Abstract
Since ancient times, even in today’s modern world, infectious diseases cause lots of people to die. Infectious organisms, pathogens, cause diseases by physical interactions with human proteins. A thorough analysis of these interspecies interactions is required to provide insights about infection strategies of pathogens. Here we analyzed the most comprehensive available pathogen–human protein interaction data including 23,435 interactions, targeting 5,210 human proteins. The data were obtained from the newly developed pathogen–host interaction search tool, PHISTO. This is the first comprehensive attempt to get a comparison between bacterial and viral infections. We investigated human proteins that are targeted by bacteria and viruses to provide an overview of common and special infection strategies used by these pathogen types. We observed that in the human protein interaction network the proteins targeted by pathogens have higher connectivity and betweenness centrality values than those proteins not interacting with pathogens. The preference of interacting with hub and bottleneck proteins is found to be a common infection strategy of all types of pathogens to manipulate essential mechanisms in human. Compared to bacteria, viruses tend to interact with human proteins of much higher connectivity and centrality values in the human network. Gene Ontology enrichment analysis of the human proteins targeted by pathogens indicates crucial clues about the infection mechanisms of bacteria and viruses. As the main infection strategy, bacteria interact with human proteins that function in immune response to disrupt human defense mechanisms. Indispensable viral strategy, on the other hand, is the manipulation of human cellular processes in order to use that transcriptional machinery for their own genetic material transcription. A novel observation about pathogen–human systems is that the human proteins targeted by both pathogens are enriched in the regulation of metabolic processes.
Keywords: pathogen–human protein–protein interactions, PHISTO, infection strategy, hub, bottleneck, gene ontology
Introduction
According to a report of World Health Organization (WHO), more than 20% of total deaths in the world are due to infectious diseases (World Health Organization, 2008). Different types of microorganisms (bacteria, fungi, protozoa, and viruses) act as pathogens for such diseases. The mechanism of infection is based on the interactions between the proteins of pathogen and host. Thanks to the developments in high-throughput protein interaction detection methods, it is possible to identify pathogen–host protein–protein interactions (PHIs) at large-scale. Infection strategies have been studied through intraspecies protein interactions of various pathogens (Flajolet et al., 2000; McCraith et al., 2000; Rain et al., 2001; LaCount et al., 2005; Uetz et al., 2006; Calderwood et al., 2007; Wang et al., 2010) as well as through interspecies protein interactions between pathogens and human (Filippova et al., 2004; Mogensen et al., 2006; Uetz et al., 2006; Calderwood et al., 2007; König et al., 2008). Notwithstanding these, a general overview of infection mechanisms of different types of pathogens is missing since there has been a lack of interspecies interactome data until very recent years.
A major step toward a complete picture of the pathogenesis of infectious diseases and consequently identifying drug targets is the cataloging of large-scale PHIs. There are few PHI-specific databases, which enable the access to PHI data for each type of pathogen from a single source (Driscoll et al., 2009; Kumar and Nanduri, 2010). Nevertheless these databases have not been updated since their first release, and miss lots of recently reported PHIs. Therefore, we have recently developed a pathogen–host interaction search tool (PHISTO), which serves as a centralized and up-to-date source for the entire available PHI data between various pathogen strains and human via a user-friendly and functional interface1.
The systemic analysis of PHI data has so far focused mainly on virus-based infections due to the scarcity of data for other types of pathogenic organisms (Uetz et al., 2006; Calderwood et al., 2007; Dyer et al., 2008). We have enough bacterial PHIs to get statistically meaningful results, providing a good opportunity to get a systemic picture of the pathogenesis of bacterial infections. In this work, we studied up-to-date PHI data reported in PHISTO with a specific focus on comparison between bacterial and viral infections of human. We constructed various sets of human proteins targeted by bacteria, fungi, protozoa, and viruses to pick out specific infection strategies of different pathogen types. On the other hand, the set of human proteins targeted by both bacteria and viruses were used to obtain common infection strategies of these pathogens. We computed degree (connectivity) and betweenness centrality distributions of the human protein sets targeted by bacteria and viruses to observe the network properties of targeted human proteins. Additionally, we computed gene ontology (GO; Ashburner et al., 2000) terms enriched in each above-mentioned protein set to find out attacked mechanisms in human. GO enrichment analysis was also performed for sets of human proteins targeted by each pathogen group included in our PHI data to decipher the pathogen group(s) manipulating specific processes in human.
Materials and Methods
Pathogen–host interaction search tool
We have developed PHISTO that presents experimentally verified pathogen–human protein interaction data in the most comprehensive and updated manner. The database provides the entirety of relevant information about the physical PHIs in a single non-redundant resource to researchers. It offers access via a user-friendly and functional web interface (see text footnote 1) with various searching, filtering, browsing, and extraction options. Results are displayed in a very clear and consistent presentation format. PHISTO enables the users to reach additional information easily by providing links in the search results to external databases. Proteins, pathogens, and publications listed in the search results are linked to UniProt, NCBI Taxonomy, and PubMed, respectively, offering users quick navigation in these informative databases.
We downloaded the pathogen–human PHIs from PHISTO in October 2011. The data cover 23,435 physical interactions occurring between 5,210 human proteins and 3,419 proteins of 257 pathogen strains of 72 pathogen groups (24 bacterial groups, 3 fungal groups, 2 protozoan groups, and 43 viral groups; Table 1). In PHISTO, pathogen strains are grouped to provide an option to present PHI results together for related strains. Bacterial groups are sets of strains of the same genus as the names of the groups are the names of the genuses. For viral groups, there are two definitions. Some viral groups are sets of strains of the same family as the names of the groups are the names coming from the families of the strains (e.g., papillomaviruses, herpesviruses, polyomaviruses). Some viral strains are grouped based on the related infections caused by them, as the names are generally coming from the diseases (e.g., HIV, hepatitis viruses, anemia viruses). Detailed PHI data for 72 groups are given in Data Sheets 1–4 in Supplementary Material.
Table 1.
