Abstract
Introduction
TGF-β1 is a cytokine with many different effects on cell proliferation, differentiation and inflammation and can protect against the development of COPD. This work aims to study the association between COPD and the TGF-β1 gene genotypes.
Material and methods
The study included 70 males: 25 smokers with COPD, 25 resistant smokers, and 20 normal non-smokers as the control. They were subjected to spirometry pre- and post-bronchodilator (FEV1, FEV1/FVC), estimation of serum level of TGF-β1 gene by PCR and RFLP.
Results
The percent of Pro-Leu was 28% in the COPD group, 84% in the resistant smokers group and 85% in the control group. There was a highly significant statistical difference in FEV1% of predicted associated with the distribution of TGF-β1 gene genotypes: 56.9 ±8.4% with Pro-Leu genotype and 35.5 ±8.8% with Leu-Leu genotype in COPD patients, 93.2 ±6.2% with Pro-Leu genotype and 86.7 ±0.9% with Leu-Leu genotype in the resistant smokers group.
Conclusions
The Pro-allele genotype is associated with increased production of TGF-β1, which has a protective role against the development of COPD and is important in preserving the decline of FEV1 in COPD patients.
Keywords: chronic obstructive pulmonary disease, leucine, proline domains
Introduction
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of death worldwide. It has been estimated that COPD will become the third leading cause of mortality in the world by the year 2020 [1].
Although cigarette smoking is the main environmental risk factor for developing COPD, only about 15% of smokers develop clinically significant disease, suggesting that there are other influences on disease expression. It was estimated that smoking contributes 15% to the variability of lung function, while genetic factors account for a further 40% [2].
Maccloskey et al. [3] suggested an interaction between genetic and environmental influences. Moreover, a study was done in the Chest Department, Cairo University in collaboration with Tsukuba University Hospital in Japan, to assess the association of CLCA1 gene polymorphisms with COPD in Japanese and Egyptian populations; the distributions tended to show a significant difference in the Egyptian and the Japanese groups. It seems likely that different ethnic populations have different components of COPD. This may be one of the reasons why prevalence of risk alleles of candidate genes for COPD differs greatly between different studies analyzing different ethnic groups [4]. On the other hand, Yoon et al. [5] could not identify significant associations between transforming growth factor β1 genetic polymorphisms and COPD among Koreans as reported previously in Caucasians, reflecting racial differences in the pathogenesis of COPD.
The genetic polymorphisms that have been identified, such as α1-antitrypsin deficiency, only account for 1-2% of COPD cases [6]. A variety of candidate genes have been assessed using case control association studies such as microsomal epoxide hydrolase, glutathione S-transferase and α1 antichymotrypsin [7].
The TGF-β1 gene is one of the genes having a number of actions that make it a candidate for a role in the pathogenesis of COPD. TGF-β1 is a cytokine with many different effects on cell proliferation, differentiation and inflammation. Some of these actions could protect against the development of COPD [8].
TGF-β1 can inhibit matrix metalloproteinase, which may contribute to the development of emphysema through the digestion of elastic fibres [9] and was found to be associated with the emphysema phenotype in the Japanese population [10]. It also promotes the formation of elastin and this could help repair damage to the lungs of individuals who are at risk of developing COPD [11].
The aim of this work is to detect the association between TGF-β1 genotypes and susceptibility to developing chronic obstructive pulmonary diseases.
