Abstract
The retrovirus XMRV (xenotropic murine leukemia virus-related virus) has been detected in human prostate tumors and in blood samples from patients with chronic fatigue syndrome, but these findings have not been replicated. We hypothesized that an understanding of when and how XMRV first arose might help explain the discrepant results. We studied human prostate cancer cell lines CWR22Rv1 and CWR-R1, which produce XMRV virtually identical to the viruses recently found in patient samples, as well as their progenitor human prostate tumor xenograft (CWR22) that had been passaged in mice. We detected XMRV infection in the two cell lines and in the later passage xenografts, but not in the early passages. Importantly, we found that the host mice contained two proviruses, PreXMRV-1 and PreXMRV-2, which share 99.92% identity with XMRV over >3.2-kb stretches of their genomes. We conclude that XMRV was not present in the original CWR22 tumor but was generated by recombination of two proviruses during tumor passaging in mice. The probability that an identical recombinant was generated independently is negligible (~10-12); our results suggest that the association of XMRV with human disease is due to contamination of human samples with virus originating from this recombination event.
Murine leukemia viruses (MLVs) are retroviruses belonging to the genus Gammaretrovirus that cause cancers and other diseases in mice, and are divided into the ecotropic, amphotropic, polytropic, and xenotropic classes on the basis of their receptor usage. Xenotropic MLVs cannot infect cells from inbred mice but can infect cells from other species, including humans. Xenotropic murine leukemia virus-related virus (XMRV) was isolated from a human prostate cancer (PC) in 2006 and has been reported to be present in 6 to 27% of human PCs (1, 2) and in the peripheral blood of 67% of chronic fatigue syndrome (CFS) patients (3). The assertion that XMRV is circulating in the human population has been challenged by several studies that have failed to detect XMRV in multiple cohorts of PC and CFS patients or healthy controls (reviewed in (4). Endogenous xenotropic MLVs can infect human tumors during passage through nude mice (5), and it has been suggested that XMRV may have arisen in this manner (5, 6). In addition, XMRV replication is highly sensitive to human APOBEC3s and tetherin (7-11), making it doubtful that XMRV replication occurred efficiently in human peripheral blood mononuclear cells of CFS patients as previously reported (3).
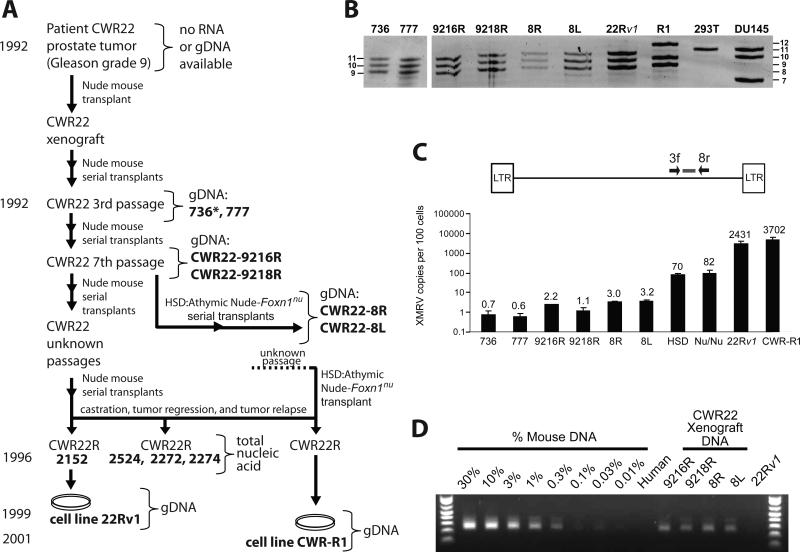
The human PC cell line CWR22Rv1 (hereafter 22Rv1) (12) produces infectious XMRV essentially identical in sequence to that obtained from patients. 22Rv1 contains ≥10 proviral copies/cell (13), and was proposed to have been derived from an XMRV-infected tumor. This cell line was derived from a xenograft (CWR22) that was established from a primary prostate tumor at Case Western Reserve University and serially passaged in nude mice (14, 15). To explore the origin of the virus in 22Rv1 cells, we analyzed various passages of the CWR22 xenograft as well as a subline of the CWR22 xenograft (2152) from which the 22Rv1 cell line was established (12), and another prostate cancer cell line, CWR-R1, which was also derived from CWR22 (16). Fig. 1A traces the timeline of the serial xenograft transplants of CWR22 up to the derivation of the cell lines 22Rv1 and CWR-R1 and indicates (bold letters) the samples that were available for analysis. Nude mouse strain(s) maintained by Charles River (NU/NU) and Harlan Laboratories (Harlan Sprague Dawley [Hsd]) are likely to have been used for in vivo passages of the xenograft (17). DNA samples from passage 3 (777, Fig. 1A) and an unknown early passage (736) were obtained along with samples from a 7th passage, CWR22-9216R and CWR22-9218R. A xenograft tumor from the early 7th passage was independently propagated at the University of California, Davis using Hsd nude mice (CWR22-8R and 8L). Total nucleic acid from relapsed androgen-independent tumors (CWR22R) 2152, 2524, 2272, and 2274 and the 22Rv1 and CWRR1 cell lines was available for analysis (14).
