Abstract
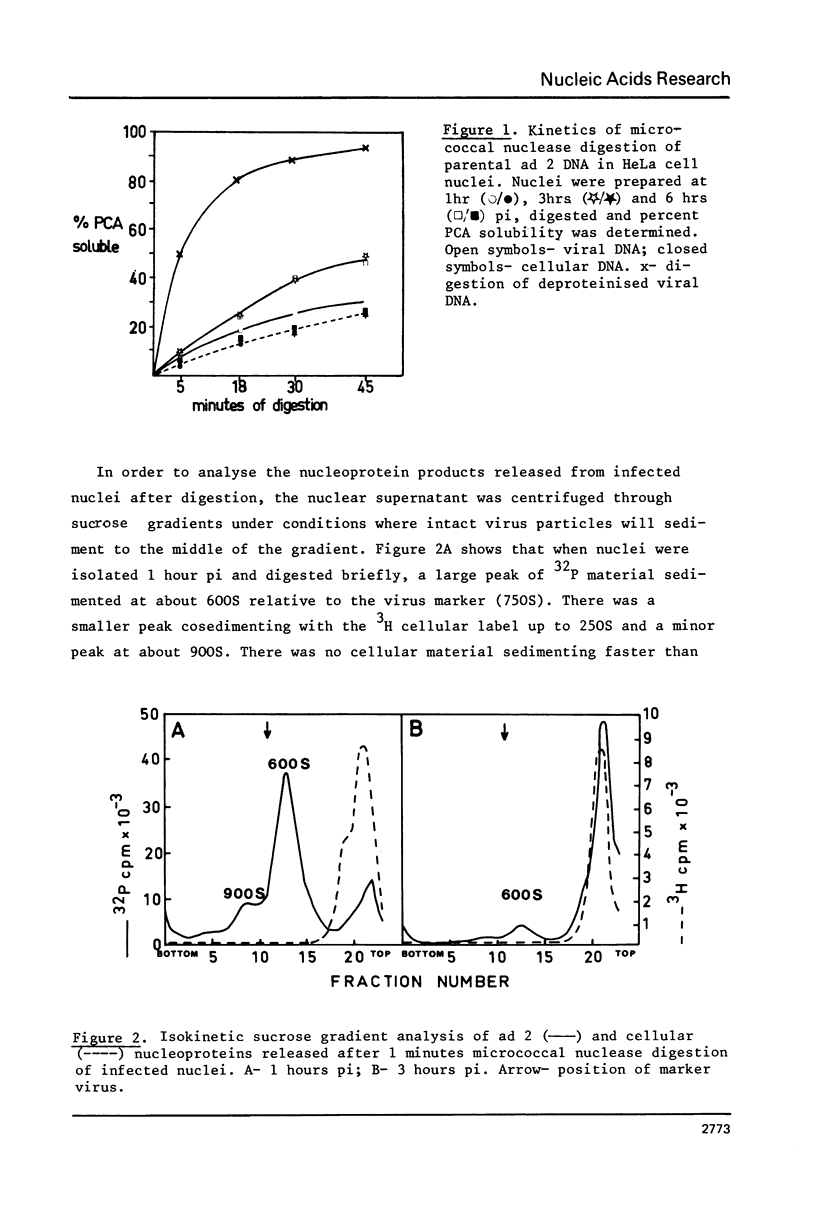
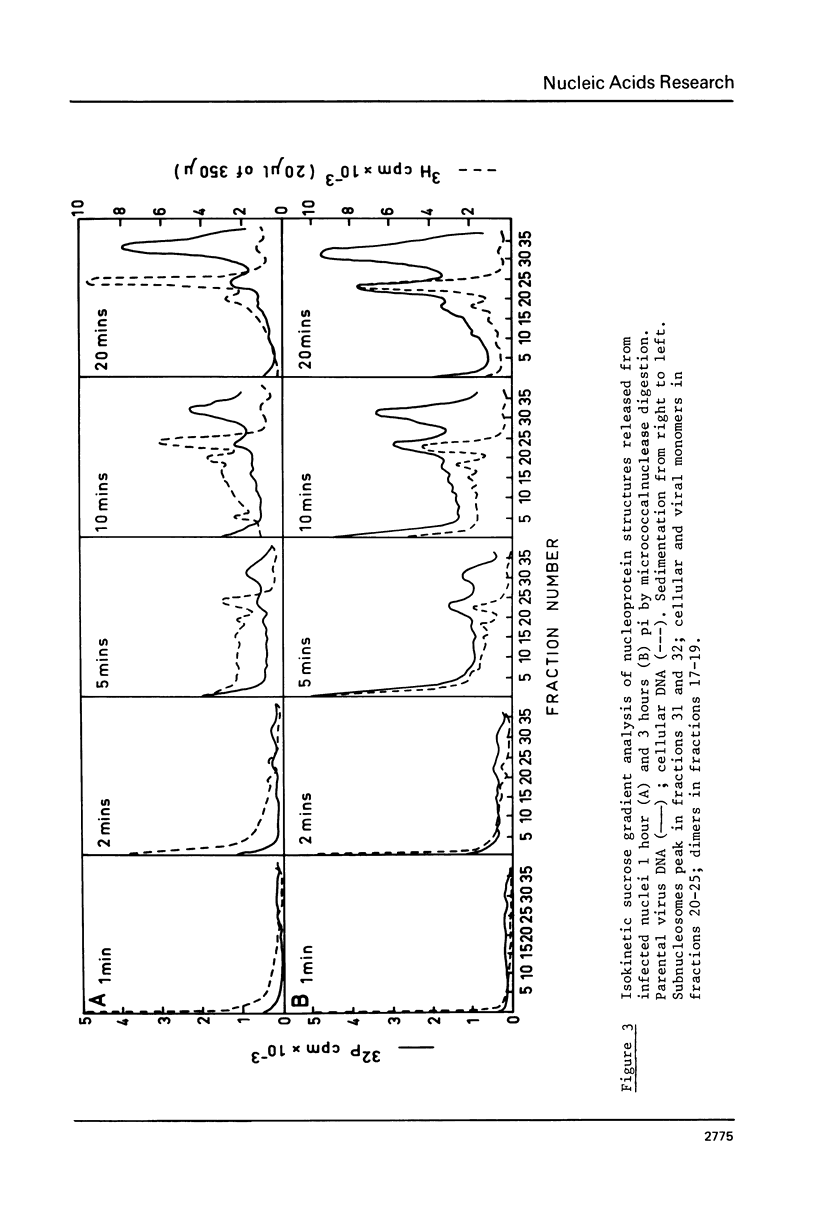
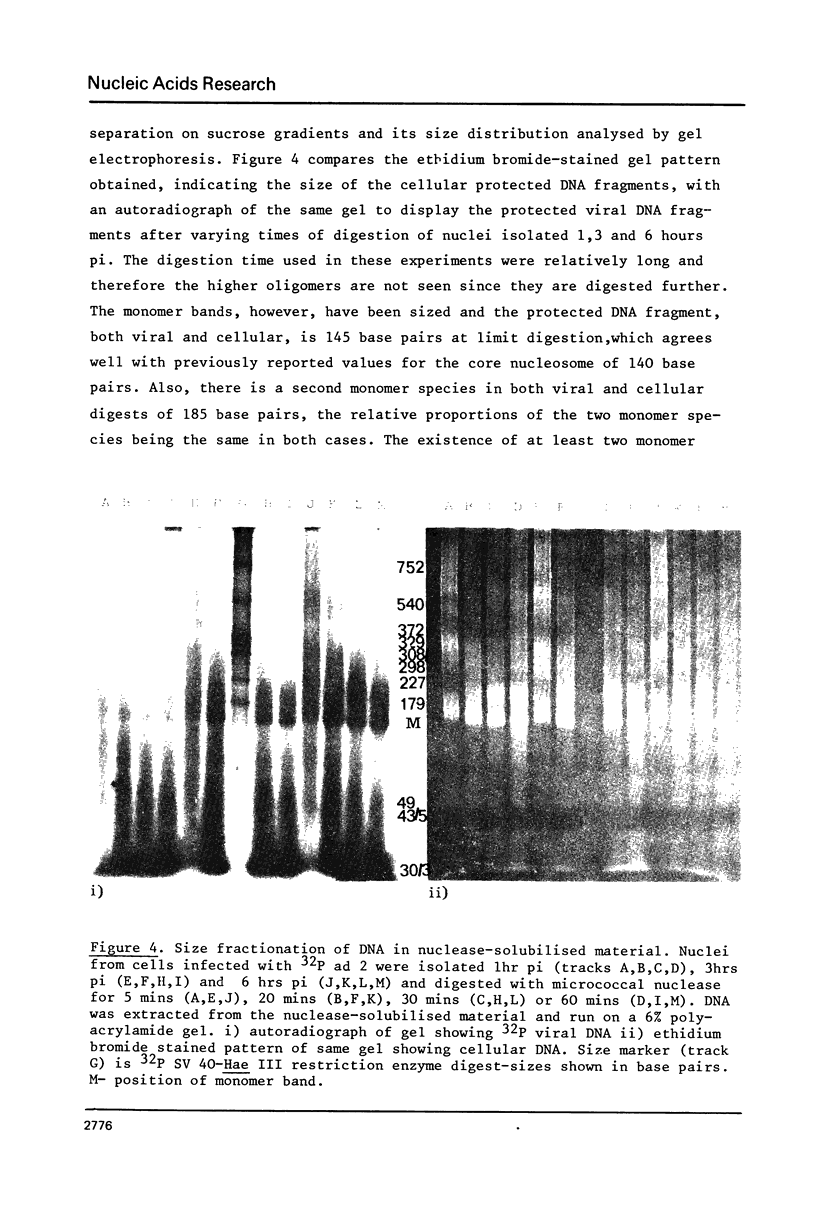
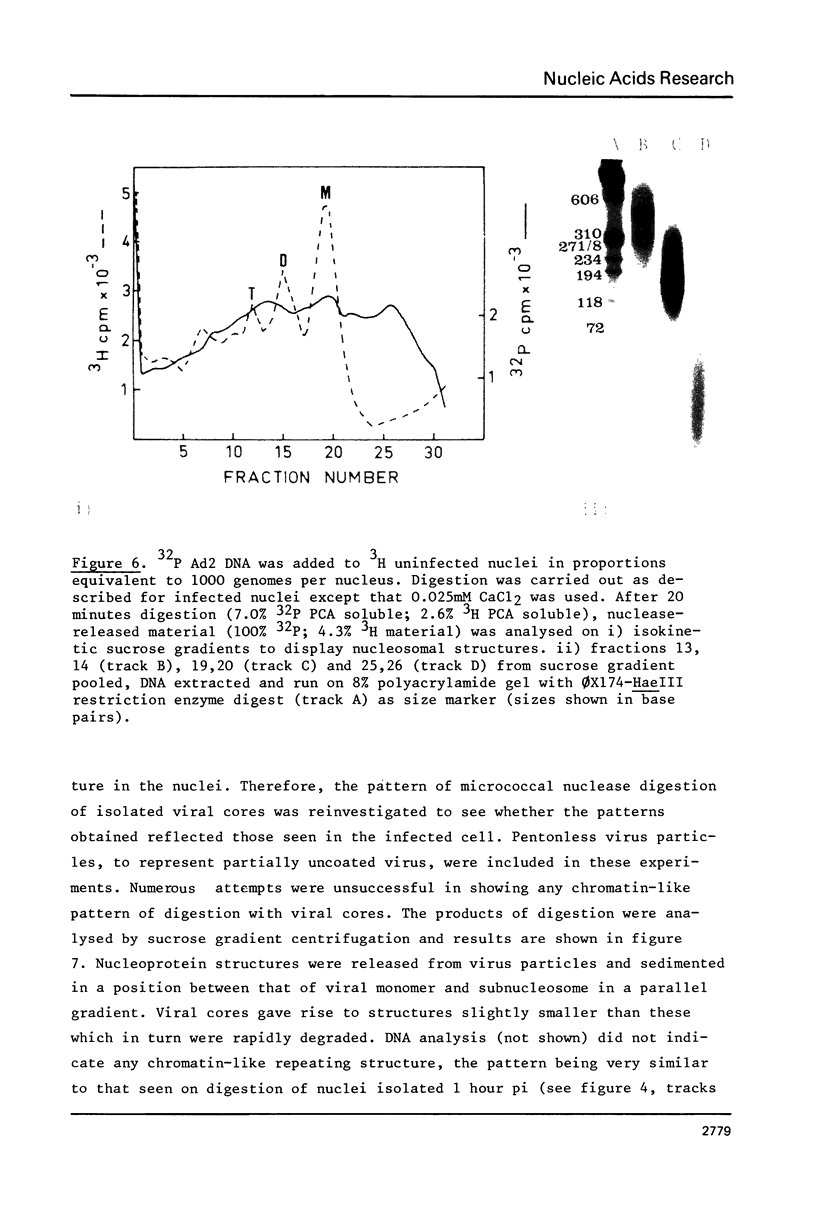
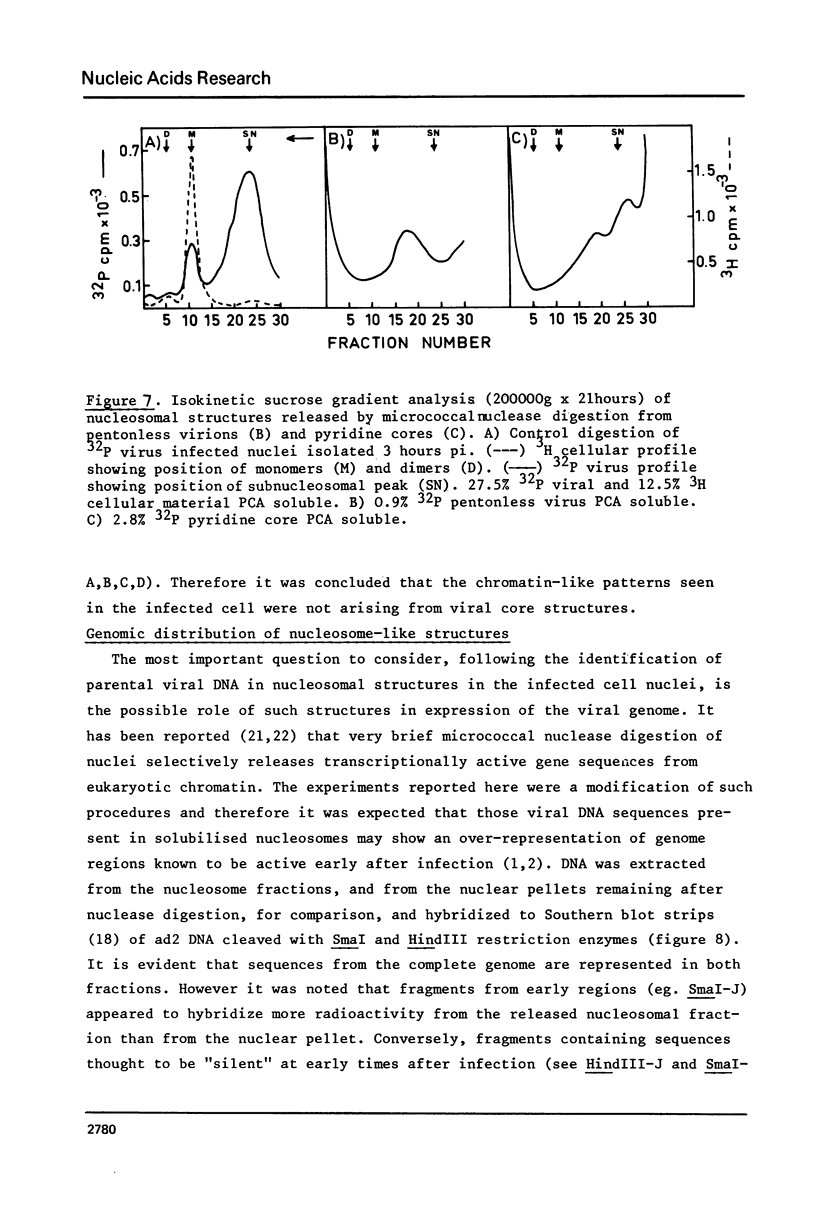
Micrococcal-nuclease digestion of adenovirus 2(ad 2) infected HeLa cell nuclei early after infection has been used to investigate the nucleoprotein nature of parental viral DNA. Viral DNA is more susceptible to nuclease digestion than cellular DNA. The pattern of digestion products changes as digestion proceeds from an indistinct pattern 1 hour post infection(pi) to a nucleosome-like pattern at 6 hours pi. The major differences between viral and cellular nucleoprotein products were i) a subnucleosome fraction from viral DNA and ii) the repeat size of DNA in viral nucleosomes was 165 base pairs and in cellular nucleosomes, 195 base pairs. Up to 50% viral DNA in nuclei 6 hours pi seems to be in nucleosome-like structures. Such patterns are not seen on digestion of partially-uncoated virus or isolated cores.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Baum P. R., Gesteland R. F. Processing of adenovirus 2-induced proteins. J Virol. 1973 Aug;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Ultraviolet mapping of the adenovirus 2 early promoters. Cell. 1977 Sep;12(1):45–55. doi: 10.1016/0092-8674(77)90184-2. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Chardonnet Y., Dales S. Early events in the interaction of adenoviruses with HeLa cells. 3. Relationship between an ATPase activity in nuclear envelopes and transfer of core material: a hypothesis. Virology. 1972 May;48(2):342–359. doi: 10.1016/0042-6822(72)90045-1. [DOI] [PubMed] [Google Scholar]
- Chardonnet Y., Dales S. Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology. 1970 Mar;40(3):462–477. doi: 10.1016/0042-6822(70)90189-3. [DOI] [PubMed] [Google Scholar]
- Corden J., Engelking H. M., Pearson G. D. Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci U S A. 1976 Feb;73(2):401–404. doi: 10.1073/pnas.73.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M., Fraser N., Ziff E., Weber J., Wilson M., Darnell J. E. The initiation sites for RNA transcription in Ad2 DNA. Cell. 1977 Nov;12(3):733–739. doi: 10.1016/0092-8674(77)90273-2. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Groneberg J., Brown D. T., Doerfler W. Uptake and fate of the DNA of adenovirus type 2 in KB cells. Virology. 1975 Mar;64(1):115–131. doi: 10.1016/0042-6822(75)90084-7. [DOI] [PubMed] [Google Scholar]
- Hodge L. D., Scharff M. D. Effect of adenovirus on host cell DNA synthesis in synchronized cells. Virology. 1969 Apr;37(4):554–564. doi: 10.1016/0042-6822(69)90273-6. [DOI] [PubMed] [Google Scholar]
- Johansson K., Pettersson U., Philipson L., Tibbetts C. Reassociation of complementary strand-specific adenovirus type 2 DNA with viral DNA sequences of transformed cells. J Virol. 1977 Jul;23(1):29–35. doi: 10.1128/jvi.23.1.29-35.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Pereira H. G., Russell W. C., Valentine R. C. Isolation of an internal component from adenovirus type 5. J Mol Biol. 1968 Nov 14;37(3):379–386. doi: 10.1016/0022-2836(68)90109-5. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg-Holm K., Philipson L. Early events of virus-cell interaction in an adenovirus system. J Virol. 1969 Oct;4(4):323–338. doi: 10.1128/jvi.4.4.323-338.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C., Rosenkranz H. S., Mednis B. Structure and development of viruses as observed in the electron microscope. V. Entry and uncoating of adenovirus. J Virol. 1969 Nov;4(5):777–796. doi: 10.1128/jvi.4.5.777-796.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Nishimoto T., Raskas H. J., Basilico C. Temperature-sensitive cell mutations that inhibit adenovirus 2 replication. Proc Natl Acad Sci U S A. 1975 Jan;72(1):328–332. doi: 10.1073/pnas.72.