Abstract
Breast cancer is a heterogeneous disease with a poorly defined genetic landscape, which poses a major challenge in diagnosis and treatment. By massively parallel mRNA sequencing, we obtained 1.2 billion reads from 17 individual human tissues belonging to TNBC, Non-TNBC, and HER2-positive breast cancers and defined their comprehensive digital transcriptome for the first time. Surprisingly, we identified a high number of novel and unannotated transcripts, revealing the global breast cancer transcriptomic adaptations. Comparative transcriptomic analyses elucidated differentially expressed transcripts between the three breast cancer groups, identifying several new modulators of breast cancer. Our study also identified common transcriptional regulatory elements, such as highly abundant primary transcripts, including osteonectin, RACK1, calnexin, calreticulin, FTL, and B2M, and “genomic hotspots” enriched in primary transcripts between the three groups. Thus, our study opens previously unexplored niches that could enable a better understanding of the disease and the development of potential intervention strategies.
Breast cancer is the leading cause of cancer death among women, accounting for 23% of the total cancer cases1. The major treatment challenge remains at the level of defining the specific types and associated biology behind the disease2,3,4. Breast cancer is known to be a heterogeneous disease with a variety of morphological features and clinical manifestations due to genetic, epigenetic, and transcriptomic alterations3,4,5,6,7. This phenotypic diversity severely affects the diagnosis and prognosis of breast cancer. The main difficulties in resolving these issues include the complexities of determining specific markers and the lack of a complete understanding of the cellular hierarchy of the mammary epithelium5,7,8,9,10,11. In addition, the remarkable variations in response to therapy12,13 also emphasise the pressing need for further understanding of breast cancer evolution, the genomic basis of heterogeneity, and the biological basis of this disease.
Numerous reports have demonstrated that the metastatic status, histological grade, tumour stage, size, and receptor expression are the main critical determinants of breast cancer treatment14,15,16,17,18. Seminal gene expression studies by Perou et al. (2000) and Sorlie et al. (2001) have established a classification of breast cancer into five broad “intrinsic phenotypic subtypes”19,20. These subtypes include Luminal A, Luminal B, Human Epidermal Growth Factor Receptor 2 (HER2)-positive, basal-like and normal breast-like breast cancers19,20,21,22. Correlating these subtypes with the traditional tumour histology provided a paradigm shift in breast cancer diagnostics. Furthermore, microarray investigations have offered an initial basis for treatment prediction22,23,24,25,26 and identification of the different breast tumour stages that are critical for breast cancer treatment27,28,11,29. However, translating molecular profiling into clinical practice has proven to be a formidable challenge as a result of complex heterogeneity30,31.
Immunohistochemically, three broad types of breast tumours have been classified by the status of therapeutically significant components, the Estrogen receptor ER, the progesterone-receptor (PR) and the HER23,32. Breast tumours lacking expression of all three receptors are defined as triple-negative breast cancer (TNBC)33,34,35,36. TNBC is often classified as basal-like breast cancer, which represents 10–25% of all tumours and is presumed to be derived from a distinct cell type and a specific developmental stage of mammary epithelial cell development19,22,34,36. In contrast, the gene expression profiles of HER2-positive (ER and PR negative) and Non-TNBC (positive for all three receptors) tumours belong to the luminal-like subgroups, representing approximately 15% of patients37,38. The main characteristics of TNBC are frequent occurrence in younger patients (<50), increased aggressiveness, significantly shorter survival periods, and higher recurrence rates compared with the Non-TNBC subtypes. These difficulties warrant an immediate and intensive focus on this difficult-to-treat type of breast cancer33,34,39.
To identify the precise genetic elements and study the exclusive nature of TNBC and the other two types of breast cancer, we employed a massively parallel in depth mRNA sequencing approach40,41. This global transcriptomic profiling can illustrate the intricate inner workings of the transcriptome at a very high resolution, enabling us to explore the distinct nature of these breast cancer subtypes, and provide a new inventory of diagnostic and therapeutic targets.
Results
Comparative transcriptome analysis strategy
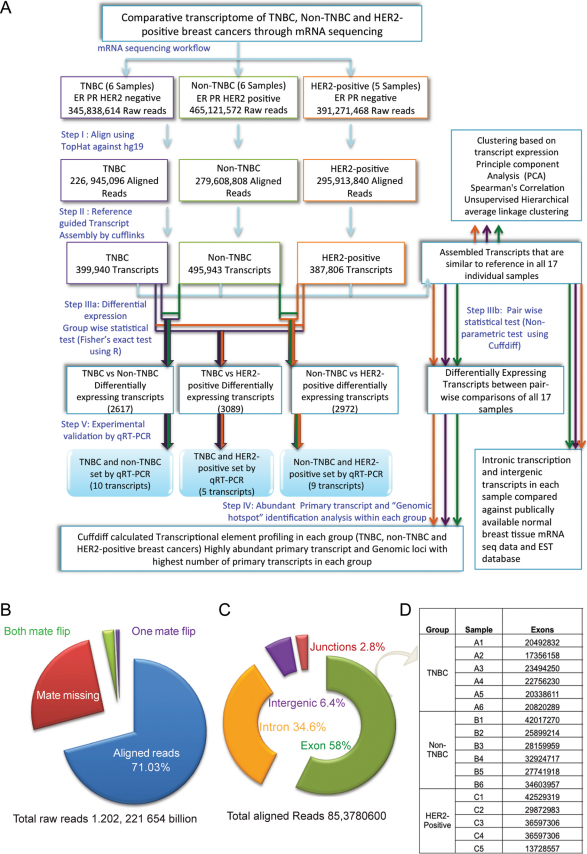
We aimed to compare the transcriptomic expression profile of TNBC, Non-TNBC and HER2-positive breast cancers. Accordingly, 17 individual human breast cancer tissues, including six TNBC samples, six Non-TNBC samples and five HER2-positive samples, were chosen. The mRNA sequencing of the samples was performed using the Illumina platform, generating a total 1.2 billion high quality raw reads (Supplementary Method, Figure 1A). In addition, we used publicly available 50-base pair paired-end single normal breast tissue RNA sequence data (http://www.ncbi.nlm.nih.gov/sra) for comparisons when applicable. The reads were aligned against the Ensembl GRCh37.62 B (hg19) reference genome using TopHat42. The reference genome-guided transcript assembly of the aligned reads was performed using cufflinks, a well-established transcript assembler43. Furthermore, all subsequent analyses were conducted using only the transcripts that were identical to the reference identified by cuffcompare. The transcripts were binned according to their abundance, and the overall relative abundances of the transcripts that were expressed in all three breast cancer types on each chromosome were compared.