Contents of pathogen–human PHI data.
| Pathogen | Number of strains | Number of PHIs | Number of targeting pathogen proteins | Number of targeted human proteins |
|---|---|---|---|---|
| BACTERIA | 41 | 8,549 | 2,591 | 3,589 |
| Aeromonas | 1 | 2 | 1 | 2 |
| Bacillus | 2 | 3,021 | 940 | 1,736 |
| Campylobacter | 1 | 3 | 1 | 3 |
| Chlamydia | 2 | 21 | 3 | 21 |
| Citrobacter | 1 | 1 | 1 | 1 |
| Clostridium | 3 | 47 | 9 | 10 |
| Corynebacterium | 1 | 1 | 1 | 1 |
| Escherichia | 4 | 30 | 14 | 27 |
| Finegoldia | 1 | 1 | 1 | 1 |
| Francisella | 1 | 1,338 | 346 | 986 |
| Helicobacter | 2 | 3 | 3 | 2 |
| Klebsiella | 1 | 1 | 1 | 1 |
| Legionella | 1 | 1 | 1 | 1 |
| Listeria | 1 | 4 | 4 | 3 |
| Moraxella | 1 | 1 | 1 | 1 |
| Mycoplasma | 1 | 2 | 1 | 2 |
| Neisseria | 1 | 17 | 1 | 17 |
| Pseudomonas | 1 | 12 | 4 | 10 |
| Salmonella | 1 | 5 | 5 | 5 |
| Shigella | 1 | 11 | 9 | 8 |
| Staphylococcus | 3 | 12 | 10 | 10 |
| Streptococcus | 5 | 15 | 12 | 9 |
| Vibrio | 1 | 1 | 1 | 1 |
| Yersinia | 4 | 3,999 | 1,221 | 2,120 |
| FUNGI | 3 | 4 | 3 | 4 |
| Candida | 1 | 1 | 1 | 1 |
| Pneumocystis | 1 | 1 | 1 | 1 |
| Radiomyces | 1 | 2 | 1 | 2 |
| PROTOZOA | 4 | 9 | 5 | 9 |
| Plasmodium | 3 | 8 | 4 | 8 |
| Toxoplasma | 1 | 1 | 1 | 1 |
| VIRUS | 209 | 14,873 | 820 | 2,398 |
| Adenovirus | 13 | 121 | 36 | 80 |
| Anemia virus | 6 | 8 | 6 | 4 |
| ASFV | 1 | 1 | 1 | 1 |
| Bacteriophage | 6 | 6 | 6 | 5 |
| Coxsackie virus | 1 | 1 | 1 | 1 |
| Dengue virus | 3 | 3 | 3 | 2 |
| Ebola virus | 1 | 1 | 1 | 1 |
| Echo virus | 2 | 3 | 3 | 1 |
| Ectromelia virus | 1 | 2 | 2 | 2 |
| Encephalitis virus | 1 | 2 | 1 | 2 |
| Foamy virus | 1 | 1 | 1 | 1 |
| Hantaan virus | 1 | 6 | 1 | 6 |
| Hendra virus | 1 | 1 | 1 | 1 |
| Hepatitis virus | 21 | 1,573 | 179 | 399 |
| Herpesvirus | 28 | 666 | 141 | 388 |
| HIV | 49 | 11,435 | 279 | 1,601 |
| Influenza virus | 9 | 523 | 27 | 182 |
| Leukemia virus | 3 | 10 | 3 | 10 |
| Measles virus | 3 | 10 | 4 | 4 |
| Molluscum virus | 1 | 1 | 1 | 1 |
| MPMV | 1 | 1 | 1 | 1 |
| Nipah virus | 1 | 1 | 1 | 1 |
| Nucleopolyhedrovirus | 1 | 1 | 1 | 1 |
| Orf virus | 2 | 2 | 2 | 1 |
| Papillomavirus | 14 | 290 | 51 | 128 |
| Parainfluenza virus | 1 | 2 | 1 | 2 |
| Parvo virus | 1 | 1 | 1 | 1 |
| Polio virus | 2 | 3 | 2 | 2 |
| Polyomavirus | 4 | 64 | 10 | 45 |
| Puumala virus | 1 | 3 | 1 | 3 |
| Rabies virus | 1 | 1 | 1 | 1 |
| Rhino virus | 1 | 1 | 1 | 1 |
| Rota virus | 4 | 8 | 5 | 6 |
| Sarcoma virus | 5 | 15 | 6 | 11 |
| SARS | 1 | 4 | 3 | 4 |
| Sendai virus | 1 | 1 | 1 | 1 |
| Seoul virus | 1 | 4 | 1 | 4 |
| SIV | 2 | 3 | 2 | 3 |
| Stomatitis virus | 3 | 7 | 3 | 6 |
| T-lymphotropic virus | 3 | 38 | 7 | 35 |
| Tula virus | 1 | 2 | 1 | 2 |
| Vaccinia virus | 5 | 46 | 20 | 33 |
| West Nile virus | 1 | 1 | 1 | 1 |
| TOTAL | 257 | 23,435 | 3,419 | 5,210 |
See Data Sheets 1–4 in Supplementary Material for detailed information.
Human PPI data
To obtain degree and betweenness centrality values of pathogen-targeted proteins, the human protein–protein interaction (PPI) network was constructed by downloading 194,006 interactions between 13,015 human proteins from BioGRID (Stark et al., 2011), DIP (Salwinski et al., 2004), IntAct (Kerrien et al., 2012), Mint (Ceol et al., 2010), and Reactome (Matthews et al., 2009; Croft et al., 2011) in April 2011.
Human protein sets
A total of 10 sets of human proteins interacting with pathogens were constructed from PHI data to analyze the properties of targeted human proteins as follows: The sets targeted by bacterial pathogens (bacteria-targeted set), fungal pathogens (fungi-targeted set), protozoan pathogens (protozoa-targeted set), and viral pathogens (virus-targeted set) were analyzed for specific infection strategies of these different pathogen types. For a deeper comparison between bacterial and viral infections, human proteins interacting with at least two bacterial groups (two-bacteria-targeted set) and two viral groups (two-viruses-targeted set) and also human proteins interacting with at least three bacterial groups (three-bacteria-targeted set) and three viral groups (three-viruses-targeted set) were used. To obtain common infection strategies of pathogens, sets of human proteins targeted by all types of pathogens (pathogen-targeted set) and by both bacteria and viruses (bacteria–virus-targeted set) were also analyzed. Finally, 72 sets of human proteins each targeted by a pathogen group reported in Table 1 were additionally used in GO enrichment analysis to investigate the human mechanisms attacked by each pathogen group in the PHI data. Totally, 82 human protein sets were constructed and analyzed.
