA Cα1 promotes distal 5′ splice-site selection of the E1A minigene in vivo 38 and that splicing factor SFRS17A is dependent on an interaction with PKA to be able to promote distal splicing of the same minigene.62 There is also a proposed PKA/CaMKIV (Ca2+/calmodulin-dependent protein kinase IV)–responsive element in RNA (CAAAAAA) that appears to regulate alternative splicing.63 The cAMP/PKA pathway is also mediating effects on splicing under the effect from dopamine and dopamine receptors.64 Finally, the recent report by Shi et al. shows that PKA promotes tau exon 10 inclusion by phosphorylation of SRSF1.27 However, to our knowledge, our results are the first to demonstrate that PKA phosphorylation of a specific site in a SR protein directly influences splicing.
To this end, several attempts have been made to obtain drugs for cancer treatment that regulate alternative splicing, such as the antitumor drug NB-506 (glycosylated indolocarbazole derivative), which inhibits topoisomerase 1 phosphorylation of SRSF1 in vitro. This prevention of phosphorylation affects the formation of the spliceosome and changes the mRNA levels and/or the splicing pattern of several tested genes in vivo.65 In non–small cell lung cancer, the phosphorylation status of the RS domain of SRSF1 promotes exon skipping in the caspase 9 pre-mRNA, changing the proapoptotic caspase 9a to the antiapoptotic caspase 9b. By manipulating the ratio between these isoforms, the sensitivity towards chemotherapeutic drugs can be enhanced.66 Interestingly, it has been shown that SRSF1 not only promotes transformation but is also involved in tumor maintenance, implying that SRSF1 inhibition theoretically could have therapeutic potential even in advanced cancers.19 Whether PKA phosphorylation of SRSF1 has potential as a target for SRSF1 inhibition will need further investigation.
Materials and Methods
Expression plasmids
Plasmids encoding native PKA Cα1 and the catalytically inactive mutant Cα1 K73M in the mammalian expression vector pEF-DEST 51 have been described previously.38,67 Native SRSF1 and SRSF1 lacking the RS domain (SRSF1 ΔRS) were PCR amplified from U2OS cell cDNA using an upper primer: 1ASF:91U19 (5′CACCGCCGCCACCATGTCGGGAGGTGGTGTGA3′) in combination with either 2ASF:813L25 (5′TTATGTACGAGAGCGAGATCTGCTA3′) or 3ASFΔ RS:648L25 (5′TTAAACTTTAACCCGGATGTAGGCAG TT3′). The PCR products were cloned into expression plasmids using the pENTR/D-TOPO Cloning Kit (cat. no. 45-0218, Invitrogen Dynal AS, Oslo, Norway) and the mammalian expression vector pEF-DEST51 (cat. no. 12285-011, Invitrogen Dynal AS) by the LR Clonase reaction (cat. no. 11791-019, Invitrogen Dynal AS) according to the manufacturer. The QuickChange mutagenesis kit (cat. no. 200524-5, Stratagene, La Jolla, CA) was used to mutate the putative PKA phosphorylation site at position 119 from serine to alanine in both pEF-DEST51 SRSF1 and pEF-DEST51 SRSF1ΔRS using the following primers: 5′CCATCCAGGCGGGCTGAAAACAGAGTG3′ and 5′CA CTCTGTTTTCAGCCCGCCTGGATGG3′. The plasmids for prokaryotic expression of SRSF1 and SRSF1 ΔRS were made by PCR amplification of the pCGT7-SF2 expression vector43 using primers introducing NcoI and HindIII restriction sites and a C-terminal 6x His-tag. The upper primer (5′AGCTCCATGGCATCGGGAGGTGGTGT GATTCGT3′) was used in combination with either 5′AGC TAAGCTTTTAGTGGTGGTGGTGGTGGTGGGTAC GAGAACGGCTGCG3′ for SRSF1 or with 5′AGCTA AGCTTATAGCTCGGGCTACGGGGCCCATC3′ for SRS F1 ΔRS. Digested PCR products were cloned into the pET24d vector (cat. no. 69752, Novagen Merck KGaA, Darmstadt, Germany). The QuickChange mutagenesis kit (cat. no. 200524-5, Stratagene) was used to mutate the putative PKA phosphorylation site at position 119 from serine to alanine in both pET24d SRSF1 and pET24d SRSF1 ΔRS using the following primers: 5′CCATCCAGGCGGGCTGAAAACAGAGTG3′ and 5′CACTCTGTTTTCAGCCC GCCTGGATGG3′. All primers were purchased from Sigma-Genosys (Haverhill, United Kingdom), and all expression plasmids were confirmed by sequencing.