Figure 1. The comparative transcriptomic profiling of TNBC, Non-TNBC and HER2-positive breast cancer mRNA sequencing.
(A) Overview of the steps involved in the mRNA sequencing analysis of TNBC, Non-TNBC and HER2-positive breast cancers. (B) The mRNA reads were mapped to the Ensembl GRCh37.62 B human genome (hg19), and the summary of the alignment statistics of the fragments mapping onto the reference genome is presented in different colours. (C) The distribution of the fragments onto the Ensembl GRCh37.62 B human genome (hg19) is shown as the percentage of reads that map onto exons, introns, intergenic regions and junctions. (D) The total number of exons assembled from the aligned reads in each sample.
From the assembled transcripts and their abundances, the following statistical and secondary analyses were performed. First, the correlation between the samples was determined using Principle Component Analysis (PCA), unsupervised hierarchical clustering and Spearman's correlation. Second, the transcripts that were differentially expressed between the three breast cancer groups and the pairwise comparison of all 17 individual samples were identified using univariate Fisher's exact test (F-test) and cuffdiff, respectively. The cuffdiff analysis employs various parametric approaches, such as the normal and negative binomial distributions. The F-test was conducted using the statistical package, R. Third, the transcriptional and post-transcriptional changes were compared in each breast cancer type. In this analysis, we focused on two critical transcriptional elements, which included the highly abundant primary transcripts and the genomic loci that comprised the highest number of primary transcripts, i.e., the genomic hotspots. Finally, we also investigated the significance of the predominant intronic reads found in all three types of breast cancer. Together, these analyses allowed us to compare the transcriptomic and post-transcriptomic profiles of TNBC, Non-TNBC and HER2-positive breast cancer.
Generation of the TNBC, Non-TNBC and HER2-positive breast cancer digital transcriptome
Among the 1.2 billion high quality raw reads, 71.03% of the reads were mapped to the human genome (Figure 1B) (Supplementary Tables 1 and 2). The distribution of these mRNA reads indicated that most reads (58%) mapped to exons. However, intriguingly, 34.6% of the reads mapped to intronic regions while 6.4% and 2.8% mapped to intergenic and junction regions, respectively (Figure 1C) (Supplementary Table 3). In each sample, the aligned reads supported an average of 80,279 transcripts (p value and FDR less than 0.05) that were identical to previously annotated transcripts (Supplementary Table 4, the transcript reassembly of HSP901B is shown as an example in Supplementary Figure 1).
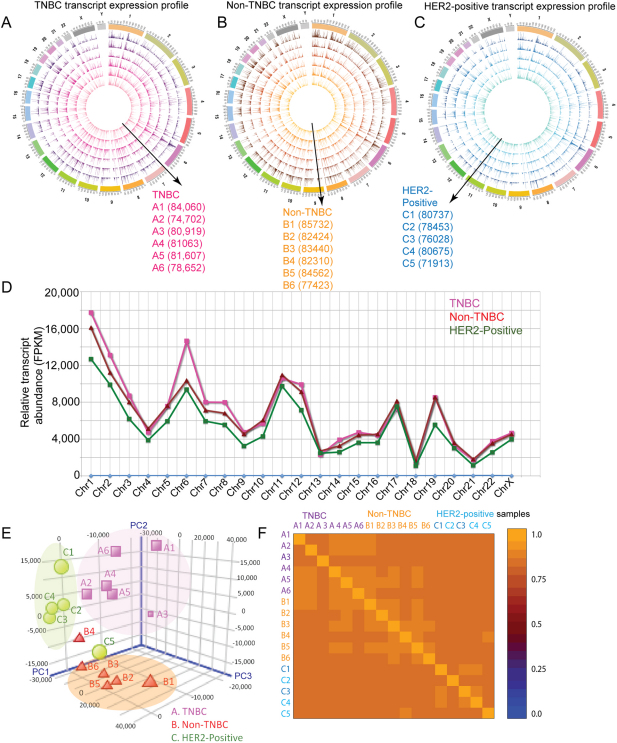
Interestingly, an average of 16,245 distinct genes per sample were detected, indicating the diversity introduced by isoforms of various genes (Supplementary Table 4). From the total of 17 sequenced breast cancer samples, 1,364,752 known transcripts were identified, comprising 5,226,535 and 11,761,893 unique and total exons, respectively (Figure 1D, Figure 2A, 2B and 2C, Supplementary Table 4, Supplementary Figure 2). The transcript abundance was calculated by estimating the fragments per kilobase of exon per million mapped fragments (FPKM)43, and all expressed transcripts were binned on the basis of their abundance (FPKM). This analysis revealed that the majority of the assembled transcripts were low in abundance, i.e., below 1 FPKM (Supplementary Method, Supplement Table 5, and Supplementary Figure 3–8). In parallel, the transcripts from all the 17 breast cancer samples were also reassembled using another reference genome (UCSC hg19), and similar transcript expression trends as described above were observed (Supplementary Table 4B).
Figure 2. The overall transcriptomic expression profile of TNBC, non-TNBC and HER2-positive breast cancers and correlation between the breast cancers.
(A) The transcriptomic expression profiles are shown in the Circos plot. The expression profile of the transcripts with FPKM (i.e., the transcript abundance measured by cufflinks using Ensembl GRCh37.62 B human genome (hg19)) of up to 200 in all six samples of the three breast cancer types was visualised in Circos48 for the (C) TNBC, (D) Non-TNBC (ER/PR and HER2-positive) and (E) HER2-positive (ER/PR negative) breast cancer types. The expression profile of each sample is represented as a single circle, and the FPKM of the individual transcripts are depicted as peaks. The order of the transcript expression profile samples is from the inner circle to the outside, as depicted by the direction of the arrow and the labels. The total number of transcripts (above FPKM 0.01) in each sample is provided in brackets next to the sample label. The abundance of an individual transcript is depicted as a peak. The expression of transcripts in several genomic loci appears similar; however, individual variations are evident at specific loci within each group. (B) The relative transcript abundance, calculated from the commonly (only transcripts expressed in all 17 samples) expressed transcripts in the three groups, shows that TNBC expressed a higher abundance of transcripts on chromosome 6. (C) PCA plots showing the clustering of the TNBC (magenta), Non-TNBC (Red) and HER2-positive (green) breast cancer samples based on the transcriptomic expression profiles. (D) The heat map of the pairwise correlation between all of the samples based on the Spearman correlation coefficient, which ranks and quantifies the degree of similarity between each pair of samples.
