Abstract
A DNA region of 2750 base pairs encompassing the genes III, VI and I of bacteriophage M13 has been sequenced by the Maxam-Gilbert procedure. By establishing the nucleotide changes introduced by several amber mutations, the coding region and the regulatory signals of each gene have been deduced. The genes appear to span 1275 base pairs (gene III; mol.wt. 44,748) 339 base pairs (gene VI; mol.wt. 12,264) and 1047 base pairs (gene I; mol.wt. 39,500). Their separating non-codogenic regions are extremely short, namely two and one base pair, respectively. The C-terminal end of gene I, however, intrudes 23 nucleotides into gene IV. From the nucleotide sequence it appears that the minor capsid protein of the phage, which is encoded by gene III, is synthesized in a precursor form containing 18 extra amino acids at its N-terminal end. Furthermore, in this capsid protein two clusters of a fourfold repeat of the sequence Glu-Gly-Gly-Gly-Ser are apparent. Gene VI appears to code for a small, extremely hydrophobic polypeptide. Its total hydrophobic amino acids content of 51% suggests that this protein can only function in the host cell membrane.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M., Blackburn E. H., Coulson A. R., Galibert F., Sanger F., Sedat J. W., Ziff E. B. Gene F of bacteriophage phiX174. Correlation of nucleotide sequences from the DNA and amino acid sequences from the gene product. J Mol Biol. 1976 Nov 15;107(4):445–458. doi: 10.1016/s0022-2836(76)80077-0. [DOI] [PubMed] [Google Scholar]
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T. S., Model P., Zinder N. D. In vitro protein synthesis directed by separated transcripts of bacteriophage f1 DNA. J Mol Biol. 1975 Dec 15;99(3):369–382. doi: 10.1016/s0022-2836(75)80132-x. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. The single-stranded DNA phages. CRC Crit Rev Microbiol. 1975 Dec;4(2):161–223. doi: 10.3109/10408417509111575. [DOI] [PubMed] [Google Scholar]
- Edens L., Konings R. N., Schoenmakers J. G. A cascade mechanism of transcription in bacteriophage M13 DNA. Virology. 1978 May 15;86(2):354–367. doi: 10.1016/0042-6822(78)90076-4. [DOI] [PubMed] [Google Scholar]
- Edens L., Konings R. N., Schoenmakers J. G. Physical mapping of the central terminator for transcription on the bacteriophage M13 genome. Nucleic Acids Res. 1975 Oct;2(10):1811–1820. doi: 10.1093/nar/2.10.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens L., Konings R. N., Schoenmakers J. G. Transcription of bacteriophage M13 DNA: existence of promoters directly preceding genes III, VI, and I. J Virol. 1978 Dec;28(3):835–842. doi: 10.1128/jvi.28.3.835-842.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens L., van Wezenbeek P., Konings N. H., Schoenmakers J. G. Mapping of promoter sites on the genome of bacteriophage M13. Eur J Biochem. 1976 Nov 15;70(2):577–587. doi: 10.1111/j.1432-1033.1976.tb11049.x. [DOI] [PubMed] [Google Scholar]
- Geider K., Beck E., Schaller H. An RNA transcribed from DNA at the origin of phage fd single strand to replicative form conversion. Proc Natl Acad Sci U S A. 1978 Feb;75(2):645–649. doi: 10.1073/pnas.75.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
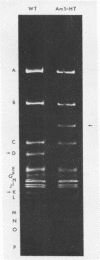
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M. E., Konigsberg W. H. Adsorption protein of the bacteriophage fd: isolation, molecular properties, and location in the virus. Biochemistry. 1977 Jun 14;16(12):2686–2694. doi: 10.1021/bi00631a016. [DOI] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Gray C. P., Sommer R., Polke C., Beck E., Schaller H. Structure of the orgin of DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1978 Jan;75(1):50–53. doi: 10.1073/pnas.75.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondel C. A., Konings R. N., Schoenmakers J. G. Regulation of gene activity in bacteriophage M13 DNA: Coupled transcription and translation of purified genes and gene-fragments. Virology. 1975 Oct;67(2):487–497. doi: 10.1016/0042-6822(75)90449-3. [DOI] [PubMed] [Google Scholar]