Protein purification
pET24d plasmids encoding SRSF1 proteins were transformed into Rosetta 2(DE)pLys bacteria. Overnight cultures representing one clone of each plasmid were grown in 500 mL LB medium at 37°C with 150-rpm agitation until OD600 reached approximately 0.7. Protein expression was induced by the addition of 1 mM IPTG and further grown for 3 hours at 30°C with 150-rpm agitation. The bacteria were lysed in Buffer A (20 mM Hepes-KOH, pH 7.9, 300 mM KCl, 5 mM imidazole, 6 M guanidine-HCl, 1x EDTA-free protease inhibitor cocktail [cat. no. 04693132001, Roche Diagnostics, Oslo, Norway], and 0.4 mM betamercaptoetanol) by sonication and purified on a crude HisTrap column (cat. no. 11-0004-58, GE Healthcare Life Sciences, Oslo, Norway) with 5 to 500 mM imidazole gradient. Peak fractions were pooled and dialyzed stepwise against Buffer D (20 mM Hepes-KOH, pH 7.9, 100 mM KCl, 20% glycerol, 200 µM EDTA, and 0.3 mM DTT) containing from 4 to 0.5 M guanidine-HCl. SR proteins were purified by double-salt precipitation as described previously.68,69
In vitro phosphorylation
The procedure was performed as previously described.38 Briefly, 2.5 ng active or heat-inactivated (10 minutes at 70°C) PKA Cα1 (cat. no. P2912, Invitrogen Dynal AS) was mixed with in vitro phosphorylation buffer (10 mM potassium phosphate, pH 7.4, 1 mM EDTA, 10 mM magnesium acetate, and 1 µCi γ-[32P]-ATP). There was 0.25 µg purified SRSF1, SRSF1 S119A, SRSF1 ΔRS, or SRSF1 ΔRS S119A added to a total volume of 20 µL. The reactions were incubated at 30°C for 30 minutes on slushy ice for 1 hour and stopped by adding SDS loading dye and boiled for 5 minutes. The proteins were resolved by SDS-PAGE and analyzed by Coomassie staining and autoradiography.
Cell culture
293T cells were maintained at 37°C in humidified air with 5% CO2 in RPMI 1640 (cat. no. R0883, Sigma-Aldrich, Oslo, Norway) supplemented with 10% fetal bovine serum (cat. no. F7524, Sigma-Aldrich), 1% nonessential amino acids (cat. no. 11140-035, Invitrogen Dynal AS), 1% L-glutamine (cat. no. G7513, Sigma-Aldrich), 1% sodium pyruvate (cat. no. 11360-039, Invitrogen Dynal AS), and 1x penicillin-streptomycin solution (cat. no. P4458, Sigma-Aldrich). U2OS cells were grown in DMEM (cat. no. D6546, Sigma-Aldrich) supplemented with 10% fetal bovine serum (cat. no. F7524, Sigma-Aldrich), 1% L-glutamine (cat. no. G7513, Sigma-Aldrich), and 1x pencillin-streptomycin solution (cat. no. P4458, Sigma-Aldrich). The cells were transfected with Fugene HD (cat. no. 04 709 705 001, Roche Diagnostics, Oslo, Norway) according to the manufacturer’s protocol. Briefly, 6 µL Fugene HD (cat. no. 04 709 705 001, Roche Diagnostics), 2 µg plasmid DNA, and 100 µL Opti-MEM (cat. no. 31985, Invitrogen Dynal AS) were mixed and added to each well (10 cm2) and incubated for 20 hours.