Material and methods
This study was performed in Ain Shams University, Chest Department and Chest Outpatient Clinic and Pulmonary Function Unit in the National Research Centre, Cairo. Seventy male subjects were included; age range was between 54 and 60 years. They were divided into group 1 which included 25 smokers with COPD, group 2 including 25 smokers who did not develop COPD proved clinically and by normal spirometry (resistant smokers), and group 3 including 20 apparently normal non-smokers, with normal spirometry, as a control group. They were subjected to full medical history taking, clinical examination, plain chest X-ray postero-anterior view, pre- and post-bronchodilator spirometric measurements of FEV1 (forced expiratory volume in the first second), FEV1/FVC (forced vital capacity) and identification of TGF-β1 genotypes in serum. All patients gave written informed consent, having obtained approval from the ethical commission. The patients were assigned to the study according to one or more of the following criteria:
age above 50 years,
smoking history above 20 pack years,
clinical and radiological examination fulfilled the diagnostic criteria of COPD [12],
staging of COPD was based on the criteria defined by GOLD [13],
normal pulmonary function test (PFT) was considered as FEV1 > 85% and FEV1/FVC > 70% of predicted,
bronchodilators stopped 24 h before PFT,
none of the patients was on current systemic corticosteroid therapy,
no other diseases were present other than COPD,
none of the patients was working in industrial areas.
Detection of serum TGF-β1 gene
Sampling
Blood samples were collected using EDTA-containing blood tubes and treated with sucrose lysis buffer and a nuclear cell pellet stored at –80°C until DNA extraction.
DNA extraction from 200 µl whole blood
Genomic DNA was extracted using a Qiagen kit following standard protocols given by the manufacturer.
Polymerase chain reaction (PCR)
Reaction mixtures of 50 µl were prepared containing 2 µl DNA, 3 µl Taq polymerase (1U taq polymerase) [Bioflux], 5 µl dNTPs (2.5 mM each) [Bioflux], 10 µl of each primer, 5 µl PCR buffer (10X), 5 µl MgCl2 (15 mM MgCl2) and 10 µl distilled water.
The forward primer sequence was ACCACACCAGCCCTGTTCGC and the reverse primer sequence was AGTAGCCACAGCAGCGGTAGCAGCT-GC [14], using a thermocycler (T1 Thermocycler 96, Biometra GmbH, Göttingen, Germany). PCR was performed with an initial denaturation at 94°C for 3 min. This was followed by 33 cycles with denaturation at 94°C for 50 s, annealing at 66°C for 1 min, then elongation at 72°C for 1 min, and a final elongation of 10 min at 72°C.
Restriction enzyme fragment length polymorphism (RFLP)
6 µl of the amplification product was digested with 4 µl PstI restriction endonuclease enzyme (Roche Diagnostics, Hague Road, IN, USA) in a 24 µl volume mixture containing: 0.4 µl restriction enzyme, 2 µl Sure/Cut Buffer H, 0.2 µl BSA, 15.4 µl distilled water and 6 µl amplification product. The reaction mixture was inoculated at 37°C for 1 h.
Restriction enzyme digestion PCR products were subjected to electrophoresis in a 2.5% agarose gel at 120 mV for 2 h in TBE buffer.
Using ultraviolet transillumination after ethidium bromide staining, the products (123 bp) were visualized and the size of the product was determined using 123 bp ladder (Figure 1).
Figure 1.
Agarose gel showing RFLP for TGF-β1 genotypes at codon 10. Lane 1 shows 123 bp DNA ladder lanes 1, 2, 3, 4, 5, 6 and 9 show Leu-Leu genotype and lanes 7, 8, 10 and 11 show Pro-Leu genotype
Statistical analysis
SPSS statistical software package (V. 15.2, Echo soft Corp., USA, 2006) was used for data analysis. Data were expressed as mean ± SD for quantitative measures and both number and percentage for categorical data. Student's t test was used for comparison between two independent mean groups for parametric data and χ2 test to study the association between each 2 variables or comparison between 2 independent groups. The probability of error at 0.05 was considered as significant and error at 0.01 and 0.001 as highly significant. Calculated relative risk assessments (calculated odds ratio, OR) measured the level risk among diseased individuals compared to non-diseased ones. They were calculated as absolute figures and as a standard error of estimate (95%).