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll H. Characterization of macromolecules by constant velocity sedimentation. Nature. 1967 Jul 22;215(5099):360–363. doi: 10.1038/215360a0. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Oliver D., Granner D., Chalkley R. Identification of a distinction between cytoplasmic histone synthesis and subsequent histone deposition within the nucleus. Biochemistry. 1974 Feb 12;13(4):746–749. doi: 10.1021/bi00701a017. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Mathews M. B. The gene and messenger RNA for adenovirus polypeptide IX. Cell. 1977 Nov;12(3):741–750. doi: 10.1016/0092-8674(77)90274-4. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Tibbetts C., Philipson L. Hybridization maps of early and late messenger RNA sequences on the adenovirus type 2 genome. J Mol Biol. 1976 Mar 15;101(4):479–501. doi: 10.1016/0022-2836(76)90241-2. [DOI] [PubMed] [Google Scholar]
- Prage L., Pettersson U., Höglund S., Lonberg-Holm K., Philipson L. Structural proteins of adenoviruses. IV. Sequential degradation of the adenovirus type 2 virion. Virology. 1970 Oct;42(2):341–358. doi: 10.1016/0042-6822(70)90278-3. [DOI] [PubMed] [Google Scholar]
- Price R., Penman S. Transcription of the adenovirus genome by an -amanitine-sensitive ribonucleic acid polymerase in HeLa cells. J Virol. 1972 Apr;9(4):621–626. doi: 10.1128/jvi.9.4.621-626.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Younghusband H. B., Bellett A. J. A circula DNA-protein complex from adenoviruses. Virology. 1973 Nov;56(1):54–69. doi: 10.1016/0042-6822(73)90287-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Gallimore P. H., Flint S. J. Mapping of adenovirus 2 RNA sequences in lytically infected cells and transformed cell lines. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):457–474. doi: 10.1101/sqb.1974.039.01.058. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tata J. R., Baker B. Enzymatic fractionation of nuclei: polynucleosomes and RNA polymerase II as endogenous transcriptional complexes. J Mol Biol. 1978 Jan 25;118(3):249–272. doi: 10.1016/0022-2836(78)90227-9. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlak J. M., Rozijn T. H., Spies F. Replication of adenovirus type 5 DNA in KB cells: localization and fate of parental DNA during replication. Virology. 1976 Jul 1;72(1):99–109. doi: 10.1016/0042-6822(76)90315-9. [DOI] [PubMed] [Google Scholar]