The overall abundances (FPKM) of commonly expressed transcripts on each chromosome were compared between the three groups. The TNBC group exhibited higher transcript expression levels on chromosome 6 compared with the Non-TNBC and HER2-positive breast cancer types (Figure 2B, Supplementary Figure 9). Frequent gains in chromosome 6p have been shown to be associated with poor prognosis in several cancers44, including invasive ductal carcinoma45,46. In general, the transcript abundance was higher in TNBC than in the other two groups. However, the overall number of transcripts and their expression profile in all three cancers followed a similar trend (Supplementary Figure 2).
Clustering based on transcript expression profile
We examined the correlation between the samples based on transcript and gene expression using Principle Component Analysis. Similar to initial immunohistochemistry and q-PCR classification, most of the samples from each breast cancer group clustered together in our analysis, except for a few outliers (sample A3 in TNBC, sample B4 in Non-TNBC, and samples C1 and C5 in HER2 group) (Figure 2C). To further explore the pairwise relationship, we calculated the Spearman's correlation, which ranks and quantifies the degree of similarity between each pair of samples (Supplementary Table 6). The results indicated variation between individual samples but no distinct outliers (Figure 2D). However, the transcript expression-based correlation showed more variation within the groups than the gene expression-based correlation (Supplementary Table 6 and Supplementary Figure 10A). In agreement with the PCA analysis, the average linkage distance matrix analyses and pairwise cuffdiff analysis also highlighted three distinct groups and similar outliers, i.e., A3, B4 (and also B6) and C5 (Supplementary Table 7, Supplementary Figure 10B and 11, Supplementary Method43). Together, these analyses illustrate the variation in individual transcript expression levels, which is possibly due to a large number of low abundance transcripts in all of the samples, a phenomenon generally observed in RNA sequencing data studies47.
Defining the transcriptomic signatures of TNBC, Non-TNBC and HER2-positive breast cancer
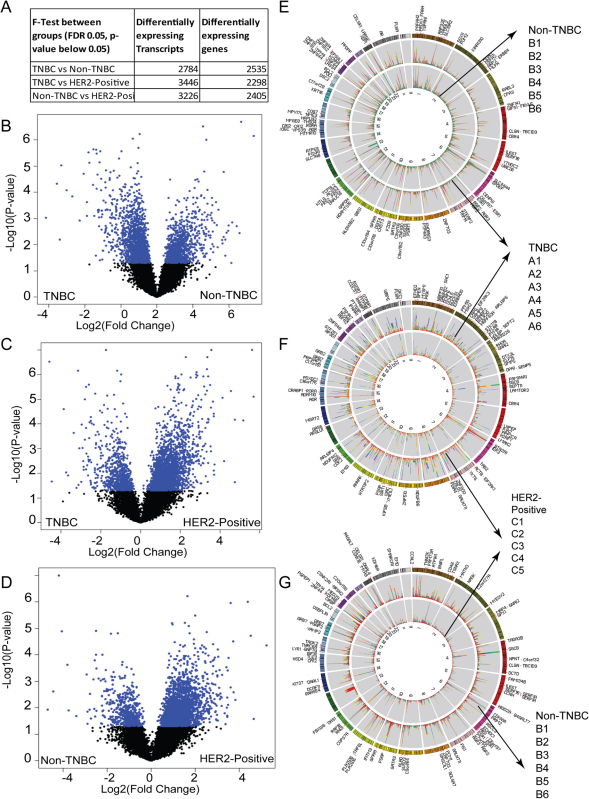
To determine the differentially expressed genes and transcripts between the three breast cancer groups, univariate F-tests were performed (Supplementary Method, Figures 3, 3A and, Supplementary Figures 12 to 15). We identified 2617 transcripts that were differentially expressed between the TNBC and Non-TNBC groups (Figure 3B) (Supporting Files 1 and 2). Among the identified transcripts, 962 transcripts exhibited higher abundance (upregulated) in the TNBC group and 1655 manifested lower relative abundance (downregulated) in the non-TNBC group. When the TNBC group was compared with the HER2-positive group (Figure 3C), 3087 transcripts were identified as differentially expressed (Supporting Files 3 and 4). Among these transcripts, 2416 were upregulated in TNBC. The Non-TNBC and the HER2-positive group comparison revealed 2971 transcripts (Figure 3C, Supporting Files 5 and 6) that were differentially expressed, and 2509 transcripts were upregulated in Non-TNBC. The comparative analyses of these three breast cancer subtypes at the isoform and gene level (Supplementary Figure 15, Supporting Files 7 to 9) provide a complete view of the global transcriptomic changes between the three groups. The distribution and abundance of the top one hundred (determined by p value) differentially expressed transcripts from the aforementioned three comparisons are shown as stacked histograms using Circos plots48 (Figures 3D, 3E and 3F).
Figure 3. Differential transcript expression between TNBC, Non-TNBC and HER2-positive cancers.
(A) The number of statistically significant differentially expressed transcripts identified from the F-test. Volcano plots show the differential expression of the statistically significant transcripts (p value less than 0.05 and FDR 0.05) between (B) TNBC vs. Non-TNBC (C) TNBC vs. HER2-positive and (D) Non-TNBC vs. HER2 positive pairwise comparisons. The Circos plots show statistically significant differences in transcript expression from the above univariate F-test between (E) TNBC vs. Non-TNBC (F) TNBC vs. HER2-positive and (G) Non-TNBC vs. HER2- positive breast cancers. The top hundred transcripts as determined by the p-value are labelled on the Circos plot. The stacked histograms represent the abundance (FPKM) associated with each sample for that specific differentially expressed transcript. The TNBC group samples A1 to A6 are coloured in red, orange, yellow, green blue and purple. The Non-TNBC group samples B1 to B6 are coloured in red, blue, green, purple, orange, yellow and the HER2-positive group samples C1 to C6 are coloured in green, orange, blue, magenta, sea green, and yellow.