Immunofluorescence
Transfected U2OS cells grown on cover slips were washed with phosphate-buffered saline (PBS) and fixed/permeabilized with 3% paraformaldehyde and 0.1% Triton X-100 in PBS. After being washed with PBS containing 0.1% Tween 20 (PBST), the cover slips were blocked with PBST containing 2% bovine serum albumin (PBST-BSA) for 15 minutes. Anti-SF2/ASF (mab 96, cat. no. 32-4500, Invitrogen Dynal AS) was diluted 1:50 in PBST-BSA and incubated for 30 minutes. The cover slips were washed extensively before incubation for 30 minutes and Alexa Fluor 594 goat anti-mouse IgG (1:400, cat. no. A11005, Invitrogen Dynal AS) secondary antibody. Finally, the cover slips were washed with PBST-BSA, and 0.5 g/mL Hoechst (cat. no. B2261, Sigma-Aldrich) was added in the last wash for DNA staining. The cover slips were mounted in Citifluor AF1 (Citifluor Ltd., London, United Kingdom) and examined using an Olympus BX61 microscope attached with an F-VIEW digital camera (Olympus, Tokyo, Japan). Photographs were acquired on the computer using analySIS (Soft Imaging Systems, Münster, Germany).
Co-immunoprecipitation (co-IP)
Four wells with co- transfected 293T cells were washed in PBS and lysed by sonication in IP buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 0.5% Triton X-100, 1x protease inhibitor cocktail [cat. no. P8340, Sigma-Aldrich], 1 mM PMSF, and 1 mM Na3VO4). The lysates were cleared by centrifugation and adjusted to equal protein concentration for each experiment (1.7-2.9 µg protein/µL) by the Bradford method (BioRad, Oslo, Norway). The lysates were precleared with Dynabeads Protein G (1:10, cat. no. 100-04D, Invitrogen Dynal AS) before input samples were collected (20 µL), and the remainder of the lysates were incubated with one of the following antibodies (2.5 µg/sample): mouse IgG (cat. no. I5381, Sigma-Aldrich), rabbit IgG (cat. no. I5006, Sigma-Aldrich), anti-SF2/ASF (cat. no. 32-4500, Invitrogen Dynal AS), or anti-PKA α cat (cat. no. SC-903, Santa Cruz Biotechnology, Santa Cruz, CA). The mixture of cell lysate and antibody was left incubated at 4°C rotating overnight before Dynabeads Protein G (1:10, cat. no. 100-04D, Invitrogen Dynal AS) was added the next day and incubated for another hour. The samples were washed with IP buffer, added with SDS loading dye, and boiled for 5 minutes before immunoblotting.
Immunoblotting
Samples were separated by SDS-PAGE and transferred to PVDF membrane by electroblotting. The membrane was blocked by drying and rehydration in methanol before incubation with primary antibodies diluted in TBST (10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20), anti-SF2/ASF (30 µg, cat. no. 32-4600, Invitrogen Dynal AS), or anti-PKAC (1:250, cat. no. 610981, BD Biosciences Transduction Laboratories, Erembodegem, Belgium). The membrane was washed in TBST before incubation with the secondary antibodies diluted in TBST, HRP-conjugated goat anti-mouse (1:2,000, cat. no. 55563, MP Biomedicals, Irvine, CA), or HRP-conjugated goat anti-rabbit (1:2,000, cat. no. 55689, MP Biomedicals). After washing with TBST, immunoreactive proteins were visualized by ECL (cat. no. 34076, Pierce, Rockford, IL) and developed using the G:BOX imaging system (Syngene, Cambridge, United Kingdom).