Degree and betweenness centrality calculations
Degree of a protein within a network is defined as its number of connections. Betweenness centrality of a protein is equal to the number of shortest paths between any pairs passing through that protein. The degree and centrality values of proteins in interaction networks provide valuable information about the role of corresponding proteins in the network’s functional organization using the topology of the interconnections. For instance, hubs (highly connected proteins) and bottlenecks (central proteins to many paths in the network) are critical players in the intraspecies protein networks for information flow (Barabasi and Oltvai, 2004; Yu et al., 2007).
The undirected human PPI network was represented as an adjacency matrix, and the degree and centrality values of each protein in the network were calculated in MATLAB environment. Betweenness centrality calculations were performed by freely available MATLAB BGL package developed by David Gleich2. The results were normalized by (n − 1)(n − 2), where n is the number of all proteins in the PPI network. Self-interactions were not taken into account in these calculations.
GO enrichment analysis
Gene Ontology (Ashburner et al., 2000) enrichments of all 82 human protein sets were performed using BiNGO plugin (ver. 2.44) of Cytoscape (ver. 2.8.1; Maere et al., 2005). Significance level was set to 0.05 meaning that only terms enriched with a p-value of at most 0.05 were considered. All three GO terms (biological process, molecular function, and cellular component) were scanned to identify the terms having significant association with each human protein set studied.
Results
Pathogen-targeted human proteins
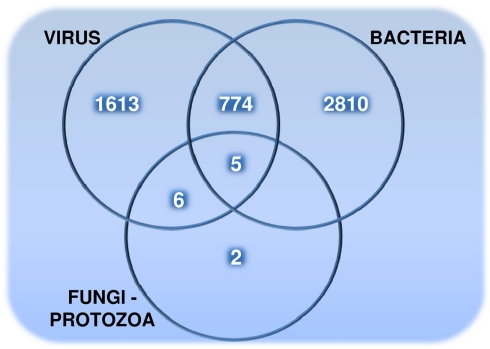
The distribution of 5,210 human proteins on their targeting pathogens are shown in the Venn diagram (Figure 1). Detailed properties of all pathogen-targeted human proteins including number and types of targeting pathogens together with degree and betweenness centrality values in the human PPI network are given in Data Sheet 5 in Supplementary Material. The most targeted human proteins are listed in Table 2. The top of this list, P53 (Tumor suppressor), DRA (HLA class II histocompatibility antigen, DR alpha chain), SUMO1 (Small ubiquitin-related modifier 1), JUN (Transcription factor AP-1), NPM (Nucleophosmin), ROA1 (Heterogeneous nuclear ribonucleoprotein A1), and UBC9 (SUMO-conjugating enzyme) and the following proteins have potential to give important insights about infections.
Figure 1.
The number of pathogen-targeted human proteins that are grouped based on their interactions with viruses, bacteria, and fungi – protozoa (targeted by fungi and/or protozoa).
Table 2.
Highly targeted human proteins.
| Protein | Degree | Betweenness centrality | Targeting bacterial groups | Targeting viral groups |
|---|---|---|---|---|
| P53 | 347 | 0.01547 | Bacillus, Escherichia, Francisella, Yersinia | Adenovirus, Hepatitis virus, HIV, Papillomavirus, Polyomavirus, SIV |
| DRA | 52 | 0.00003 | Bacillus, Francisella, Mycoplasma, Staphylococcus, Yersinia | Herpesvirus, HIV, Influenza virus |
| SUMO1 | 103 | 0.00366 | Bacillus | Herpesvirus, HIV, Papillomavirus, Puumala virus, SARS, Tula virus, Vaccinia virus |
| JUN | 122 | 0.00335 | Bacillus, Francisella, Yersinia | Hepatitis virus, HIV, Papillomavirus, Vaccinia virus |
| NPM | 137 | 0.00166 | Bacillus, Francisella, Yersinia | Adenovirus, Hepatitis virus, Herpesvirus, HIV |
| ROA1 | 246 | 0.00262 | Bacillus, Francisella, Yersinia | Herpesvirus, HIV, Influenza virus, SARS |
| UBC9 | 134 | 0.00410 | Yersinia | Hantaan virus, Herpesvirus, HIV, Influenza virus, Papillomavirus, Seoul virus |
| IGHG1 | 57 | 0.00219 | Bacillus, Francisella, Staphylococcus, Streptococcus, Yersinia | Herpesvirus |
| RAC1 | 239 | 0.00279 | Bacillus, Clostridium, Pseudomonas, Salmonella, Yersinia | HIV |
| CDC42 | 232 | 0.00405 | Bacillus, Francisella, Salmonella, Yersinia | HIV, T-lymphotropic virus |
| DRB5 | – | – | Bacillus, Francisella, Streptococcus, Yersinia | Herpesvirus, HIV |
| LCK | 147 | 0.00202 | Bacillus, Francisella, Yersinia | Hepatitis virus, Herpesvirus, HIV |
| XRCC6 | 131 | 0.00445 | Bacillus, Francisella, Yersinia | Herpesvirus, HIV, Polyomavirus |
| KPYM | 76 | 0.00041 | Bacillus, Francisella, Yersinia | Hepatitis virus, Herpesvirus, Papillomavirus |
| ROA2 | 189 | 0.00069 | Bacillus, Francisella, Yersinia | Herpesvirus, Influenza virus, Vaccinia virus |
| P85A | 402 | 0.00914 | Bacillus, Francisella, Yersinia | Anemia virus, HIV, Influenza virus |
| STAT3 | 77 | 0.00133 | Bacillus, Francisella, Yersinia | Hepatitis virus, Herpesvirus, HIV |
| STAT1 | 71 | 0.00104 | Bacillus, Francisella, Yersinia | Adenovirus, Herpesvirus, HIV |
| GBLP | 93 | 0.00265 | Bacillus, Francisella, Yersinia | Adenovirus, Herpesvirus, HIV |
| PARP4 | 1 | 0.00000 | Bacillus, Francisella, Yersinia | Hepatitis virus, Herpesvirus, HIV |
| RB | 149 | 0.00282 | Yersinia | Adenovirus, Herpesvirus, HIV, Papillomavirus, Polyomavirus |
| SP1 | 103 | 0.00268 | Yersinia | Adenovirus, Herpesvirus, HIV, Polyomavirus, T-lymphotropic virus |
| TAF1 | 58 | 0.00025 | Bacillus | Adenovirus, Hepatitis virus, HIV, Papillomavirus, Polyomavirus |
| CDK2 | 151 | 0.00220 | Shigella | Herpesvirus, HIV, Papillomavirus, Polyomavirus, T-lymphotropic virus |
| TF2B | 69 | 0.00020 | Bacillus | Hepatitis virus, Herpesvirus, HIV, Papillomavirus, Polyomavirus |
| EP300 | 123 | 0.00245 | Bacillus | Adenovirus, Hepatitis virus, HIV, Papillomavirus, Polyomavirus |
| CBP | 147 | 0.00304 | Yersinia | Adenovirus, Hepatitis virus, HIV, Papillomavirus, Polyomavirus |
| TBP | 147 | 0.00241 | – | Adenovirus, Hepatitis virus, Herpesvirus, HIV, Papillomavirus, Polyomavirus |
The targeting pathogenic proteins for each human protein can be obtained from Data Sheets 1–4 in Supplementary Material.