Results
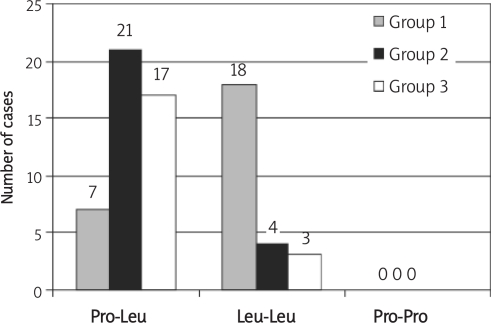
Seventy male subjects were enrolled in the study. The age and the smoking history were matched between groups (p > 0.05). There was a highly statistically significant decrease in the mean value of the post-bronchodilator FEV1% of predicted in group 1 (COPD), 41.5%, compared to that of group 2 (resistant smokers), 92.2%, and of group 3 (control), 92.6% (p < 0.001). There was no statistically significant difference in the mean value of the post-bronchodilator FEV1% of predicted between groups 2 and 3 (p > 0.05) (Table I). The distribution of the genotypes of the TGF-β1 gene in the studied groups is shown in Figure 2. The proline-proline (Pro-Pro) genotype was not detected in the study. There was a highly statistically significant increase of the leucine-leucine (Leu-Leu) genotypes distribution in group 1, 72% of COPD cases, compared to group 2, 16% of resistant smokers, and group 3, 15% of controls, p < 0.001. There was no statistically significant difference between group 2 and group 3 in the genotype distribution of the TGF-β1 gene. There was a highly statistically significant reduction in the mean value of the post-bronchodilator FEV1% of predicted in the case of Leu-Leu genotypes compared to that in the case of Pro-Leu genotypes in group 1 (COPD), 35.5 and 56.9% respectively (p < 0.001) and that in group 2 (resistant smokers), 86.7 and 93.2% respectively (p < 0.001). There was no statistically significant difference of mean value of the post-bronchodilator FEV1% of predicted in group 3 (control) according to the genotype distribution (p > 0.05) (Table II). There was a highly statistically significant difference between the distributions of TGF-β1 gene genotypes in group 1 (COPD) according to the stages of COPD. From 7 cases with the Pro-Leu genotype, 5 (71.4%) were in stage II and 2 (28.6%) in stage III, compared to one case from 18 (5.5%) with the Leu-Leu genotype in stage II, 12 (66.7%) in stage III and 5 (27.7%) in stage IV (p < 0.01) (Table III). Smokers with the Pro-Leu allele are at low risk of developing COPD (OR < 1) while smokers with Leu-Leu allele are at high risk of developing COPD (OR > 1) (Table IV).
Table I.
Statistically significant difference of the mean values of age (years), smoking history (pack years) and percent of predicted FEV1 post-bronchodilator between the 3 groups
| Group | N° | Mean age±SD [year] | Mean smoking history±SD | Mean post-bronchodilator FEV1% of predicted ± SD |
|---|---|---|---|---|
| 1 | 25 | 57.28 ±1.99 | 50 ±6.8 | 41.5 ±13 |
| 2 | 25 | 57.32 ±1.46 | 48.72 ±8.57 | 92.2 ±6.1 |
| 3 | 25 | 56.95 ±1.63 | – | 92.6 ±6.2 |
| 1 vs. 2 | T | −0.081 | 0.585 | −17.56 |
| P | > 0.05 | > 0.05 | < 0.001 | |
| 1 vs. 3 | T | 0.598 | – | −17.31 |
| P | > 0.05 | – | < 0.001 | |
| 2 vs. 3 | T | 0.802 | – | −0.24 |
| P | > 0.05 | – | > 0.05 |
Figure 2.
Distribution of genotypes of TGF-β 1 gene in the studied groups
Table II.