Furthermore, the differential transcript expressions that occurred in more than one comparison were separated from the “bona-fide” transcripts that were specific to one comparison, i.e., the transcripts specific for TNBC vs. Non-TNBC (1000 transcripts, 945 genes), TNBC vs. HER2-positive (1316 transcripts, 885 genes) and Non-TNBC vs. HER2-positive (1011 transcripts, 763 genes) (Supporting Files 10–12). Interestingly, 78 transcripts, including oncogenic Ras family member, RAB21, and TMEM219, were expressed differentially in all three comparisons and consequently qualified to be common breast cancer modulators (Supporting File 13).
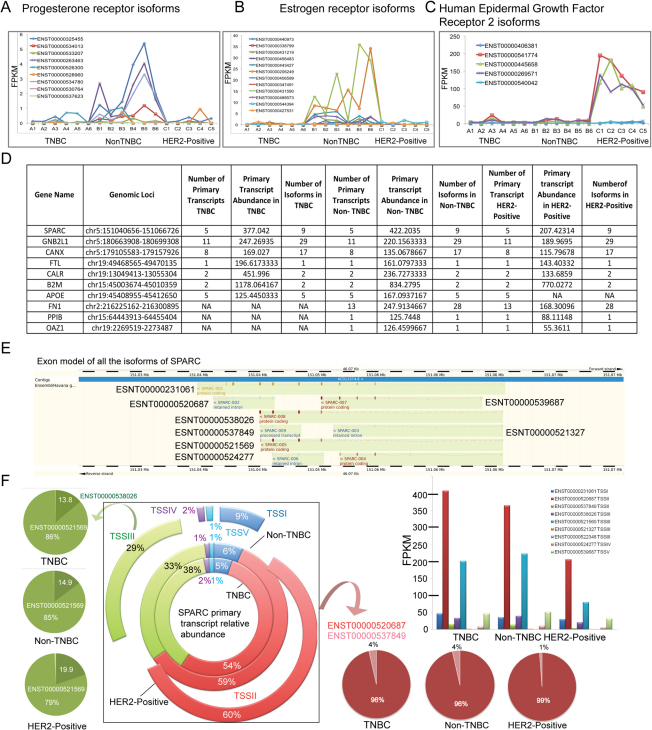
We next compared the new transcript atlases between the three groups with the previously reported breast cancer-associated targets derived from Online Mendelian Inheritance in Man (OMIM) database. About 3 to 6% of the known targets were represented in our dataset, highlighting new isoforms that were identified in this study (Supporting Excel File 14). Notably, the reduced expression of all three receptors in the TNBC group and high abundance of the three isoforms in the HER2 group were observed in all five samples of HER2-positive cancer samples (Figures 4A, 4B and 4C). These observations indicate the accuracy of the mRNA sequencing-based transcript reconstruction and abundance calculation.
Figure 4. The top five highly abundant primary transcripts are common in all three breast cancers.
The transcript expression profiles of all expressed isoforms of (A) the Progesterone receptor, (B) the Oestrogen receptor and (C) the Human epidermal growth factor receptor 2 in all 17 samples. (D) The table presents the six most common highly abundant primary transcripts and all of the associated information derived from the cuffdiff and cufflinks analyses. The bottom four lines of the table show the primary transcript expression profiles specific for the TNBC and Non-TNBC (APOE) and HER2-positive (FN1, PP1B and OAZ1) groups. However, the primary transcript abundance of FN1, PP1B and OAZ1 indicates that they are among the ten most highly expressed primary transcripts within each group. (E) The exon model of all of the nine isoforms that belong to SPARC. The exons are shown as coloured blocks, and the introns are shown as dotted lines. (F) The broken pie chart shown in the middle represents the relative abundance of the SPARC primary transcript groups in the TNBC (inner circle), Non-TNBC (the middle circle) and HER2-positive (the outer circle) breast cancer groups. The five commonly expressed SPARC primary transcripts are labelled as TSS1 to TSS5. Their relative abundances are represented by different colours, and the relative expression levels as percentages are indicated on the circle. The expression of specific isoforms and the changes in abundance are indicated for primary transcript groups II and III. The lime (on the top left) and salmon (bottom right) coloured arrows point to the isoforms that originate from the TSSII and TSSIII primary transcripts, respectively. The relative abundances of isoforms that belong to TSSII and TSSIII in TNBC, Non-TNBC and HER2-positive cancers are presented as pie chart in shades of colours similar to their primary transcripts. The bar chart shows the abundance of the all of the SPARC isoforms estimated by cufflinks.
Common transcriptional regulatory elements in TNBC, Non-TNBC and HER2-positive breast cancer
Several critical regulatory elements, such as splicing, and different promoter usage have been established to be the major components that contribute to transcriptional dynamics49. From the assembled transcripts and their abundances, the transcriptional and post-transcriptional dynamics of the genes can be deciphered. The difference in the expression of various isoforms of a given gene reflects the post-transcriptional regulation, whereas the relative abundances of the primary transcripts and the genomic loci comprising the highest number of primary transcripts indicate transcriptional regulatory elements. The comprehensive transcriptome analysis using cuffdiff allowed us to estimate the number of primary transcripts in each breast cancer group. The primary transcript group in this study was defined as the group of transcripts that share the same Transcription Start Site (TSS). Typically, each TSS group comprised several specific isoforms that originated from a particular TSS. The TSS group relative abundances were estimated from the sum of the FPKMs of all of the isoforms belonging to an individual primary transcript. We first investigated the common high confidence primary transcripts and their abundances and associated genes in each breast cancer group (Supporting Files 19 to 21).
Surprisingly, the six most abundant primary transcripts were common among all three breast cancer groups (Figure 4D). These primary transcript pools appear highly active in generating abundant isoforms in breast cancer. These transcripts encode genes including secreted protein acidic and rich in cysteine (osteonectin, SPARC), guanine nucleotide binding protein beta polypeptide 2-like 1 (RACK1 or GNB2L), calnexin (CANX), ferritin L subunit (FTL), calreticulin (CALR) and beta-2 microglobulin (B2M). In each primary transcript group, only selected isoforms were preferentially expressed at high abundance, possibly because of their functional role. The top six primary transcripts have been reported to be critical players in breast cancer because of their roles in extracellular matrix remodelling and cell motility. For instance, the cell surface proteins SPARC, RACK1, B2M, and FTL are well associated with breast cancer50,51,52,53. In addition, B2M, FTL, and RACK1 are reported to be tumour prognostic markers52,53,54. Calnexin and calreticulin, which promote the correct folding of proteins that enter the secretory pathway, have also been linked to the breast cancer metastatic phenotype55. More interestingly, the highly abundant primary transcripts identified in normal breast tissue (DNAJB1, SCGB2A2, MUCL1, SCGB1D2 and ACTG1) were different from those in the three breast cancer groups (Supporting File 22). Thus, our study identifies a primary transcript group that is breast cancer-specific and could therefore provide potential common markers upon validation in a larger sample set.