- Hulsebos T., Schoenmakers J. G. Nucleotide sequence of gene VII and of a hypothetical gene (IX) in bacteriophage M13. Nucleic Acids Res. 1978 Dec;5(12):4677–4698. doi: 10.1093/nar/5.12.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Jacob E., Hofschneider P. H. Replication of the small coliphage M13: evidence for long-living M13 specific messenger RNA. Nature. 1970 Jul 4;227(5253):59–60. doi: 10.1038/227059a0. [DOI] [PubMed] [Google Scholar]
- Konings R. N., Hulsebos T., Van den Hondel C. A. Identification and characterization of the in vitro synthesized gene products of bacteriophage M13. J Virol. 1975 Mar;15(3):570–584. doi: 10.1128/jvi.15.3.570-584.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N. S., Pratt D. Role of bacteriophage M13 gene 2 in viral DNA replication. J Mol Biol. 1972 Dec 14;72(1):37–49. doi: 10.1016/0022-2836(72)90066-6. [DOI] [PubMed] [Google Scholar]
- Martin J., Webster R. E. The in vitro translation of a terminating signal by a single Escherichia coli ribosome. The fate of the subunits. J Biol Chem. 1975 Oct 25;250(20):8132–8139. [PubMed] [Google Scholar]
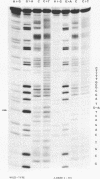
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. Studies on bacteriophage fd DNA. II. Localization of RNA initiation sites on the cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):33–44. doi: 10.1016/0022-2836(75)90333-2. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Horiuchi K., Zinder N. D. Nucleotide sequences near the origin of replication of bacteriophage f1. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4219–4222. doi: 10.1073/pnas.74.10.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch J., Horiuchi K., Model P. Mapping of bacteriophage f1 ribosome binding sites to their cognate genes. Virology. 1977 Sep;81(2):341–351. doi: 10.1016/0042-6822(77)90150-7. [DOI] [PubMed] [Google Scholar]
- Rivera M. J., Smits M. A., Quint W., Schoenmakers J. G., Konings R. N. Expression of bacteriophage M13 DNA in vivo. Localization of the transcription initiation and termination signal of the mRNA coding for the major capsid protein. Nucleic Acids Res. 1978 Aug;5(8):2895–2912. doi: 10.1093/nar/5.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits M. A., Simons G., Konings R. N., Schoenmakers J. G. Expression of bacteriophage M13 dna in vivo. I. Synthesis of phage-specific RNA and protein in minicells. Biochim Biophys Acta. 1978 Nov 21;521(1):27–44. doi: 10.1016/0005-2787(78)90246-0. [DOI] [PubMed] [Google Scholar]
- Steege D. A. 5'-Terminal nucleotide sequence of Escherichia coli lactose repressor mRNA: features of translational initiation and reinitiation sites. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4163–4167. doi: 10.1073/pnas.74.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Sugisaki H., Okamoto T., Takanami M. Studies on bacteriophage fd DNA. IV. The sequence of messenger RNA for the major coat protein gene. J Mol Biol. 1977 Apr 25;111(4):487–507. doi: 10.1016/s0022-2836(77)80065-x. [DOI] [PubMed] [Google Scholar]
- Van Den Hondel C. A., Pennings L., Schoenmakers J. G. Restriction-enzyme-cleavage maps of bacteriophage M13. Existence of an intergenic region on the M13 genome. Eur J Biochem. 1976 Sep;68(1):55–70. doi: 10.1111/j.1432-1033.1976.tb10764.x. [DOI] [PubMed] [Google Scholar]
- Van Den Hondel C. A., Weijers A., Konings R. N., Schoenmakers J. G. Studies on bacteriophage M13 DNA. 2. The gene order of the M13 genome. Eur J Biochem. 1975 May 6;53(2):559–567. doi: 10.1111/j.1432-1033.1975.tb04099.x. [DOI] [PubMed] [Google Scholar]