RNA IP
The method is according to Rogne et al.45,70 In brief, one sample is the extract of four 10-cm2 wells of transfected 293T cells. Culture medium was replaced by PBS before the cells were UV irradiated (254 nm, 200 mJ/cm2, twice), harvested, washed in PBS, and lysed by sonication in 300 µL IPB buffer (10 mM HEPES, pH 7.5, 2 mM EDTA, 10 mM KCl, 1% TX-100, 1 mM PMSF, and 1x protease inhibitor cocktail [cat. no. P8340, Sigma-Aldrich]) supplemented with 150 mM NaCl. Lysates were cleared by centrifugation before addition of T1 RNase (final concentration of 0.025 U/µL, cat. no. EN0541, Fermentas, St. Leon-Rot, Germany) and incubation for 6 minutes at 37°C with 1,000-rpm agitation. The lysates were adjusted to equal protein concentrations by the Bradford method and precleared with Dynabeads Protein G (1:10, cat. no. 100.04D, Invitrogen Dynal AS) for 30 minutes at 4°C rotating. Samples for immunoblotting were collected, added with SDS loading dye, and boiled for 5 minutes. Dynabeads Protein G (1:10, cat. no. 100.04D, Invitrogen Dynal AS) was conjugated with anti-SF2/ASF (2.5 µg, cat. no. 32-4600, Invitrogen Dynal AS) or mouse IgG (2.5 µg, cat. no. I5381, Sigma-Aldrich) for 2 hours at 4°C followed by 3 washes in IPB buffer. Supernatants were incubated with 30 µL conjugated beads for 2 hours at 4°C rotating. The immunoprecipitated samples were washed 4 times by alternating between 1 mL RIPA (1x PBS, 0.05% SDS, 0.1% deoxycholate, 1% NP-40, 1x protease inhibitor cocktail [cat. no. P8340, Sigma-Aldrich]) and RIPA 1000 (as RIPA buffer but supplied with 1 mM EDTA and 1 M NaCl). The immunoprecipitated samples were dephosphorylated by calf intestinal phosphatase (final concentration of 0.038 U/µL, cat. no. 10713023001, Roche Diagnostics) for 10 minutes and washed 3 times with Buffer C (50 mM Tris HCl, pH 7,4, 10 mM MgCl2, 0.5% NP-40, 1x protease inhibitor cocktail [cat. no. P8340, Sigma-Aldrich]), followed by labeling with γ-[32P]-ATP (2.1 µCi, cat. no. NEG502H, Perkin Elmer, Maanstraat, Germany) by polynucleotide kinase phosphorylation (0.5 U/µL, cat. no. M0201L, New England Biolabs Inc., Ipswich, MA) and further washed 3 times with Buffer C. Samples were resolved on 6% denaturing polyacrylamide gels. Incorporated phosphate was visualized by autoradiography. Unsaturated images from the Typhoon 9410 phosphoimager (GE Healthcare Life Sciences) were quantified using the histogram function in Adobe Photoshop (San Jose, CA). The lanes were manually defined with identical frames. The statistical analysis was performed with a paired Student t test in Prism GraphPad (La Jolla, CA).
Transcript synthesis and in vitro splicing
Radiolabeled Minx pre-mRNA was generated by T7 RNA polymerase transcription of a BamHI-cut PCR-amplified product from pMINX MS271 as previously described.72 Nuclear and S100 extracts were prepared from HeLa spinner cells as previously described.72 The total splicing reaction volume was 25 µL and contained approximately 20 to 25 fmol 32P-labeled Minx transcript, 8 µL nuclear or S100 extracts, 2.6% polyvinyl alcohol, 12% glycerol, 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 2 mM ATP, 20 mM creatine phosphate, 2.5 mM MgCl2, and 0.3 mM DTT. Reactions were incubated at 30°C for 2 hours, treated with proteinase K, and phenol extracted. After sodium acetate/ethanol precipitation, the RNA was resolved on 6% denaturing polyacrylamide gels, followed by autoradiography. Unsaturated images from the Typhoon 9410 phosphoimager (GE Healthcare Life Sciences) were quantified using Scion Image (Scion Corporation) and adjusted for background. The statistical analysis was performed with a paired Student t test in Prism GraphPad.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
This work was supported by The Norwegian Cancer Society (AKA and AKK), University of Oslo (SE and BSS), and Uppsala University (GA).