Degree and centrality distributions
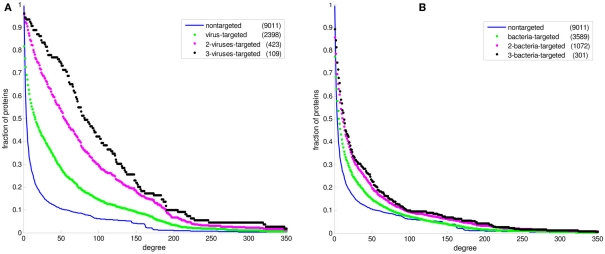
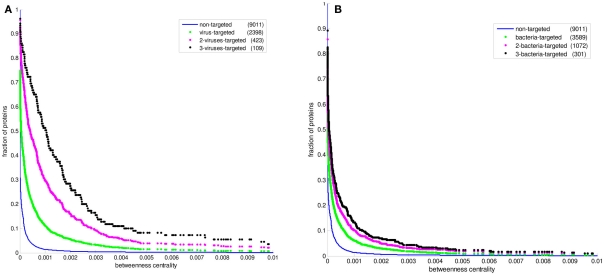
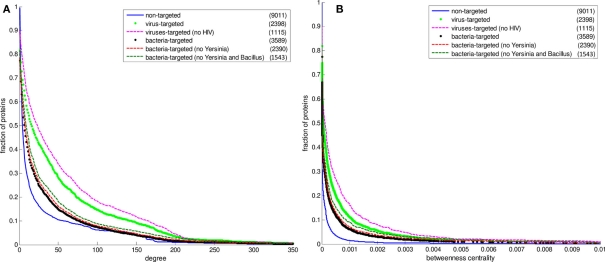
Figure 2 displays the comparison between the degree distributions of non-targeted proteins in the human PPI network and bacteria and virus-targeted sets. For both cases of bacteria and virus-targeted sets, it is observed that pathogen-targeted human proteins have generally higher degree values than non-targeted ones. However a difference is observed in trends of degree distributions of multibacteria and multiviruses targeted sets. For bacteria-targeted cases, the increase in degree values of human proteins with increasing number of targeting pathogen groups is not as clear as those of virus-targeted cases (Figure 2). Very similar trends are obtained for centrality distributions of human proteins (Figure 3). In order to justify these global trends, the same analyses was then repeated with human protein sets excluding the overrepresented pathogens, i.e., Bacillus, Yersinia, and HIV which target the largest number of human proteins (Table 1). Similar results are still obtained when major pathogen groups are eliminated (Figure 4).
Figure 2.
The cumulative degree distributions of human protein sets. The distribution of all proteins in the PPI network is given in comparison with (A) the bacteria-targeted sets, and (B) the virus-targeted sets. The number of proteins in each set is given in the parentheses. The fraction of proteins at a particular value of degree is the number of proteins having that value and greater divided by the number of proteins in the set.
Figure 3.
The cumulative betweenness centrality distributions of human protein sets. The distribution of all proteins in the PPI network is given in comparison with (A) the bacteria-targeted sets, and (B) the virus-targeted sets. The number of proteins in each set is given in the parentheses. The fraction of proteins at a particular value of betweenness centrality is the number of proteins having that value and greater divided by the number of proteins in the set.
Figure 4.
The cumulative distributions of degree and betweenness centrality of human proteins excluding Yersinia and HIV data. The number of proteins in each set is given in the parentheses. (A) The degree distributions (B) the betweenness centrality distributions. The fraction of proteins at a particular value of degree is the number of proteins having that value and greater divided by the number of proteins in the set.
GO enrichment analysis
All enriched GO terms for each human protein set are available in Data Sheets 6–10 in Supplementary Material for further detailed analyses. Special attention should be paid to the results of sets of human proteins interacting with three and more bacterial groups and three and more viral groups for a comparison between their infection strategies. The human proteins targeted by more pathogen groups reflect more specificity to infection mechanism of the corresponding pathogen (bacteria or virus). The enriched GO terms in human proteins interacting with both bacterial and viral pathogens are also important to highlight common infection mechanisms. The first 20 enriched GO process terms for three-bacteria-targeted-set, three-viruses-targeted-set, and bacteria–virus-targeted set are listed in Tables 3– 5 to point out the human processes that are attacked by pathogens.
Table 3.