Furthermore, the variation in abundances of isoforms originating from the same primary transcript is exemplified by the primary SPARC transcripts. All of the eight known SPARC isoforms that were expressed in the breast cancer samples (Figure 4E) originate from five different primary transcripts (Figure 4F shown as TSSI to TSSV). Interestingly, the highly abundant transcript from the primary transcript group I of SPARC, ENST00000520687, is defined as a non-expressed protein in the hg19 reference genome, but the abundance of that specific isoform was high in our samples of all three breast cancer types (Figures 4E and 4F). In primary transcript group III, the SPARC splice variant 5 (ENST00000521569) expresses 79% to 86% whereas SPARC splice variant 8 (ENST00000538026) expresses 13.8 to 20% out of total SPARC gene expression in all three breast cancers. These results reveal that the ENST00000521569 isoform of SPARC is most highly expressed in all three breast cancers.
Furthermore, some highly abundant primary transcript pools were unique to each of the breast cancer subtypes (Figure 4D). For example, the primary transcript abundance of apolipoprotein E (APOE) was high in the TNBC and Non-TNBC groups but not in the HER2-positive group. In the case of the HER2-positive group, primary transcripts for protein phosphatase 1B (PP1B) and ornithine decarboxylase antizyme 1 (OAZ1) were highly abundant and unique to this group compared with the other two groups. From this primary transcript analysis, we could unravel the transcriptional dynamics to the finer details, i.e., the expression and significance of the specific primary transcript pools and the precise isoforms that are abundant in specific breast cancer type.
“Genomic Hotspots”: the genomic loci enriched in primary transcripts are identical in all three groups
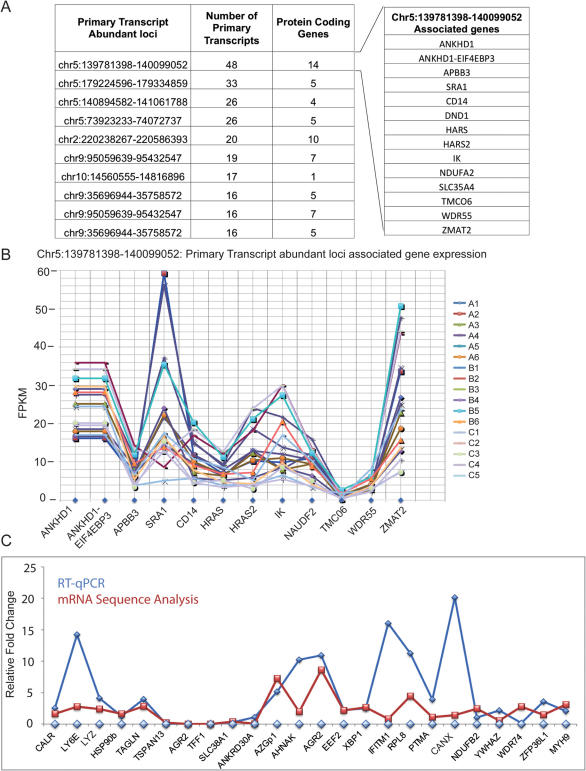
Next, we searched for the genomic loci that are enriched in primary transcripts, i.e. the “genomic hotspots”, in each type of breast cancer analysed in this study. These genomic regions containing highly abundant primary transcripts could steer the cellular machinery towards oncogenic processes through predominant splicing. Surprisingly, the top eight “genomic hotspots” that produced the highest number of primary transcripts are common in TNBC, Non-TNBC and HER2-positive breast cancers. They are presented in Figure 5A, along with the number of protein-coding genes that are encoded from one of these loci (Supporting Files 23, 24 and 25 comprising the top 20 genomic hotspots). For example, the genomic locus that has the highest primary transcript enrichment, Chr5: 139781398–140099052, encodes 12 genes (Figure 5B), including HRAS, HRAS2, SLC35A4 and NDUFA2. The expression levels of these genes, calculated as the sum of all of the isoforms associated with the genes in all 17 samples, are presented in Figure 5C. These findings clearly illustrate that these loci are highly transcriptionally active and participate in robust splicing to produce several isoforms. These loci are strikingly different from the normal breast tissue (Supporting File 26).
Figure 5. The “genomic hotspots”, the highly spliced loci in all three cancers are conserved, and experimental validation by RT-qPCR confirms the accuracy of the analysis.
(A) The genomic loci that comprise the highest numbers of primary transcripts in TNBC, Non-TNBC and HER2-positive breast cancers, along with the number of primary transcripts identified from these loci and the number of genes encoded in all seventeen samples, are presented in a table. The separate panel shows the genes associated with genomic loci chr5:139781398–140099052, which encodes the largest number of genes in all three breast cancers. (B) The abundance of genes (FPKM) that belong to genomic loci chr5:139781398–140099052 was estimated by cufflinks in all 17 samples. (C) A comparison of the RT-qPCR and RNA sequence expression analysis. The isoforms differentially expressed at statistically significant levels were selected randomly from the TNBC vs. Non-TNBC, TNBC vs. HER2-positive and Non-TNBC vs. HER2-positive pairwise comparisons. RT-qPCR of the isoforms in all of the samples from the pairwise comparisons was performed. The average fold change was calculated from the mean of the experimentally calculated mRNA levels of the isoform within one group divided by the levels in the other. Supporting Document S23 presents the individual RT-qPCR validation bar chart for each isoform in all of the tested samples. For the RNA sequencing analysis, the average fold change was calculated from the mean FPKM of the isoform in all samples in one group divided by the mean FPKM of the other.