References
- 1. Butcher SE, Brow DA. Towards understanding the catalytic core structure of the spliceosome. Biochem Soc Trans. 2005;33:447-9 [DOI] [PubMed] [Google Scholar]
- 2. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336 [DOI] [PubMed] [Google Scholar]
- 3. Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386-98 [DOI] [PubMed] [Google Scholar]
- 4. Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24: 2343-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342-57 [DOI] [PubMed] [Google Scholar]
- 8. Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647-54 [DOI] [PubMed] [Google Scholar]
- 9. House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283:1217-21 [DOI] [PubMed] [Google Scholar]
- 10. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24:1073-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597-607 [DOI] [PubMed] [Google Scholar]
- 12. Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179-89 [DOI] [PubMed] [Google Scholar]
- 13. Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15-27 [DOI] [PubMed] [Google Scholar]
- 14. Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613-5 [DOI] [PubMed] [Google Scholar]
- 15. Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727-38 [DOI] [PubMed] [Google Scholar]
- 17. Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223-7 [DOI] [PubMed] [Google Scholar]
- 18. Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karni R, de Stanchina E, Lowe SW, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313-22 [DOI] [PubMed] [Google Scholar]
- 21. Ezponda T, Pajares MJ, Agorreta J, et al. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clin Cancer Res. 2010;16:4113-25 [DOI] [PubMed] [Google Scholar]
- 22. Thorsen K, Mansilla F, Schepeler T, et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol Cell Proteomics. 2011;10:M110002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678-82 [DOI] [PubMed] [Google Scholar]
- 24. Colwill K, Pawson T, Andrews B, et al. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265-75 [PMC free article] [PubMed] [Google Scholar]
- 25. Rossi F, Labourier E, Forne T, et al. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80-2 [DOI] [PubMed] [Google Scholar]
- 26. Shi J, Zhang T, Zhou C, et al. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J Biol Chem. 2008;283:28660-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi J, Qian W, Yin X, et al. Cyclic AMP-dependent protein kinase regulates the alternative splicing of tau exon 10: a mechanism involved in tau pathology of Alzheimer disease. J Biol Chem. 2011;286:14639-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway: differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. 2000;5:D678-93 [DOI] [PubMed] [Google Scholar]
- 29. Skalhegg BS, Funderud A, Henanger HH, et al. Protein kinase A (PKA): a potential target for therapeutic intervention of dysfunctional immune cells. Curr Drug Targets. 2005;6:655-64 [DOI] [PubMed] [Google Scholar]
- 30. Pidoux G, Tasken K. Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J Mol Endocrinol. 2010;44:271-84 [DOI] [PubMed] [Google Scholar]
- 31. Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci U S A. 2005;102:349-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skroblin P, Grossmann S, Schafer G, Rosenthal W, Klussmann E. Mechanisms of protein kinase A anchoring. Int Rev Cell Mol Biol. 2010;283:235-330 [DOI] [PubMed] [Google Scholar]
- 33. Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. Protein kinase A binds and activates heat shock factor 1. PLoS One. 2010;5:e13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han I, Xue Y, Harada S, et al. Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol Cell Biol. 2002;22:2136-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rannels SR, Corbin JD. Two different intrachain cAMP binding sites of cAMP-dependent protein kinases. J Biol Chem. 1980;255:7085-8 [PubMed] [Google Scholar]
- 36. Nigg EA, Hilz H, Eppenberger HM, Dutly F. Rapid and reversible translocation of the catalytic subunit of cAMP-dependent protein kinase type II from the Golgi complex to the nucleus. EMBO J. 1985;4:2801-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harootunian AT, Adams SR, Wen W, et al. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol Biol Cell. 1993;4:993-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kvissel AK, Orstavik S, Eikvar S, et al. Involvement of the catalytic subunit of protein kinase A and of HA95 in pre-mRNA splicing. Exp Cell Res. 2007;313:2795-809 [DOI] [PubMed] [Google Scholar]
- 39. Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351-62 [DOI] [PubMed] [Google Scholar]
- 40. Velazquez-Dones A, Hagopian JC, Ma CT, et al. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761-8 [DOI] [PubMed] [Google Scholar]
- 41. Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ngo JC, Chakrabarti S, Ding JH, et al. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20: 77-89 [DOI] [PubMed] [Google Scholar]
- 43. Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413-24 [DOI] [PubMed] [Google Scholar]