First 20 enriched GO process terms in human proteins targeted by at least three bacterial groups (three-bacteria-targeted set).
| GO process term | p-Value |
|---|---|
| I-kappaB kinase/NF-kappaB cascade | 9.64E−13 |
| Regulation of biological process | 9.69E−10 |
| Biological regulation | 2.59E−09 |
| Negative regulation of biological process | 4.89E−09 |
| Positive regulation by organism of immune response of other organism involved in symbiotic interaction | 6.64E−09 |
| Modulation by organism of immune response of other organism involved in symbiotic interaction | 6.64E−09 |
| Modulation by symbiont of host immune response | 6.64E−09 |
| Positive regulation by symbiont of host immune response | 6.64E−09 |
| Response to immune response of other organism involved in symbiotic interaction | 6.64E−09 |
| Response to host immune response | 6.64E−09 |
| Positive regulation by organism of defense response of other organism involved in symbiotic interaction | 6.64E−09 |
| Positive regulation by symbiont of host defense response | 6.64E−09 |
| Positive regulation by organism of innate immunity in other organism involved in symbiotic interaction | 6.64E−09 |
| Modulation by organism of innate immunity in other organism involved in symbiotic interaction | 6.64E−09 |
| Pathogen-associated molecular pattern dependent modulation by organism of innate immunity in other organism involved in symbiotic interaction | 6.64E−09 |
| Modulation by organism of defense response of other organism involved in symbiotic interaction | 6.64E−09 |
| Pathogen-associated molecular pattern dependent induction by organism of innate immunity of other organism involved in symbiotic interaction | 6.64E−09 |
| Modulation by symbiont of host defense response | 6.64E−09 |
| Pathogen-associated molecular pattern dependent induction by symbiont of host innate immunity | 6.64E−09 |
| Modulation by symbiont of host innate immunity | 6.64E−09 |
See Data Sheet 10 in Supplementary Material for the whole list and the human proteins corresponding to each GO term.
Table 5.
First 20 enriched GO process terms in human proteins targeted by both bacterial and viral groups (bacteria–virus-targeted set).
| GO process term | p-Value |
|---|---|
| Interspecies interaction between organisms | 1.64E−52 |
| Multi-organism process | 1.01E−47 |
| Positive regulation of biological process | 5.62E−47 |
| Regulation of biological process | 1.32E−42 |
| Biological regulation | 2.66E−40 |
| Positive regulation of cellular process | 4.59E−40 |
| Negative regulation of biological process | 6.32E−37 |
| Regulation of cellular process | 2.00E−36 |
| Negative regulation of cellular process | 4.84E−32 |
| Regulation of protein metabolic process | 7.68E−30 |
| Regulation of macromolecule metabolic process | 3.21E−29 |
| Regulation of cellular protein metabolic process | 1.39E−28 |
| Regulation of cell death | 1.74E−28 |
| Positive regulation of macromolecule metabolic process | 1.91E−28 |
| Positive regulation of cellular metabolic process | 7.04E−28 |
| Regulation of programmed cell death | 1.13E−27 |
| Cellular macromolecule metabolic process | 1.94E−27 |
| Positive regulation of metabolic process | 2.92E−27 |
| Negative regulation of macromolecule metabolic process | 5.31E−27 |
| Regulation of apoptosis | 6.54E−27 |
See Data Sheet 10 in Supplementary Material for the whole list and the human proteins corresponding to each GO term.
Discussion
In this study, we aim to provide a general overview of infection strategies used by different pathogens based on the comprehensive PHI data in PHISTO. Although large-scale pathogen–human protein interaction data have been identified in the last few years, the data for fungal and protozoan systems are still scarce (Table 1) to extract significant conclusions about their infection mechanisms. On the other hand, interspecies protein interaction networks for bacterial and viral pathogens with human have been identified, enough to provide some insights about their strategies to subvert human cellular processes during infection.
With increasing PHI data of bacterial and viral pathogens, studies have been performed to enlighten specific bacteria–human (Mogensen et al., 2006; Dyer et al., 2010) and virus–human (Filippova et al., 2004; Uetz et al., 2006; Calderwood et al., 2007; König et al., 2008) interaction systems. Although some studies provided global views of infection strategies of viruses (Dyer et al., 2008) and bacteria (Dyer et al., 2010) separately, they do not provide a direct comparison between bacterial and viral infections. In fact, only <2% of the PHI data of Dyer et al. (2008) are for bacteria–human interactions whereas it is more than 36% in our database of PHISTO. Hence, our study constitutes the first extensive comparison between bacteria–human and virus–human interspecies protein interaction networks to retrieve information about infection strategies specific to each system and then common to both systems. Our findings should be interpreted with caution since the protein interaction networks between pathogens and human are not complete yet.
Special infection strategies
In recent studies it has been suggested that viral proteins (Calderwood et al., 2007; Dyer et al., 2008; Itzhaki, 2011) and bacterial proteins (Dyer et al., 2010) have evolved to preferentially interact with hubs and bottlenecks in the human PPI network. The degree and betweenness centrality distributions of the bacteria-targeted and virus-targeted human protein sets are displayed in comparison with non-targeted proteins in the human PPI network in Figures 2 and 3. We observe that the degree and centrality values of human proteins increase with increasing number of targeting bacterial and viral groups, confirming the previous results with the most comprehensive PHI data. A novel finding by our comparative analysis is that the increase in degree and centrality values with increasing number of pathogen groups is much more pronounced in virus-targeted cases than bacteria-targeted cases (Figures 2 and 3). Therefore we can conclude that attacking to hub and bottleneck proteins in the human interaction network is more specific to viral infections.