Fig. 1.
Characterization of CWR22 xenografts and XMRV-related sequences. (A) Genesis of 22Rv1 and CWR-R1 cell lines. Bold letters indicate samples from which genomic DNA (gDNA) or total nucleic acid was available for analysis. XMRV-positive samples are boxed. *, unknown early passage. (B) Short tandem repeat (STR) analysis. Representative D7S280 allele pattern of xenografts, 22Rv1 and CWR-R1 cell lines, along with analysis of six additional loci (Fig. S1). An allelic ladder is shown on left and right of gel. (C) Quantitative real-time PCR to detect XMRV env sequences. Calculated copies/100 cells are indicated above each bar. (D) IAP assay to quantitate the amount of mouse DNA present in the xenograft gDNAs.
We verified that the xenograft samples (736, 777, 9216R, 9218R, 8R and 8L) and the 22Rv1 or CWR-R1 cell lines were all derived from the same person by performing short tandem repeat (STR) analysis at 7 loci (Fig. 1B and S1). The probabilities that the xenografts and the two cell lines have the same allele patterns for these loci by chance are 1.6 × 10-13 and 6.3 × 10- 13, respectively.
To quantify the amount of XMRV DNA in the CWR22 xenografts, we developed a real-time PCR primer-probe set that specifically detected XMRV env and excluded murine endogenous proviruses present in BALB/c and NIH3T3 genomic DNA (Fig. 1C). We used quantitative PCR of 22Rv1 DNA to estimate 20 proviruses/cell and used the 22Rv1 DNA to generate a standard curve. The CWR22 xenografts had significantly fewer copies of XMRV env (<1 -3 copies/100 cells) compared to the 22Rv1 cells (2000 copies/100 cells). The CWR-R1 cell line had ~3000 copies/100 cells, and the NU/NU and Hsd nude mice, thought to have been used to passage the CWR22 xenograft, had 58 and 68 copies/100 cells, respectively. Since xenograft tumors are expected to contain a mixture of human and mouse cells, we quantified the amount of mouse DNA by analyzing mouse intracisternal A-type particle (IAP) DNA as previously described (18, 19). Approximately 0.3-1% of the total DNA from all 6 xenografts consisted of mouse DNA (Fig. 1D); this result is consistent with the <1 - 3 XMRV env sequences/100 cells detected in the same samples (Fig. 1C).
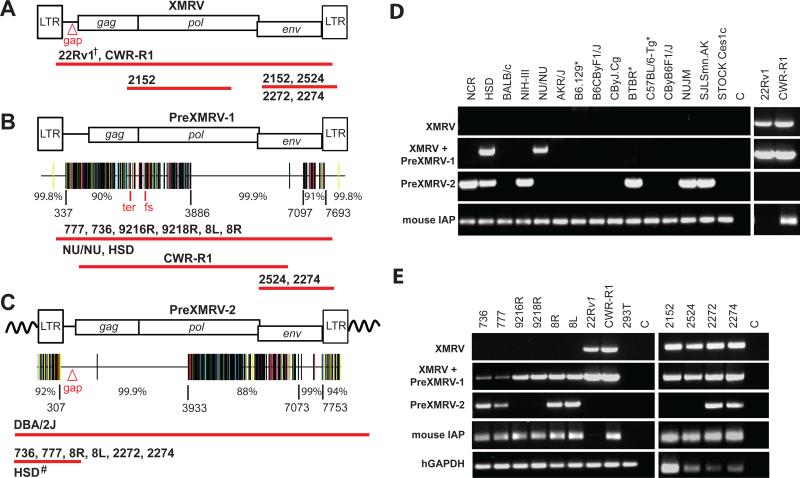
We characterized XMRV and related sequences in the xenografts, cell lines, and nude mouse strains by PCR and DNA sequencing (Fig. 2). Using primers previously used to clone and sequence XMRV from 22Rv1 cells (8), we determined that all the XMRV proviruses in the CWRR1 and 22Rv1 cell lines are identical in sequence, with the exception of some rare hypermutated proviruses (Fig. 2A and Figs. S2 and S3). Next, we developed several primer sets to specifically amplify XMRV sequences and exclude endogenous murine retroviruses (Fig. S2). Primers that specifically amplified XMRV were used to perform PCR on DNA from the late-passage xenografts 2152, 2524, 2272 and 2274; sequencing confirmed the presence of these XMRV sequences in these tumors (Fig. 2A and Fig. S3A; boxed in Fig. 1A).