- 45. Rogne M, Stokka AJ, Tasken K, Collas P, Kuntziger T. Mutually exclusive binding of PP1 and RNA to AKAP149 affects the mitochondrial network. Hum Mol Genet. 2009;18:978-87 [DOI] [PubMed] [Google Scholar]
- 46. Lisitsky I, Schuster G. Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res. 1995;23: 2506-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Idriss H, Kumar A, Casas-Finet JR, et al. Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry. 1994;33:11382-90 [DOI] [PubMed] [Google Scholar]
- 48. Hamilton BJ, Burns CM, Nichols RC, Rigby WF. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes: the roles of cytoplasmic location, transcription, and phosphorylation. J Biol Chem. 1997; 272:28732-41 [DOI] [PubMed] [Google Scholar]
- 49. Tintaru AM, Hautbergue GM, Hounslow AM, et al. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Allemand E, Gattoni R, Bourbon HM, et al. Distinctive features of Drosophila alternative splicing factor RS domain: implication for specific phosphorylation, shuttling, and splicing activation. Mol Cell Biol. 2001;21:1345-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101:2381-411 [DOI] [PubMed] [Google Scholar]
- 52. Diggle TA, Subkhankulova T, Lilley KS, et al. Phosphorylation of elongation factor-2 kinase on serine 499 by cAMP-dependent protein kinase induces Ca2+/calmodulin-independent activity. Biochem J. 2001;353:621-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anthonsen MW, Ronnstrand L, Wernstedt C, Degerman E, Holm C. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J Biol Chem. 1998;273:215-21 [DOI] [PubMed] [Google Scholar]
- 54. Stralfors P, Bjorgell P, Belfrage P. Hormonal regulation of hormone-sensitive lipase in intact adipocytes: identification of phosphorylated sites and effects on the phosphorylation by lipolytic hormones and insulin. Proc Natl Acad Sci U S A. 1984;81: 3317-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garton AJ, Campbell DG, Cohen P, Yeaman SJ. Primary structure of the site on bovine hormone-sensitive lipase phosphorylated by cyclic AMP-dependent protein kinase. FEBS Lett. 1988;229:68-72 [DOI] [PubMed] [Google Scholar]
- 56. Nara M, Dhulipala PD, Wang YX, Kotlikoff MI. Reconstitution of beta-adrenergic modulation of large conductance, calcium-activated potassium (maxi-K) channels in Xenopus oocytes: identification of the camp-dependent protein kinase phosphorylation site. J Biol Chem. 1998;273:14920-4 [DOI] [PubMed] [Google Scholar]
- 57. Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971-1005 [DOI] [PubMed] [Google Scholar]
- 58. Iyer GH, Garrod S, Woods VL, Jr., Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110-22 [DOI] [PubMed] [Google Scholar]
- 59. Iyer GH, Moore MJ, Taylor SS. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J Biol Chem. 2005;280:8800-7 [DOI] [PubMed] [Google Scholar]
- 60. Taylor SS, Kim C, Vigil D, et al. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25-37 [DOI] [PubMed] [Google Scholar]
- 61. Cho S, Hoang A, Sinha R, et al. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A. 2011;108:8233-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jarnaess E, Stokka AJ, Kvissel AK, et al. Splicing factor arginine/serine-rich 17A (SFRS17A) is an A-kinase anchoring protein that targets protein kinase A to splicing factor compartments. J Biol Chem. 2009;284:35154-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li H, Liu G, Yu J, et al. In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J Biol Chem. 2009;284:16191-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Berke JD, Sgambato V, Zhu PP, et al. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron. 2001;32:277-87 [DOI] [PubMed] [Google Scholar]
- 65. Pilch B, Allemand E, Facompre M, et al. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61:6876-84 [PubMed] [Google Scholar]
- 66. Shultz JC, Goehe RW, Murudkar CS, et al. SRSF1 regulates the alternative splicing of caspase 9 via a novel intronic splicing enhancer affecting the chemotherapeutic sensitivity of non-small cell lung cancer cells. Mol Cancer Res. 2011;9:889-900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Orstavik S, Funderud A, Hafte TT, et al. Identification and characterization of novel PKA holoenzymes in human T lymphocytes. FEBS J. 2005;272:1559-67 [DOI] [PubMed] [Google Scholar]
- 68. Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6: 837-47 [DOI] [PubMed] [Google Scholar]
- 69. Huang TS, Nilsson CE, Punga T, Akusjarvi G. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 2002;3:1088-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rogne M, Landsverk HB, Van EA, et al. The KH-Tudor domain of a-kinase anchoring protein 149 mediates RNA-dependent self-association. Biochemistry. 2006;45:14980-9 [DOI] [PubMed] [Google Scholar]
- 71. Deckert J, Hartmuth K, Boehringer D, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26:5528-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Muhlemann O, Akusjarvi G. Preparation of soluble extracts from adenovirus-infected cells for studies of RNA splicing. Methods Mol Med. 2007;131:33-46 [DOI] [PubMed] [Google Scholar]