Abstract
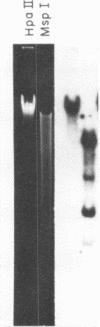
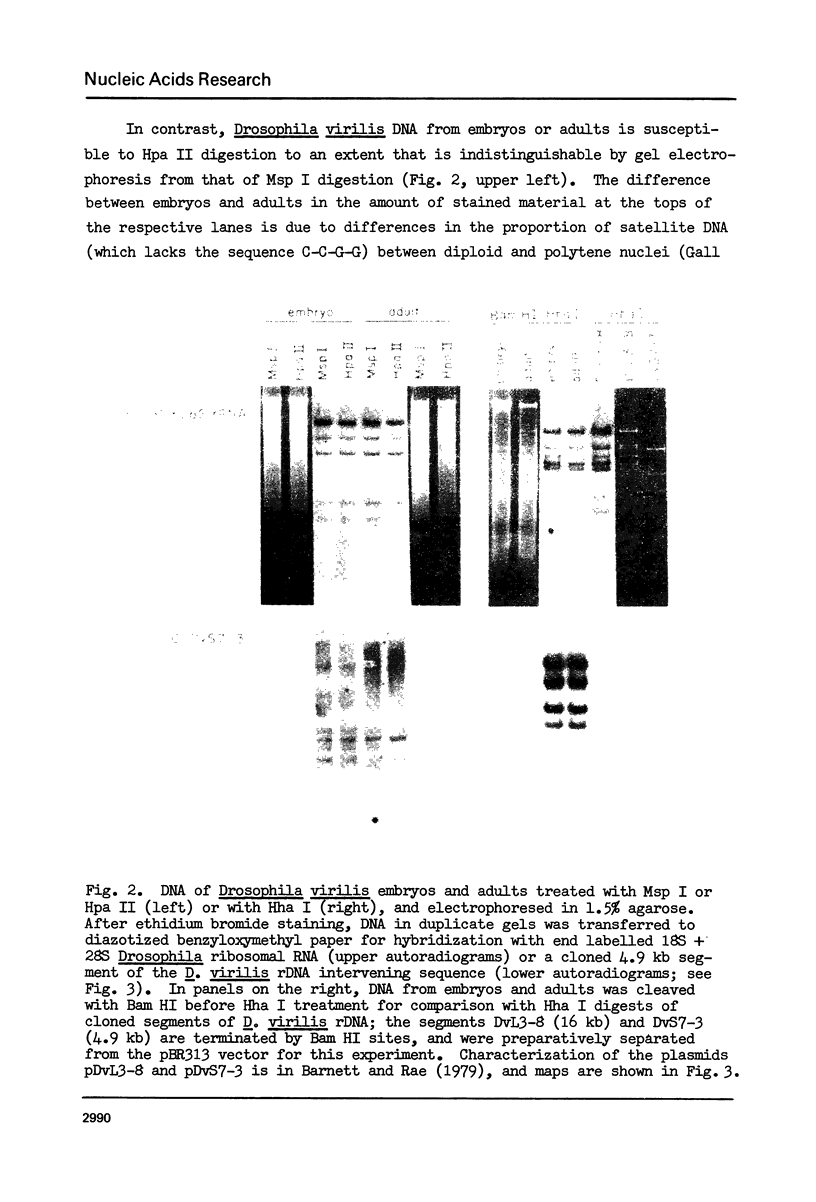
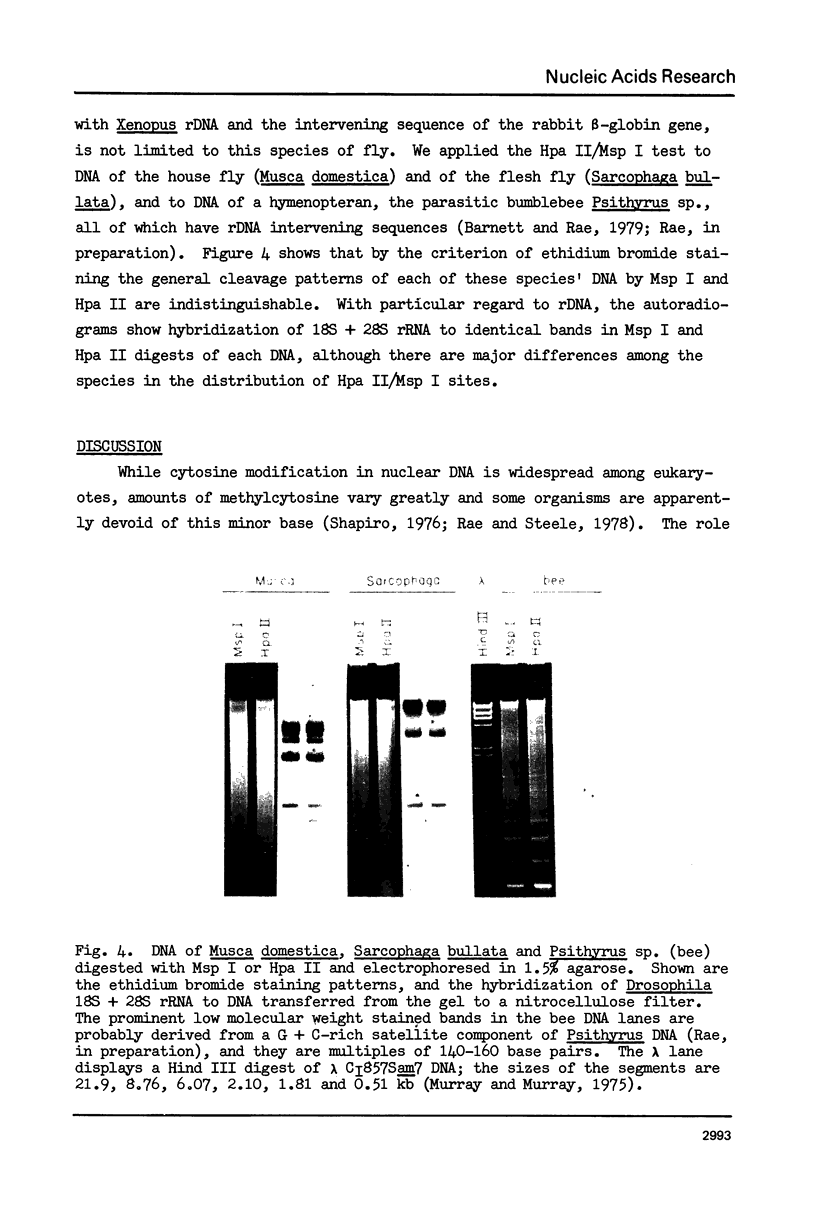
Cytosine residues in C-G dinucleotides are frequently methylated in eukaryote DNA. In DNA of the dinoflagellate C. cohnii, the sequence C-MeC-G-G apparently renders Hpa II (C-C-G-G) incapable of digesting whole cell DNA in general, and rDNA in particular. Msp I, which also recognizes C-C-G-G but cleaves irrespective of methylation, degrades C. cohnii DNA and produces rDNA segments of 10.2 to 1.4 kb. We have applied this Hpa II/Msp I test to unfractionated DNA, and to rDNA and the rDNA intervening sequence of Drosophila virilis embryos and adults. There is no evidence of C-MeC-G-G sequences in either developmental stage of this species. Absence of G-MeC-G-C from coding and intervening sequences of rDNA was shown in comparisons of Hha I (G-C-G-C) cleavage patterns of unfractionated DNA and cloned (unmodified) segments of rDNA. Comparisons of Hpa II and Msp I cleavage products of DNA from the house fly, the flesh fly and a bumblebee also revealed no internal cytosine methylation in the sequence C-C-G-G. Because amounts of McC in C-G dinucleotides vary greatly among species, from apparent nonexistence to substantial proportions, no inference may yet be drawn about the role of such base modifications in DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARGYRAKIS M. P., BESSMAN M. J. Analysis of the base composition of the DNA from Drosophila melanogaster. Biochim Biophys Acta. 1963 May 28;72:122–124. [PubMed] [Google Scholar]
- Barnett T., Rae P. M. A 9.6 kb intervening sequence in D. virilis rDNA, and sequence homology in rDNA interruptions of diverse species of Drosophila and other diptera. Cell. 1979 Apr;16(4):763–775. doi: 10.1016/0092-8674(79)90092-8. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Brown D. D., Reeder R. H. Composition and structure of chromosomal and amplified ribosomal DNA's of Xenopus laevis. J Mol Biol. 1970 Jul 28;51(2):341–360. doi: 10.1016/0022-2836(70)90147-6. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Atherton D. D. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974 Jan 5;85(4):633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Glover D. M., Hogness D. S. A novel arrangement of the 18S and 28S sequences in a repeating unit of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):167–176. doi: 10.1016/0092-8674(77)90212-4. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Rae P. M. 5-Hydroxymethyluracil in the DNA of a dinoflagellate. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1141–1145. doi: 10.1073/pnas.70.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Modified bases in the DNAs of unicellular eukaryotes: an examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978 Apr;10(1-2):37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- SINSHEIMER R. L. The action of pancreatic desoxyribonuclease. I. Isolation of mono- and dinucleotides. J Biol Chem. 1954 May;208(1):445–459. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]