Fig. 2.
PCR and sequencing analysis of XMRV and XMRV-related sequences from xenografts, cell lines, and nude mouse strains. Using specific primer sets (Fig. S2), cloned PCR products from the xenografts, 22Rv1, CWR-R1, or mouse strains were sequenced. Approximate length and location of sequences determined from samples that were positive for XMRV (A), PreXMRV-1 (B) and PreXMRV-2 (C) are shown as red bars beneath each provirus. Details of primers and numbers of cloned products sequenced are shown in Figs. S2 and S3. Hypermut plots (see Fig. S3 for details), which indicate nucleotide mismatches relative to XMRV as color-coded vertical lines, are shown for PreXMRV-1 (B) and PreXMRV-2 (C), together with the percent identity to consensus XMRV for different regions of each provirus (nucleotide numbers refer to the 22Rv1 XMRV sequence [FN692043]). PreXMRV-1 has a 16-nt deletion (Δ16) in gag and a frameshift (fs) in pol making it replication defective while PreXMRV-2 gag, pol, and env reading frames are open. Mouse strains (D) and xenograft and PC cell lines (E) were analyzed by PCR for the presence of XMRV, PreXMRV-1 and PreXMRV-2. Mouse IAP and human GAPDH serve as positive controls for the presence of mouse and human DNA, respectively. For both (D) and (E) the primer set used to detect PreXMRV-1 can also detect XMRV. For ease of comparison, the 22Rv1 and CWR-R1 gel lanes from (E), which were run in parallel, are duplicated in (D). DNAs in (D) and (E) were all amplified with the same PCR primer master mix. †We previously determined the full-length sequence of XMRV from 22Rv1 cells (8). Δgap refers to the 24-bp deletion in the gag leader characteristic of XMRV. All mouse strains shown in (D) are nudes except for those indicated with *.
We used the same XMRV-specific primer sets to amplify and sequence DNA from early passage xenografts (736, 777, 8L, 8R, 16R, and 18R; Fig. 2B); the results showed that XMRV env, but not gag sequences were present (sequencing coverage summarized in Fig. S3), indicating that the early xenografts did not contain XMRV. However, we did find that early xenografts contained a previously undescribed XMRV-related provirus that we have named PreXMRV-1 (Fig. 2B). The complete sequence of PreXMRV-1 was determined from the early passage xenografts, the NU/NU and Hsd strains, and the CWR-R1 cell line. PreXMRV-1 and consensus XMRV differed by only one base in a 3211-nt stretch of the genome encoding the 3' half of pol and the 5' 2/3 of env. In addition, the LTRs were nearly identical; PreXMRV-1 had a single adenine deletion relative to XMRV in a run of 6 adenines. The two genomes differed by 10% over the remaining 3.5-kb stretch of gag-pro-pol and by 9% in a 600-nt stretch at the 3' end of env. PreXMRV-1 is replication defective because of a 16-nt deletion in gag and a +1 frameshift mutation in pol. Late-passage xenografts 2524 and 2274, but not 2152 and 2272, also contained PreXMRV-1. The detection of low levels of XMRV env sequence in the early xenografts (Fig. 1C) can be attributed to the PreXMRV-1 proviruses present in the contaminating mouse DNA. Overall, these results indicate that PreXMRV-1 is an endogenous murine provirus that is present in the NU/NU and Hsd strains, but neither of these strains contains XMRV (the PCR and sequencing coverage are detailed in Figs. S3A and S3B).