In our PHI data, some pathogen groups are overrepresented with their larger number of reported interactions with human (Table 1). As most of these large-scale data have been produced with high-throughput detection methods, which are prone to experimental biases and errors, it was necessary to check whether the distributions of degree and centrality values of the pathogen-targeted human proteins would be same without the groups with large number of interacting partners of human proteins (i.e., Bacillus, Yersinia, and HIV). Hence, we performed the above-mentioned analyses with human protein sets excluding these major pathogen groups. 1,199 human proteins targeted by only Yersinia strains were excluded from the bacteria-targeted set, and 1,283 human proteins targeted by only HIV strains were excluded from the virus-targeted set to obtain the degree and betweenness centrality values of the remaining human proteins. We also analyzed the human proteins targeted by bacteria other than Bacillus and Yersinia to exclude the effect of large-scale data of the two. 1,199 only Yersinia-targeted and 847 only Bacillus-targeted human proteins were excluded from the bacteria-targeted set. The behavior of the remaining human proteins can be observed in Figure 4 resulting in similar trends with the global case. Additionally, a direct comparison of the degree and centrality between bacteria and virus-targeted interaction partners with respect to non-targeted human proteins is also given in Figure 4. The difference in the behavior of the bacteria- and virus-targeted sets are clear especially in degree distributions (Figure 4A). The degree values of bacteria-targeted human proteins with or without Bacillus and Yersinia are nearly same. On the other hand, attack of viruses to more connected human proteins is more clear when HIV data are excluded.
From the enriched GO process terms in human proteins targeted by at least three bacterial groups (Table 3), we can conclude that bacteria may have adapted to attack proteins involved generally in human immunity pathways. Therefore, the most specific bacterial infection strategy is through evading or suppressing human immune responses as also concluded previously (Lai et al., 2001; Park et al., 2002; Zhang et al., 2005; Dyer et al., 2010). The human immune system is manipulated by bacterial proteins attacking human proteins functioning in innate and adaptive immunity (i.e., TLR4 and TLR7), inflammation (i.e., NF-κB and BCL6), and activation of T cells (i.e., CXCR4 and LCK; Zhang and Ghosh, 2000; Alonso et al., 2004; Oda and Kitano, 2006; Dyer et al., 2010). In our PHI data it is observed that Yersinia bacteria attack all these human defense mechanisms targeting all mentioned human proteins. Proteins of Bacillus and Francisella interact with NF-κB and LCK (Dyer et al., 2010) aiming to disrupt the mechanisms of inflammation and T cell responses. On the other hand, proteins of Chlamydia, Escherichia, and Neisseria interact with crucial players of innate and adaptive immunity, toll-like receptors (TLR4 and TLR7; Croft et al., 2011) to collapse the human immune system. There are several other bacteria-targeted human proteins involved in the immune system. Their interactions with bacterial proteins should be investigated carefully for a complete understanding of bacterial strategies targeting human defense mechanism during infection.
Viruses attack human cellular processes (Table 4) enabling themselves to proliferate in human during infection. All viruses use this mechanism since they need host’s transcriptional machinery for viral genetic material transcription. Even the human proteins targeted by only one viral group are enriched in GO process terms relevant to regulation of cellular mechanisms (Data Sheet 10 in Supplementary Material). Viruses manipulate human cellular mechanisms by interacting with various proteins functioning in cell cycle (i.e., DLG1, PTMA, and EP300), with human transcription factors to promote their own genetic material transcription (i.e., E2F1 and TAF1), with key proteins controlling apoptosis (i.e., P53), and with nuclear membrane proteins for transporting their genetic material across the nuclear membrane (i.e., RAN, and SUMO1; Lechner and Laimins, 1994; Thompson et al., 1997; Carrillo et al., 2004; Thomas et al., 2005; Dyer et al., 2008). In our PHI data, Adenoviruses, HIV, Papillomaviruses, and Polyomaviruses are observed to target one or more proteins in each of four groups; cell cycle proteins, transcription factors, apoptosis regulator, and nuclear membrane proteins. Proteins of Hepatitis viruses interact with PTMA, EP300, TAF1, and p53 while proteins of Herpesviruses interact with PTMA and SUMO1. On the other hand, viral groups of Influenza, Puumala, Tula, SARS, and Vaccinia are observed to target nuclear membrane proteins. The other virus-targeted human proteins involved in cellular mechanisms should be investigated comprehensively for a complete understanding of viral strategies targeting human cellular mechanism.
Table 4.
First 20 enriched GO process terms in human proteins targeted by at least three viral groups (three-viruses-targeted set).
| Go process term | p-Value |
|---|---|
| Interspecies interaction between organisms | 1.89E−40 |
| Multi-organism process | 1.19E−27 |
| Positive regulation of cellular process | 1.12E−17 |
| Positive regulation of biological process | 1.06E−16 |
| Cellular macromolecule metabolic process | 1.12E−15 |
| Nucleic acid metabolic process | 4.49E−14 |
| Positive regulation of macromolecule metabolic process | 4.60E−14 |
| Cell cycle process | 6.72E−14 |
| Positive regulation of gene expression | 1.49E−13 |
| Cell cycle | 2.06E−13 |
| Positive regulation of metabolic process | 3.79E−13 |
| Positive regulation of transcription | 4.37E−13 |
| Macromolecule metabolic process | 8.51E−13 |
| Positive regulation of cellular metabolic process | 3.89E−12 |
| Cellular response to stimulus | 6.61E−12 |
| Positive regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 7.26E−12 |
| Positive regulation of macromolecule biosynthetic process | 1.32E−11 |
| Positive regulation of nitrogen compound metabolic process | 1.41E−11 |
| Positive regulation of transcription, DNA-dependent | 1.44E−11 |
| Regulation of cell cycle | 1.47E−11 |
See Data Sheet 10 in Supplementary Material for the whole list and the human proteins corresponding to each GO term.
We can conclude that the main infection strategies of bacteria and viruses are through attacking human immune system and cellular processes, respectively. However, there are some exceptions such that some bacterial groups target human proteins functioning in cellular mechanisms whereas some viral groups target human proteins functioning in defense mechanisms. In the case of bacteria, the difference might arise from the life-style, e.g., intracellular bacteria like Chlamydia, Listeria, and Mycoplasma are able to grow and reproduce only within the host cells just like viruses (Kaufmann, 1993). Therefore, human protein sets targeted by these intracellular bacterial groups are enriched in GO process terms related to the cellular mechanisms (e.g., regulation of cellular processes, regulation of transcription) in addition to the immune sytem (Data Sheet 6 in Supplementary Material). On the other hand, viruses like herpes and pox (ectromelia, molluscum, orf, vaccinia) viruses as well as HIV have the ability to evade human immune system (Alcami and Koszinowski, 2000) as observed in our results (Data Sheet 9 in Supplementary Material).