Novel intronic predominance highlights common new transcripts in TNBC, Non-TNBC and HER2-positive breast cancer
Apart from the known transcripts, we also investigated the significance of intronic reads found in all three types of breast cancer. We compared the read distribution in publicly available single normal tissue against all 17 breast cancer samples. This comparison allowed us to investigate whether the phenomena were found in the breast tissue or specific to the cancer samples. In the normal breast tissue sample, 80% of the reads mapped to exons while 16% and 0.1% of reads corresponded to introns and intergenic regions, respectively. These results indicate that the intronic predominance was specific to the cancer samples, possibly because of intron retention and splicing modifications. Further, close inspection of this phenomenon revealed the expression of intronic regions in several annotated transcripts. The normal breast tissue comprises 10,337 defined exons in the place of introns in known transcripts (high confident transcripts with FPKM above 0.1). In contrast, an average of 96,215 exons, i.e., ∼10-fold more novel exons, were found within introns in the cancer tissue samples (Supplementary Table 8). Furthermore, the number of annotated transcripts comprising these exons in normal tissue was 1211. In contrast, an average of 15,142 known transcripts were found to encompass exons in the place of introns in cancer tissues. When we compared these novel transcripts against the EST (Expressed Sequence Tags) database, we found that about 93–97% of the transcripts mapped to similar locations as reported for ESTs. Therefore, these exons may be expressed in the three breast cancer types. Moreover, within each breast cancer group, there were a few hundred common transcripts (Supplementary Table 8 and Supporting Files 15 to 17) that underwent such changes as well as 78 common transcripts in all three breast cancer groups (Supporting File 18). Given these described predominant modifications in several transcripts, it is not surprising that we found a high intronic read count in these samples when compared with hg19.
Novel intergenic unannotated transcripts in the breast cancer transcriptome
In addition to introns, the intergenic region also appears to comprise an average of 24,540 exons that could assemble into high confidence, previously unknown transcripts in all of the breast cancer samples. Several (average 723) unannotated transcripts (with FPKM above 0.1) could be assembled from reads that were recovered from the intergenic region of each sample, which indicates the existence of possible new transcripts in breast cancer conditions (Supplementary Table 8). Notably, some transcripts comprised more than one exon, but the singletons were the predominantly identified transcripts (Supplementary Table 9). A similar phenomenon was found in normal breast tissue. Therefore, these previously unknown transcriptional changes identified in this study reveal several new elements that occur specifically in breast cancer. However, future mRNA sequencing of several normal breast and other tissue samples are needed to establish the breast tissue specificity of the alterations of these newly discovered transcripts.
Identifying common critical pathways in breast cancer
The pathway analysis was performed using the identified differentially expressed transcripts obtained from the TNBC vs. Non-TNBC, TNBC vs. HER2-positive and Non-TNBC vs. HER2-positive comparisons. In all three pairwise comparisons, cell motility, development, apoptosis and cancer-related signalling pathways are influenced, that highlights the common critical functions that are “hijacked” in breast cancer (Supplementary Figures 16 to 21). Moreover, the differentially expressed transcripts identified in the TNBC vs. Non-TNBC comparison are also involved in small GTPase-mediated signal transduction, proteosomal ubiquitin-dependent protein catabolism, negative regulation of apoptosis, cytoskeleton protein binding and oxidoreductase activity (Supplementary Figures 16 and 17). The transcripts that were differentially expressed in the Non-TNBC vs. HER2-positive comparison appear to affect the regulation of mammary gland and epithelial proliferation, apoptotic mitochondrial changes, mitotic cell cycle, cell division and enzymatic activity (Supplementary Figure 18). Furthermore, canonical pathways, such as oncostatin M signalling, PI3K/MAPK signalling and functional pathways involving cancer, cell cycle and cell death, are influenced by all three groups of differentially expressed transcripts (TNBC vs. Non-TNBC, TNBC vs. HER2-positive and Non-TNBC vs. HER2-positive) (Supplementary Figures 19, 20 and 21). In contrast, oestrogen receptor signalling, cellular growth and modification are specifically modulated by transcripts that arise from the Non-TNBC vs. HER2-positive differential expression comparison (Supplementary Figure 21). In summary, this analysis highlights the prevalence of breast cancer signalling-associated pathways that are commonly influenced in all three subtypes analysed here.
qRT-PCR validation of the mRNA sequencing analysis
The mRNA sequence analysis was validated by experimental confirmation of the levels of differentially expressed transcripts in TNBC, Non-TNBC and HER2-positive samples. We used qRT-PCR to validate the expression levels of 28 transcripts (randomly selected across all the three bins shown in Supplementary Figures 3 to 8). The transcripts were selected from pairwise differential expression comparisons of all of the samples of the relevant group (Supplementary Methods). We observed a high correlation and a similar expression trend between the mRNA-sequence based abundance estimation (FPKM) and qRT-PCR assays revealing the accuracy of the analysis (Figure 5C and Supporting File with primers 27, Supporting File 28 shows individual RT qPCR data of all the transcripts that were validated).
Discussion
For the first time, we report the transcriptional and post-transcriptional profiling of TNBC, Non-TNBC and HER2-positive breast cancers using a deep mRNA sequencing approach (35, 36). In breast cancer, microarrays have been extensively used for molecular subtyping and identification of breast cancer-specific gene signatures. Although mRNA sequencing covers the total transcript expression compared with the preselected microarray approach, which focuses on specific gene sets, we compared our mRNA sequencing-based differentially expressed transcripts against the previously reported gene signatures of TNBC, Non-TNBC and HER2-positive breast cancer. When the signature gene lists from Perou et al., 200019, Sorlie et al., 200120,56, Hu et al., 200657 and Parker et al., 200926 were compared, only few overlapping genes were identified (Supplementary Method and Supplementary Table 12), because these studies classify the samples on the basis of five intrinsic subtypes and identify genes that are modulated in all the five subtypes from a preselected gene set. In contrast, our mRNA sequencing captures all the expressed transcripts which are ∼75,000 transcripts per sample compared to the preselected gene sets of microarray platform. Moreover, this study classifies the samples on the basis of receptor status. Therefore the differentially expressing transcripts from this study will be different to the previously identified gene signatures of microarray. However, we performed parallel comparative microarray analyses using publicly available datasets for which the receptor status can be derived (Supplementary Method and Supplementary Table 10, 11 and Supplementary Figure 22). The results (Supplementary Table 11 and Supporting file 29 to 31) show several genes overlapping and 90 to 100% agreement in the expression trend of genes that were identified in common in the abovementioned pairwise comparisons (e.g., TNBC vs. Non-TNBC) in the microarray and mRNA sequencing. Although mRNA sequencing identifies previously reported breast cancer modulators as well as several new players, further many larger scale mRNA sequencing studies will be needed to draw firm conclusions on mRNA-seq based gene signatures.