To screen for the presence of endogenous XMRV in mouse strains, we developed an XMRV-specific PCR assay based on sequence differences in the LTR and gag leader regions that excluded all known endogenous murine retroviruses (Fig. S2). A survey of 45 laboratory mouse strains and 44 wild mice failed to detect XMRV (Fig. S4). In a search for proviruses that might contain XMRV-specific sequence features, we found a second previously undescribed endogenous provirus that we named PreXMRV-2 (Fig. 2C). A portion of PreXMRV-2 corresponds to an 1124-nt sequence of an endogenous provirus from the 129X1/SvJ mouse genome (Acc. No. AAHY0159188.1) (6, 20). The sequence of PreXMRV-2 revealed that gag, pol, and env reading frames are open and can potentially express functional proteins. A 3.6-kb stretch encompassing the gag leader region and gag-pro-pol differs by one base from the consensus XMRV (99.9% identity); in addition, a ~700-nt region of env is 99% identical to XMRV; however, the LTRs and the remaining viral genome differ by 6-12% from consensus XMRV. Phylogenetic analysis indicates that PreXMRV-1 groups with xenotropic viruses whereas PreXMRV-2 appears to be a recombinant, grouping with polytropic and modified polytropic viruses for certain stretches of its genome (Fig. S5).
We screened 15 mouse strains, which included 12 nude mice, for the presence of XMRV, PreXMRV-1, and PreXMRV-2 using XMRV-specific primers, primers that amplified XMRV or PreXMRV-1, and PreXMRV-2-specific primers (Fig. 2D and S2). None of the mouse strains contained XMRV and only the Hsd and the NU/NU outbred nude strains contained PreXMRV-1 (Fig. 2D and S6). Six of the 15 mouse strains contained PreXMRV-2, but only the NU/NU and Hsd mice contained both PreXMRV-1 and PreXMRV-2 (Fig. 2D and S6). It should be noted that since the Hsd and the NU/NU are outbred strains, individual mice differ in their endogenous proviruses. NU/NU mice showed variation in the presence of these two endogenous proviruses, and two out of five animals tested contained both (Fig. S6). The 22Rv1 cell line contained only XMRV as confirmed by sequence analysis; however the CWR-R1 cell line contained both XMRV and PreXMRV-1. The CWR-R1 cell line has been reported to contain contaminating mouse cells (21) (and see IAP signal, Fig 2D), which is likely to be the source of the PreXMRV-1 sequences.
We used the same specific primer sets to determine the distribution of XMRV, PreXMRV-1 and PreXMRV-2 in early and late xenografts (Fig. 2E). None of the early xenografts (736, 777, 9216R, 9218R, 8R and 8L), but all of the late xenografts (2152, 2524, 2272, and 2274) and both cell lines were positive for XMRV. The primers used to detect PreXMRV-1 could also detect XMRV; sequencing analysis of the PCR products from all of the early xenografts detected only PreXMRV-1, but both XMRV and PreXMRV-1 were detected from the late xenografts 2524 and 2274 (Fig. 2B). Amplification with PreXMRV-2-specific primers revealed the presence of this provirus in early xenografts 736, 777, 8R and 8L, and late xenografts 2272 and 2274 (Fig. 2C, 2E and S3C). The variable detection level of PreXMRV-2 in the late xenografts could be due to individual differences in the outbred mice, and by extension, in the mouse DNA in these samples.