For more specific infection strategies of pathogen groups, the results of GO enrichment analysis for the human protein sets targeted by each of the 72 groups in the PHI data can be used (Data Sheets 6–9 in Supplementary Material). Additionally, intranetworks of pathogenic proteins in each pathogen group should be analyzed for drug target identification after a thorough understanding of pathogenesis via interspecies protein interactions.
Common infection strategies
In spite of the difference in the trends of distributions of degree and centrality values of human proteins in bacteria-targeted and virus-targeted sets, the tendency to attack human proteins that are highly connected (hubs) and central to shortest paths (bottlenecks) is common to all types of pathogens. We observed in our PHI data that the degree and centrality values of pathogen-targeted human proteins are generally greater than non-targeted ones. This infection strategy of pathogens, attacking more connected and central nodes in the human PPI network, is probably due to enabling themselves to control and disrupt essential complexes and pathways more easily. With scale-free nature, the human PPI network is robust to attacks on random nodes. However, the selective attacks to even a small number of nodes of high degree can dramatically change the topology and functionality of the network (Albert et al., 2000; Li et al., 2006).
Although bacteria and viruses have a tendency to interact with different human proteins (Figure 1), they together target those (779 human proteins) enriched in the regulation of metabolic processes in addition to cellular processes (Table 5). For instance, a pyruvate kinase isozyme, KPYM, functions in glycolysis catalyzing the transfer of a phosphoryl group from phosphoenolpyruvate (PEP) to ADP, generating ATP. This metabolic human protein is targeted by three bacterial (Bacillus, Francisella, and Yersinia) and three viral groups (Hepatitis, Herpesviruses, and Papillomaviruses) in the PHI data. Alpha-enolase is another bacteria–virus-targeted enzyme which functions in glycolysis, just before KPYM enzyme, converting 2-phospho-glycerate to PEP. A metabolic step operating again around lower glycolysis is the production of lactate from pyruvate. Both isoenzymes (LDHA, LDHB) are found to be a target for bacterial and viral groups. In addition to lower glycolysis, some enzymes functioning in lipid metabolism (ACSA, ACOT9, CPT1A) were identified as common targets of bacteria and viruses. Interestingly, two enzymes functioning for protection against oxidative-stress are in our common-target list: catalase (CATA) and glutathione peroxidase 3 (GPX3). These enzymes remove H2O2, which is a reactive oxygen species (ROS) harmful for the cell.
To our knowledge, the human proteins targeted by both bacteria and viruses have not been investigated in any previous study. Through our analyses using large-scale PHI data we can conclude that both bacteria and viruses attack to the proteins functioning in human metabolic processes as a common infection strategy. All bacteria–viruses-targeted human proteins involved in metabolic processes should be investigated carefully for a complete picture of commonalities in bacterial and viral infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Microbial_Immunology/10.3389/fmicb.2012.00046/abstract
Data Sheets 1–4| Pathogen–human PHIs.
Bacteria–human PHIs.
Fungi–human PHIs.
Protozoa–human PHIs.
Virus–human PHIs.
Properties of pathogen-targeted set.
Data Sheets 6–10| Gene ontology enrichment results.
Gene ontology enrichment results of sets of human proteins targeted by each bacterial group.
Gene ontology enrichment results of sets of human proteins targeted by each fungal group.
Gene ontology enrichment results of sets of human proteins targeted by each protozoan group.
Gene ontology enrichment results of sets of human proteins targeted by each viral group.
Gene ontology enrichment results of other sets of human proteins (pathogen-targeted, virus-targeted, two-virus-targeted, three-virus-targeted, bacteria-targeted, two-bacteria-targeted, three-bacteria-targeted, fungi-targeted, protozoa-targeted, and bacteria–virus-targeted).
Acknowledgments
We thank Ali Semih Sayilirbaş for his invaluable contribution to the development of PHISTO database. The financial support for this research was provided by the Research Funds of Boğaziçi University and TÜBİTAK through projects 5554D and 110M428, respectively. The scholarship for Saliha Durmuş Tekir is provided by TÜBİTAK, is gratefully acknowledged.