Statistically significant difference of mean value of post-bronchodilator FEV1% of predicted in each of the studied groups according to the distribution of TGF-β1 genotypes
| Groups | Genotype of TGFβ1 gene | Mean±SD of post-bronchodilator FEV1% of predicted | T value | P value |
|---|---|---|---|---|
| COPD N=25 | Pro-Leu N=7 (28%) | 56.9 ±8.4 | 5.61 | < 0.001 |
| Leu-Leu N = 18 (72%) | 35.5 ±8.8 | |||
| Resistant smokers N = 25 | Pro-Leu N = 21 (84%) | 93.2 ±6.2 | 4.5 | < 0.001 |
| Leu-Leu N = 4 (16%) | 86.7 ±0.9 | |||
| Control N = 20 | Pro-Leu N = 17 (85%) | 92.8 ±6.5 | 0.4 | > 0.05 |
| Leu-Leu N=3 (15%) | 91.6 ±4.1 |
Table III.
Statistically significant difference of the distribution of TGF-β1 genotypes in group 1 (COPD) according to COPD stages
| Group 1 (COPD) | Genetic polymorphism of TGF-β1 gene | X2=12.63 | |||
|---|---|---|---|---|---|
| Pro-Leu No. = 7 | Leu-Leu No. = 18 | ||||
| Stage II | No. | 5 | 1 | p < 0.01 | |
| % | 71.4 | 5.5 | |||
| Stage III | No. | 2 | 12 | ||
| % | 28.6 | 66.7 | |||
| Stage IV | No. | 0 | 5 | ||
| % | 0 | 27.7 | |||
Table IV.
Calculated odds ratio between Pro-Leu and Leu-Leu genotypes of TGF-β1 gene in group 1 (COPD) and group 2 (resistant smokers)
| Value | 95% confidence interval | ||
|---|---|---|---|
| Lower | Upper | ||
| For cohort Pro-Leu | 0.333 | 0.174 | 0.639 |
| For cohort Leu-Leu | 4.5 | 1.77 | 11.41 |
| Number of valid cases | 50 | ||
Discussion
TGF-β1 can inhibit matrix metalloproteinase, which may contribute to the development of emphysema through the digestion of elastic fibres. It also promotes the formation of elastin and this could help repair damage to the lungs of individuals who are smokers and who are at risk of developing COPD. TGF-β1 plays a role in maintaining the integrity of the pulmonary vasculature [11].
Alveolar macrophages from COPD patients release less TGF-β1 than smokers with normal lung function and non-smokers, which may reduce the anti-inflammatory and anti-elastolytic responses in COPD patients, subsequently contributing to progressive extracellular matrix destruction [15].
This study selected randomized samples of subjects of the same sex and race. The mean ages of the subjects and their smoking index were statistically matched (Table I). All subjects were of Caucasian origin; it was found that the frequency of an allele can vary in different racial groups and that the Pro allele of the TGF-β1 gene was significantly more common in African Americans than in white subjects [16].
This study included patients with a heavy smoking history of more than 20 pack years [17]. Several studies [14, 18] have detected TGF-β1 genotype and increased susceptibility to COPD in subjects with mean smoking index of 45 pack years, which is nearly equal to the smoking index of the studied subjects, ± 50 pack years (Table I).
Three genotypes have been demonstrated for the TGF-β1 gene: Pro-Pro, Pro-Leu and Leu-Leu [14]. In our study the Pro-Pro genotype was not detected. This may be explained by the racial differences in the frequency of the Pro allele [16, 19].
In the COPD patients (group 1), the incidence of the Pro-Leu allele of TGF-β1 was low (28% of the total number of COPD cases). This result agreed with that of Wu et al. [14] who detected the Pro allele in 33% of COPD cases. There was a highly statistically significant increase in the percent of cases with the Pro-Leu allele in group 2 (resistant smokers) and group 3 (control) compared to group 1, 84, 85 and 28% respectively, p < 0.001 (Figure 2). On the other hand, there was no statistically significant difference in the incidence of the Pro-Leu allele between groups 2 and 3, p > 0.05 (Table IV). These findings agreed with other studies which reported that the frequency of the Pro allele is lower in COPD cases than in resistant smokers and control subjects [14, 20]. However, a study done on 36 patients with COPD, 27 resistant smokers and 60 healthy non-smokers with normal pulmonary function did not find any statistically significant differences in the distribution of genotypes of the TGF-β1 gene between the three studied groups and did not demonstrate a role of the TGF-β1 genotypes in COPD [21].