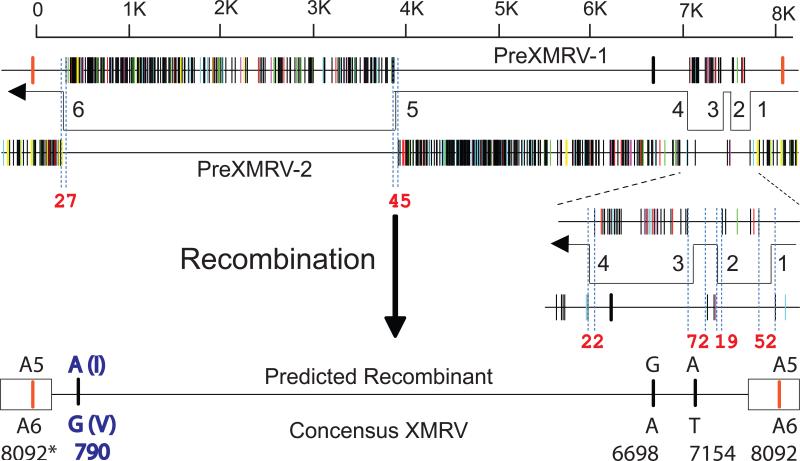
Comparison of the PreXMRV-1 and PreXMRV-2 sequences revealed that the regions of near identity to XMRV are reciprocal and largely non-overlapping. We therefore hypothesized that recombination between these two retroviruses resulted in the formation of XMRV. As shown in Fig. 3A, reverse transcriptase template switching events during minus-strand DNA synthesis can form a recombinant that is essentially identical to the sequences of all of the XMRVs reported to date, and differing from the consensus XMRV by only 4 nucleotides. The six switching events occurred in 20 - 73 nucleotide stretches that are identical between PreXMRV-1 and PreXMRV-2 (Fig. 3A, red numbers; Fig. S7A). Of the four nucleotide differences between the predicted recombinant and consensus XMRV, only the A>G change at position 790 results in a conservative valine-to-isoleucine amino acid substitution; the other 3 substitutions are silent. The 22Rv1 and CWR-R1 cell lines as well as VP42 have an A at position 790, whereas all other XMRV isolates have a G at position 790. The insertion of an A at position 8092 occurred within a run of 6 adenines; frameshift mutations commonly occur in such homopolymers during retroviral replication (22). A comparison of the predicted recombinant to the available XMRV sequences is shown in Fig. S7B. The available XMRV sequences all have the same six recombination junctions predicted in the hypothetical recombinant, and differ from the consensus XMRV by 3 - 14 nucleotides. These differences may be the result of errors during PCR or sequencing, or mutations during the passage of XMRV in another cell line. Phylogenetic analysis supports the predicted recombinant virus as the precursor of the virus in the CWR22 xenografts, the 22Rv1 and CWR-R1 cell lines, and all XMRVs isolated and sequenced from patients (Fig. 3B and ref. 23).
Fig. 3.
Predicted recombinant between PreXMRV-1 and PreXMRV-2 is nearly identical to XMRV. (A) Alignment of Hypermut plots of PreXMRV-1 and PreXMRV-2 reveals the reciprocal and largely nonoverlapping regions of near identity to XMRV. The direction of minus-strand DNA synthesis catalyzed by reverse transcriptase, and predicted template switching events (numbered 1-6) are shown. The lengths of nucleotide identity within the presumed template switching regions are indicated in red numbers. The predicted recombinant and the 4 nucleotide differences with consensus XMRV are shown. The nucleotide numbers refer to numbers of the 22Rv1 XMRV (Acc. No. FN692043). Note that nucleotide 8092 is within the U3 region, and is present in both LTRs (boxes). A5 and A6 refer to homopolymeric runs of 5 and 6 adenines, respectively. The A>G change at 790 results in an isoleucine (I) to valine (V) substitution. (B) Phylogenetic tree of all full-length XMRV sequences to date and the predicted recombinant implicates the predicted recombinant as the ancestor of all sequenced XMRV isolates. The tree shown is an enlargement of the XMRV-specific portion of the complete endogenous MLV tree (See Fig. S5A and ref 23).
Our findings indicate that virus derived from two previously undescribed murine endogenous retroviruses, PreXMRV-1 and PreXMRV-2, most likely underwent retroviral recombination to generate XMRV during in vivo passaging of the CWR22 xenograft in nude mice. The fact that both parental endogenous proviruses were present in some of the nude mouse strains used for in vivo passaging of the xenografts indicates that there were opportunities for this recombinant to form and spread in the tumor cells that were the progenitors of the 22Rv1 and CWR-R1 cell lines. Only 6 template switching events, which is close to the average of 4 template switches per replication cycle (24), are needed to generate a recombinant that is both essentially identical and ancestral to all XMRV sequences characterized to date from cell lines and patients (Fig. 3B). We have estimated the probability that the exact set of template switching events occurred independently is 1.3 × 10-12 (Fig. S8 and ref 23), making it very likely that contamination of human samples with XMRV originating from the relapsed CWR22 xenografts or either of the two cell lines, perhaps through other intermediate cell lines, contributed to its reported association with PC and CFS. Our results and conclusions relate to XMRV detection by isolation of virus of this specific sequence (1-3), and do not directly address detection of antibodies or viral antigens (25, 26), or PCR detection of related but distinct MLV sequences (27). We note, however, that most “XMRV-specific” PCR assays may detect PreXMRV-1 or -2 proviruses in contaminating mouse DNA, and that specific detection of XMRV requires the use of primers that flank a crossover site.