Compared with the microarray approach, the emerging mRNA sequencing-centred technology41,58,59 allows us to de-convolute the transcriptional and post-transcriptional elements of breast cancer at a higher resolution. The fundamental theme that emerges from this detailed mRNA-Seq analysis is the similarity observed at the transcriptional level (i.e., primary transcripts and abundantly splicing genomic hotspots), which differs from the heterogeneity in post-transcriptional isoform levels. This heterogeneity appears to be mainly due to post-transcriptional regulation that fine-tunes the splicing and abundances of various isoforms specific for each cancer. This notion is supported by the identified primary transcript pool of SPARC, GNB2L1, CANX, CALR and B2M, which have been proven to be significant players in breast cancer onset and progression by modulating cell migration, motility, anti-apoptosis and cellular stress management50,60,61,62. In fact, two of the highly abundant primary transcripts, B2M and FTL, have also been confirmed to be markers of cancer progression and thus raise new therapeutic possibilities that could arise from this study. To achieve the final frontier of using the transcriptomic data in clinical settings, it will also be important to design a prospective analysis involving a larger series of patient cohorts.
Methods
Human Patient Samples
Dr. Suzanne Fuqua (Baylor College of Medicine) provided the human breast cancer tissue RNA samples. All of the human samples were used in accordance with the IRB procedures of Baylor College of Medicine. The breast tumour types, TNBC, Non-TNBC and HER2-positive, were classified on the basis of immunohistochemical and RT-qPCR classification (data not shown).
Illumina Genome sequencing RNA sequencing library preparation
Enrichment of rRNA-free transcriptome RNA: Whole transcriptome RNA was extracted from total RNA by removing large and small ribosomal RNA (rRNA) using RiboMinus Eukaryote Kit (Invitrogen, Carlsbad, CA). Five micrograms of total RNA was hybridised to rRNA-specific biotin labelled probes at 70°C for 5 minutes. The rRNA-probe complexes were then removed by streptavidin-coated magnetic beads. The rRNA-free transcriptome RNA was concentrated by ethanol precipitation.
cDNA synthesis and DNA library construction from transcriptome RNA
cDNA was synthesised from the isolated RNAas described in the supplementary method. Double-stranded cDNA was treated with a mix of T4 DNA polymerase, Klenow large fragment and T4 polynucleotide kinase to create blunt-ended DNA, to which a single A base was subsequently added at the 3′ end using Klenow fragment (3′ to 5′ exo-) and dATP. The A-tailed DNA was ligated with paired end adaptors using T4 DNA ligase, provided by the Illumina RNA-Seq kit (Illumina, San Diego, CA). Size selection (200 base pair) of the adaptor ligated DNA was performed by cutting the target fragment out of a 4–12% acrylamide gel. The amplified DNA library containing ideal fragment sizes was obtained by in-gel PCR using the Phusion High-Fidelity system (New England Biolabs). Each library that was prepared was sequenced, and image analysis and base calling were performed with Illumina pipeline version 1.3.2.
Read alignment and transcript assembly
We have aligned the pair end reads using TopHat version 1.1.442, allowing two mismatches in the alignment. The aligned reads were assembled into transcripts using cufflinks43. The alignment quality and distribution of the reads were estimated using SAMtools63, and the PCA plot and sample clustering were performed using (imported genes and transcript expression profiles generated from cufflinks assembled transcripts that were identical to the reference) the Avadis NGS program.
The cuffcompare program was used to identify the transcripts that were identical to the reference human genome for further analysis. To examine differential expression of genes/transcripts between the three groups of samples belonging to the three different subtypes of breast cancer, we first performed cuffcompare and cuffdiff43.
The high confidence transcripts associated with FPKMs above 0.01 in all of the samples were isolated. Next, we calculated the average expression of an individual gene/transcript from the FPKMs obtained using cufflinks. The samples were arranged in groups without preference to the order within the group. The geometric averages of the FPKMs were calculated for each group of samples. The values were not calculated using a reference. Next, univariate F-tests were performed (after log2 conversion of the transcript expression abundance, FPKM) between the three groups, using a p value cut off of 0.05 a false discovery rate below 0.05, with statistical package R.
Author Contributions
RK directed all aspects of the breast cancer transcriptome project. JE and RK designed the experiments and wrote the main manuscript text. JE, DC, PM and SG designed experiments, generated results and analyzed the data. SDR and SBP performed RT-qPCR validation experiments and provided reagents. SAWF provided reagents and LF and SSN provided technical assistance for the data analyses.
Supplementary Material
Supplementry Tables and Figures
Supporting Information
Supplementary_Datafiles_excelsheets
Acknowledgments
We acknowledge Krishna S Ghanta for technical assistance and Dr. Andriy Morgun for fruitful discussions and critical reading of the manuscript.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T. & Rosen J. M. Modelling breast cancer: one size does not fit all. Nat Rev Cancer 7, 659–672 (2007). [DOI] [PubMed] [Google Scholar]
- Reis-Filho J. S. & Lakhani S. R. Breast cancer special types: why bother? J Pathol 216, 394–398 (2008). [DOI] [PubMed] [Google Scholar]
- Geyer F. C., Marchio C. & Reis-Filho J. S. The role of molecular analysis in breast cancer. Pathology 41, 77–88 (2009). [DOI] [PubMed] [Google Scholar]
- Weigelt B. & Reis-Filho J. S. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol 6, 718–730 (2009). [DOI] [PubMed] [Google Scholar]
- Geyer F. C., Lopez-Garcia M. A., Lambros M. B. & Reis-Filho J. S. Genetic characterization of breast cancer and implications for clinical management. J Cell Mol Med 13, 4090–4103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger H. et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 189, 521–526 (1999). [DOI] [PubMed] [Google Scholar]
- Buerger H. et al. Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol 194, 165–170 (2001). [DOI] [PubMed] [Google Scholar]
- Stingl J. & Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer 7, 791–799 (2007). [DOI] [PubMed] [Google Scholar]
- Brenton J. D., Carey L. A., Ahmed A. A. & Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 23, 7350–7360 (2005). [DOI] [PubMed] [Google Scholar]
- Sjoblom T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006). [DOI] [PubMed] [Google Scholar]
- Oakman C., Santarpia L. & Di Leo A. Breast cancer assessment tools and optimizing adjuvant therapy. Nat Rev Clin Oncol 7, 725–732 (2010). [DOI] [PubMed] [Google Scholar]