The results of our study showed that there were 7 cases with the Pro allele in the COPD group. The mean value of the percent of predicted FEV1 of these 7 cases was statistically higher (56.9 ±8.4%) compared to that of the other 18 cases of COPD with Leu-Leu genotype (35.5 ±8.8%) p < 0.01 (Table II). This may be attributed to the protective role of the Pro allele against further decline in FEV1, which is in concordance with 2 studies on patients with cystic fibrosis [22] and on 590 smokers [23].
Moreover, the relation between genetic polymorphism of the TGF-β1 gene and the stages of COPD [13] were studied in group 1. There were 7 cases with Pro-Leu genotype; 5 cases were in stage II COPD and 2 cases in stage III. There were 18 cases with Leu-Leu genotype: one case in stage II, 12 in stage III and 5 in stage IV (Table III). These results demonstrated that the Pro allele of the TGF-β1 gene has a protective effect on the lung function of COPD cases and decreases the rapid decline of FEV1, in agreement with other studies [22, 23].
In group 2, resistant smokers, the mean value of the percent of predicted FEV1 in subjects with Pro-Leu genotype (21 cases) was 93.2 ±6.2%, and 86.7 ±0.9% in the subjects with Leu-Leu genotype (4 cases) p > 0.05 (Table II). In spite of the presence of Leu-Leu genotype (4 cases) in this group, the mean value of the FEV1 percent of predicted was still within the normal range but statistically significantly lower than that of the subjects with Pro-Leu genotype (21 cases), p < 0.001. This may be attributed to other genetic factors which protect against COPD in resistant smokers. These results agreed with those of Arkwright et al. [22], who concluded that the Pro allele has a protective effect on lung function and decreases the rapid decline of FEV1.
The calculated odds ratio (OR) showed a low risk of developing COPD in smokers with the Pro-Leu allele (OR = 0.33), and a high risk of developing COPD in smokers with the Leu-Leu allele, while smokers with the Leu-Leu allele are at high risk of developing COPD (OR = 4.5) (Table IV).
This result was in parallel to that found by Wu et al. [14],who observed that the Pro allele occurred less frequently in subjects with COPD than in resistant smokers, and the OR was 0.59 for smokers with the Pro allele and 3.2 for smokers with the Leu allele.
In spite of the limited number of patients, this case control study gave an idea about the distribution of TGF-β1 genotypes in Egyptians and demonstrated its protective role in COPD. Moreover, absence of the Pro-Pro allele in all cases of the study denotes high predisposition to COPD in Egyptian smokers.
In conclusion, although cigarette smoking is the primary risk factor in COPD, genetic risk factors likely influence its development. Moreover, the proline allele of the TGF-β1 gene is expressed more commonly in resistant smokers than in smokers who develop COPD, demonstrating a protective role against COPD and an important effect in preserving further decline in FEV1.