Footnotes
References
- Albert R., Jeong H., Barabasi A. L. (2000). Error and attack tolerance of complex networks. Nature 406, 378–382 10.1038/35019019 [DOI] [PubMed] [Google Scholar]
- Alcami A., Koszinowski U. H. (2000). Viral mechanisms of immune evasion. Immunol. Today 21, 447–455 10.1016/S0167-5699(00)01632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Bottini N., Bruckner S., Rahmouni S., Williams S., Schoenberger S. P., Mustelin T. (2004). Lck dephosphorylation at Tyr-394 and inhibition of T cell antigen receptor signaling by Yersinia phosphatase YopH. J. Biol. Chem. 279, 4922–4928 10.1074/jbc.M403442200 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A. L., Oltvai Z. N. (2004). Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007). Epstein–Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 10.1073/pnas.0702332104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo E., Garrido E., Gariglio P. (2004). Specific in vitro interaction between papillomavirus E2 proteins and TBP-associated factors. Intervirology 47, 342–349 10.1159/000080878 [DOI] [PubMed] [Google Scholar]
- Ceol A., Catr Aryamontri A., Licata L., Peluso D., Briganti L., Perfetto L., Castagnoli L., Cesareni G. (2010). MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 38, D532–D539 10.1093/nar/gkp983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., Jupe S., Kalatskaya I., Mahajan S., May B., Ndegwa N., Schmidt E., Shamovsky V., Yung C., Birney E., Hermjakob H., D’Eustachio P., Stein L. (2011). Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697 10.1093/nar/gkq1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T., Dyer M. D., Murali T. M., Sobral B. W. (2009). PIG – the pathogen interaction gateway. Nucleic Acids Res. 37, D647–D650 10.1093/nar/gkp371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer M. D., Murali T. M., Sobral B. W. (2008). The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 4, e32. 10.1371/journal.ppat.0040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer M. D., Neff C., Dufford M., Rivera C. G., Shattuck D., Bassaganya-Riera J., Murali T. M., Sobral B. W. (2010). The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS ONE 5, e12089. 10.1371/journal.pone.0012089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova M., Parkhurst L., Duerksen-Hughes P. J. (2004). The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 279, 25729–25744 10.1074/jbc.M401012200 [DOI] [PubMed] [Google Scholar]
- Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspe G., Tiollais P., Transy C., Legrain P. (2000). A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242, 369–379 10.1016/S0378-1119(99)00511-9 [DOI] [PubMed] [Google Scholar]
- Itzhaki Z. (2011). Domain-domain interactions underlying herpes virus human protein-protein interaction networks. PLoS ONE 6, e21724. 10.1371/journal.pone.0021724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H. E. (1993). Immunity to intracellular bacteria. Annu. Rev. Immunol. 11, 129–163 10.1146/annurev.iy.11.040193.001021 [DOI] [PubMed] [Google Scholar]
- Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., Jandrasits C., Jimenez R. C., Khadake J., Mahadevan U., Masson P., Pedruzzi I., Pfeiffenberger E., Porras P., Raghunath A., Roechert B., Orchard S., Hermjakob H. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–D846 10.1093/nar/gkr1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008). Global analysis of host-pathogen interactions that regulate early stage HIV-1 replication. Cell 135, 49–60 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Nanduri B. (2010). HPIDB – a unified resource for host-pathogen interactions. BMC Bioinformatics 11, S16. 10.1186/1471-2105-11-S6-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005). A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107 10.1038/nature04104 [DOI] [PubMed] [Google Scholar]
- Lai X. H., Golovliov I., Sjostedt A. (2001). Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69, 4691–4694 10.1128/IAI.69.7.4691-4694.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M. S., Laimins L. A. (1994). Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J. Virol. 68, 4262–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li J., Ouyang S., Wang J., Wu S., Wan P., Zhu Y., Xu X., He F. (2006). Protein interaction networks of Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster: large-scale organization and robustness. Proteomics 6, 456–461 10.1002/pmic.200500228 [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D’Eustachio P. (2009). Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37, D619–D622 10.1093/nar/gkn863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S., Holtzman T., Moss B., Fields S. (2000). Genome-wide analysis of vaccinia virus protein–protein interactions. Proc. Natl. Acad. Sci. U.S.A. 97, 4879–4884 10.1073/pnas.080078197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T. H., Paludan S. R., Kilian M., Østergaard L. (2006). Live Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis activate the inflammatory response through toll-like receptors 2, 4, and 9 in species-specific patterns. J. Leukoc. Biol. 80, 267–277 10.1189/jlb.1105626 [DOI] [PubMed] [Google Scholar]
- Oda K., Kitano H. (2006). A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2, 20060015. 10.1038/msb4100057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Greten F. R., Li Z.-W., Karin M. (2002). Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science 297, 2048–2051 10.1126/science.1074111 [DOI] [PubMed] [Google Scholar]
- Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001). The protein–protein interaction map of Helicobacter pylori. Nature 409, 211–215 10.1038/35051615 [DOI] [PubMed] [Google Scholar]
- Salwinski L., Miller C. S., Smith A. J., Pettit F. K., Bowie J. U., Eisenberg D. (2004). The database of interacting proteins: 2004 update. Nucleic Acids Res. 32, D449–D451 10.1093/nar/gkh086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B. J., Chatr-Aryamontri A., Boucher L., Oughtred R., Livstone M. S., Nixon J., Van Auken K., Wang X., Shi X., Reguly T., Rust J. M., Winter A., Dolinski K., Tyers M. (2011). The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 39, D698–D704 10.1093/nar/gkq1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Massimi P., Navarro C., Borg J. P., Banks L. (2005). The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24, 6222–6230 10.1038/sj.onc.1208757 [DOI] [PubMed] [Google Scholar]
- Thompson D. A., Belinsky G., Chang T. H.-T., Jones D. L., Schlegel R., Münger K. (1997). The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene 15, 3025–3035 10.1038/sj.onc.1201495 [DOI] [PubMed] [Google Scholar]
- Uetz P., Dong Y., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S., Roupelieva M., Rose D., Fossum E., Haas J. (2006). Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 10.1126/science.1116804 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cui T., Zhang C., Yang M., Huang Y., Li W., Zhang L., Gao C., He Y., Li Y., Huang F., Zeng J., Huang C., Yang Q., Tian Y., Zhao C., Chen H., Zhang H., He Z. G. (2010). Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome. Res. 9, 6665–6677 10.1021/pr100808n [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2008). The Global Burden of Disease: 2004 Update. Geneva: WHO Press [Google Scholar]
- Yu H., Kim P. M., Sprecher E., Trifonov V., Gerstein M. (2007). The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 3, e59. 10.1371/journal.pcbi.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Ghosh S. (2000). Molecular mechanisms of NF-κB activation induced by bacterial lipopolysaccharide through toll-like receptors. J. Endotoxin Res. 6, 453–457 10.1177/09680519000060060701 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ting A. T., Marcu K. B., Bliska J. B. (2005). Inhibition of MAPK and NF-kappa B pathways is necessary for rapid apoptosis in macrophages infected with Yersinia. J. Immunol. 174, 7939–7949 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacteria–human PHIs.
Fungi–human PHIs.
Protozoa–human PHIs.
Virus–human PHIs.
Properties of pathogen-targeted set.
Gene ontology enrichment results of sets of human proteins targeted by each bacterial group.
Gene ontology enrichment results of sets of human proteins targeted by each fungal group.
Gene ontology enrichment results of sets of human proteins targeted by each protozoan group.
Gene ontology enrichment results of sets of human proteins targeted by each viral group.
Gene ontology enrichment results of other sets of human proteins (pathogen-targeted, virus-targeted, two-virus-targeted, three-virus-targeted, bacteria-targeted, two-bacteria-targeted, three-bacteria-targeted, fungi-targeted, protozoa-targeted, and bacteria–virus-targeted).