The alternative possibility is that recombination between PreXMRV-1 and PreXMRV-2 occurred during mouse evolution, giving rise to an endogenous XMRV provirus that is present in mice and can occasionally infect humans. We think this possibility is remote because analysis of the early xenografts, which contained contaminating nude mouse DNAs, failed to detect XMRV. Furthermore, we were unable to detect XMRV in a screen of 89 inbred and wild-derived mouse strains including 17 individual nude mice (Fig. S4 and ref 23).
We conclude that XMRV was generated as a result of a unique recombination event between two endogenous MLVs that took place around 1993-1996 in a nude mouse carrying the CWR22 PC xenograft. Since the probability that the same recombination event could occur independently by random chance is essentially negligible, any XMRV isolates with the same or nearly the same sequences identified elsewhere originated from this event (23).
Supplementary Material
Acknowledgments
We thank W. Shao for analysis of MLV diversity, and E. Freed and S. Hughes for helpful discussions. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was also supported in part by Bench-to-Bedside Award to VKP, research grant R37 CA 089441 to JMC and RO1CA150197 to HJK. JMC was a Research Professor of the American Cancer Society with support from the FM Kirby Foundation.
References and Notes
- 1.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106:16351. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Urisman A, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Lombardi VC, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 4.Van der Kuyl AC, Cornelissen M, Berkhout B. Of mice and men: on the origin of XMRV. Frontiers in Microbiology. 2011;1 doi: 10.3389/fmicb.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss RA. A cautionary tale of virus and disease. BMC Biol. 2010;8:124. doi: 10.1186/1741-7007-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hue S, et al. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology. 2010;7:111. doi: 10.1186/1742-4690-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groom HC, Yap MW, Galao RP, Neil SJ, Bishop KN. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc Natl Acad Sci U S A. 2010;107:5166. doi: 10.1073/pnas.0913650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paprotka T, et al. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J Virol. 2010;84:5719. doi: 10.1128/JVI.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stieler K, Fischer N. Apobec 3G efficiently reduces infectivity of the human exogenous gammaretrovirus XMRV. PLoS One. 2010;5:e11738. doi: 10.1371/journal.pone.0011738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd HP, Zhang F, Bieniasz PD, Cullen BR. Human APOBEC3 proteins can inhibit xenotropic murine leukemia virus-related virus infectivity. Virology. 2011;410:234. doi: 10.1016/j.virol.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaipan C, et al. Severe Restriction of Xenotropic Murine Leukemia Virus-Related Virus Replication and Spread in Cultured Human Peripheral Blood Mononuclear Cells. J Virol. 2011;85:4888. doi: 10.1128/JVI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sramkoski RM, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 13.Knouf EC, et al. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J Virol. 2009;83:7353. doi: 10.1128/JVI.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagabhushan M, et al. CWR22: the first human prostate cancer xenograft with strongly androgen-dependent and relapsed strains both in vivo and in soft agar. Cancer Res. 1996;56:3042. [PubMed] [Google Scholar]
- 15.Pretlow TG, et al. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85:394. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- 16.Gregory CW, Johnson RT, Jr., Mohler JL, French FS, Wilson EM. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 2001;61:2892. [PubMed] [Google Scholar]
- 17.Materials and Methods are available as supporting material in Science Online.
- 18.Oakes B, et al. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology. 2010;7:109. doi: 10.1186/1742-4690-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MJ, et al. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology. 2010;7:108. doi: 10.1186/1742-4690-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courgnaud V, Battini J-L, Sitbon M, M. A. L. Mouse retroviruses and chronic fatigue syndrome: Does X (or P) mark the spot? . Proc Natl Acad Sci U S A. 2010;107:15666. doi: 10.1073/pnas.1007944107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Bokhoven A, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 22.Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990;87:6019. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Further discussion in support of the unique origin of XMRV and its role as the source of XMRV in PC and CFS patients is available as supporting material in Science Online.
- 24.Zhuang J, Mukherjee S, Ron Y, Dougherty JP. High rate of genetic recombination in murine leukemia virus: implications for influencing proviral ploidy. J Virol. 2006;80:6706. doi: 10.1128/JVI.00273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold RS, et al. XMRV infection in patients with prostate cancer: novel serologic assay and correlation with PCR and FISH. Urology. 2010;75:755. doi: 10.1016/j.urology.2010.01.038. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, et al. Characterization of antibodies elicited by XMRV infection and development of immunoassays useful for epidemiologic studies. Retrovirology. 2010;7:68. doi: 10.1186/1742-4690-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo SC, et al. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc Natl Acad Sci U S A. 2010;107:15874. doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.