References
- 1.Ghanei M, Aslani J, AzizAbadi-Farahani M, Assari S, Saadat AR. Clinical research Logistic regression model to predict chronic obstructive pulmonary disease exacerbation. Arch Med Sci. 2007;3:360–6. [Google Scholar]
- 2.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–52. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 3.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of patients with severe obstruction pulmonary disease have a significant risk of airflow obstruction. Am J Resp Crit Care Med. 2001;164:1419–24. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 4.Hegab AE, Sakamoto T, Uchida Y, et al. CLCA1 gene polymorphisms in chronic obstructive pulmonary disease. J Med Genet. 2004;41:e27. doi: 10.1136/jmg.2003.012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon HI, Silverman EK, Lee HW, et al. Lack of association between COPD and transforming growth factor-beta1 (TGFB1) genetic polymorphisms in Koreans. Int J Tuberc Lung Dis. 2006;10:504–9. [PubMed] [Google Scholar]
- 6.Hoidal J. Genetics of COPD: present and future. Eur Resp. 2001;18:741–3. doi: 10.1183/09031936.01.00268501. [DOI] [PubMed] [Google Scholar]
- 7.Lemos D, Silverman E. The genetics of chronic obstructive pulmonary disease. Respir Res. 2001;2:20–6. doi: 10.1186/rr34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annu Rev Immunol. 1998;16:137–61. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 9.Eickelberg O, Köhler E, Reichenberger F. Extracellular matrix deposition by primary human lung fibroblasts in response to TGF-beta1 and TGF-beta3. Am J Physiol. 1999;276:L814–24. doi: 10.1152/ajplung.1999.276.5.L814. [DOI] [PubMed] [Google Scholar]
- 10.Ito M, Hanaoka M, Droma Y, et al. The association of transforming growth factor beta 1 gene polymorphisms with the emphysema phenotype of COPD in Japanese. Intern Med. 2008;47:1387–94. doi: 10.2169/internalmedicine.47.1116. [DOI] [PubMed] [Google Scholar]
- 11.Kucich U, Rosenbloom JC, Abrams WR, Rosenbloom J. Transforming growth factorbeta stabilizes elastin mRNA by a pathway requiring active Smads, protein kinase C-delta and p38. Am J Respir Cell Mol Biol. 2002;26:183–8. doi: 10.1165/ajrcmb.26.2.4666. [DOI] [PubMed] [Google Scholar]
- 12.American Thoracic Society (AST): COPD, criteria of diagnosis. Am J Resp Crit Case Med. 1995 152S77. [Google Scholar]
- 13. Global Initiative for chronic obstructive pulmonay disease (GOLD): Global strategy for the diagnosis, management and prevention of Chronic Obstructive Pulmonary Disease. National Institute of Health, National Heart, Lung and blood Institute and World Health Organization 2006. Available at: http://www.goldcopd.com.
- 14.Wu L, Chau J, Young RP, et al. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax. 2004;59:126–9. doi: 10.1136/thorax.2003.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pons AR, Sauleda J, Noguera A, et al. Decreased macrophage release of TGF-beta and TIMP-1 in chronic obstructive pulmonary disease. Eur Respir J. 2005;26:60–6. doi: 10.1183/09031936.05.00045504. [DOI] [PubMed] [Google Scholar]
- 16.Suthanthiran M, Li B, Song JO, Ding R, Sharma VK, Schwartz JE. Transforming growth factor-beta 1 hy-perexpression in African-American hypertensives: A novel mediator of hypertension and/or target organ damage. Proc Natl Acad Sci U S A. 2000;97:3479–84. doi: 10.1073/pnas.050420897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.British Thoracic Society. BTS guidelines for the management of chronic obstructive pulmonary disease. Thorax. 1997;52(Suppl 5):S1–28. [PMC free article] [PubMed] [Google Scholar]
- 18.Su ZG, Wen FQ, Feng YL, Xiao M, Wu XL. Transforming growth factor-beta 1 gene polymorphisms associated with chronic obstructive pulmonary diseases in Chinese population. Acta Pharmacol Sin. 2005;26:714–20. [PubMed] [Google Scholar]
- 19.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 20.Celedon JC, Lange C, Raby BA, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13:1649–56. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 21.Liebhart J, Polak M, Dobek R, et al. TGF-beta1 gene polymorphism in chronic obstructive pulmonary disease. Pneumonol Alergol Pol. 2005;73:216–20. [PubMed] [Google Scholar]
- 22.Arkwright PD, Laurie S, Super M. TGF-beta1 genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax. 2000;55:459–62. doi: 10.1136/thorax.55.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa E, Ruan J, Connett JE, Anthonisen NR, Paré PD, Sandford AJ. Transforming growth factor-beta1 polymorphisms, airway responsiveness and lung function decline in smokers. Respir Med. 2007;101:938–43. doi: 10.1016/j.rmed.2006.09.008. [DOI] [PubMed] [Google Scholar]