- Andre F. & Pusztai L. Heterogeneity of breast cancer among patients and implications for patient selection for adjuvant chemotherapy. Pharm Res 23, 1951–1958 (2006). [DOI] [PubMed] [Google Scholar]
- Elston C. W., Ellis I. O. & Pinder S. E. Pathological prognostic factors in breast cancer. Crit Rev Oncol Hematol 31, 209–223 (1999). [DOI] [PubMed] [Google Scholar]
- Soerjomataram I., Louwman M. W., Ribot J. G., Roukema J. A. & Coebergh J. W. An overview of prognostic factors for long-term survivors of breast cancer. Breast Cancer Res Treat 107, 309–330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S. J., Provenzano E. & Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer 45 Suppl 1, 27–40 (2009). [DOI] [PubMed] [Google Scholar]
- Rakha E. A. et al. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol 26, 3153–3158 (2008). [DOI] [PubMed] [Google Scholar]
- Weigelt B., Geyer F. C. & Reis-Filho J. S. Histological types of breast cancer: how special are they? Mol Oncol 4, 192–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- Sorlie T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98, 10869–10874 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T. et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100, 8418–8423 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha E. A., Reis-Filho J. S. & Ellis I. O. Basal-like breast cancer: a critical review. J Clin Oncol 26, 2568–2581 (2008). [DOI] [PubMed] [Google Scholar]
- van de Vijver M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- van 't Veer L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002). [DOI] [PubMed] [Google Scholar]
- van 't Veer L. J. et al. Expression profiling predicts outcome in breast cancer. Breast Cancer Res 5, 57–58 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005). [DOI] [PubMed] [Google Scholar]
- Naderi A. et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26, 1507–1516 (2007). [DOI] [PubMed] [Google Scholar]
- Sgroi D. C. Preinvasive breast cancer. Annu Rev Pathol 5, 193–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B., Baehner F. L. & Reis-Filho J. S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol 220, 263–280 (2010). [DOI] [PubMed] [Google Scholar]
- Pusztai L., Mazouni C., Anderson K., Wu Y. & Symmans W. F. Molecular classification of breast cancer: limitations and potential. Oncologist 11, 868–877 (2006). [DOI] [PubMed] [Google Scholar]
- Bauer K. R., Brown M., Cress R. D., Parise C. A. & Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109, 1721–1728 (2007). [DOI] [PubMed] [Google Scholar]
- Carey L. A. et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13, 2329–2334 (2007). [DOI] [PubMed] [Google Scholar]
- Foulkes W. D., Smith I. E. & Reis-Filho J. S. Triple-negative breast cancer. N Engl J Med 363, 1938–1948 (2010). [DOI] [PubMed] [Google Scholar]
- Finnegan T. J. & Carey L. A. Gene-expression analysis and the basal-like breast cancer subtype. Future Oncol 3, 55–63 (2007). [DOI] [PubMed] [Google Scholar]
- Dent R. et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13, 4429–4434 (2007). [DOI] [PubMed] [Google Scholar]
- Gusterson B. A. Identification and interpretation of epidermal growth factor and c-erbB-2 overexpression. Eur J Cancer 28, 263–267 (1992). [DOI] [PubMed] [Google Scholar]
- Gusterson B. A. et al. Prognostic importance of c-erbB-2 expression in breast cancer. International (Ludwig) Breast Cancer Study Group. J Clin Oncol 10, 1049–1056 (1992). [DOI] [PubMed] [Google Scholar]
- Badve S. et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol 24, 157–167 (2011). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Haas B. J. & Zody M. C. Advancing RNA-Seq analysis. Nat Biotechnol 28, 421–423 (2010). [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos G. C., Zielenska M., Prasad M. & Squire J. A. Chromosome 6p amplification and cancer progression. J Clin Pathol 60, 1–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard F. et al. Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer 89, 305–310 (2000). [PubMed] [Google Scholar]
- Seute A. et al. Clinical relevance of genomic aberrations in homogeneously treated high-risk stage II/III breast cancer patients. Int J Cancer 93, 80–84 (2001). [DOI] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D. D. & Darnell R. B. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11, 75–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins G., Douglas-Jones A., Bryce R., Mansel R. E. & Jiang W. G. Increased levels of SPARC (osteonectin) in human breast cancer tissues and its association with clinical outcomes. Prostaglandins Leukot Essent Fatty Acids 72, 267–272 (2005). [DOI] [PubMed] [Google Scholar]
- Morabito A. et al. Analysis and clinical relevance of human leukocyte antigen class I, heavy chain, and beta2-microglobulin downregulation in breast cancer. Hum Immunol 70, 492–495 (2009). [DOI] [PubMed] [Google Scholar]
- Ricolleau G. et al. Surface-enhanced laser desorption/ionization time of flight mass spectrometry protein profiling identifies ubiquitin and ferritin light chain as prognostic biomarkers in node-negative breast cancer tumors. Proteomics 6, 1963–1975 (2006). [DOI] [PubMed] [Google Scholar]
- Cao X. X. et al. RACK1: A superior independent predictor for poor clinical outcome in breast cancer. Int J Cancer 127, 1172–1179 (2010). [DOI] [PubMed] [Google Scholar]
- Moore L. E. et al. Proteomic biomarkers in combination with CA 125 for detection of epithelial ovarian cancer using prediagnostic serum samples from the prostate, lung, colorectal, and ovarian (PLCO) cancer screening trial. Cancer (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. B. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci 119, 615–623 (2006). [DOI] [PubMed] [Google Scholar]
- Sorlie T. et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics 7, 127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7, 96 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Grabherr M. G., Guttman M. & Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8, 469–477 (2011). [DOI] [PubMed] [Google Scholar]
- Ozsolak F. & Milos P. M. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12, 87–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins G., Martin T. A., Bryce R., Mansel R. E. & Jiang W. G. Gamma-Linolenic acid regulates the expression and secretion of SPARC in human cancer cells. Prostaglandins Leukot Essent Fatty Acids 72, 273–278 (2005). [DOI] [PubMed] [Google Scholar]
- Cao X. X. et al. RACK1 promotes breast carcinoma migration/metastasis via activation of the RhoA/Rho kinase pathway. Breast Cancer Res Treat 126, 555–563 (2011). [DOI] [PubMed] [Google Scholar]
- Cao X. X. et al. RACK1 promotes breast carcinoma proliferation and invasion/metastasis in vitro and in vivo. Breast Cancer Res Treat 123, 375–386 (2010). [DOI] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementry Tables and Figures
Supporting Information
Supplementary_Datafiles_excelsheets