Abstract
The 3' flanking sequences of all five histone genes have been sequenced in the histone DNA clone h19 of the sea urchin Psammechinus miliaris. A large (23 bp) and a small (10 bp) conserved sequence was found by sequence comparison, some 29-40 bp downstream from the termination codon. 12 bases of the larger homology block show a dyad symmetry. The available sequences of clone h22 of the same species and those of the histone clones pSp2 and pSp17 of Strongylocentrotus purpuratus, another sea urchin species, fit well into this comparison. Two types of sequences are involved in the dyad symmetry; one is H1, H3 and H4 specific, the other is H2A and H2B specific. If these conserved sequences are transcribed, a hairpin loop could form in the RNA molecules. This secondary structure might serve as a recognition signal for a regulatory protein.
Full text
PDF


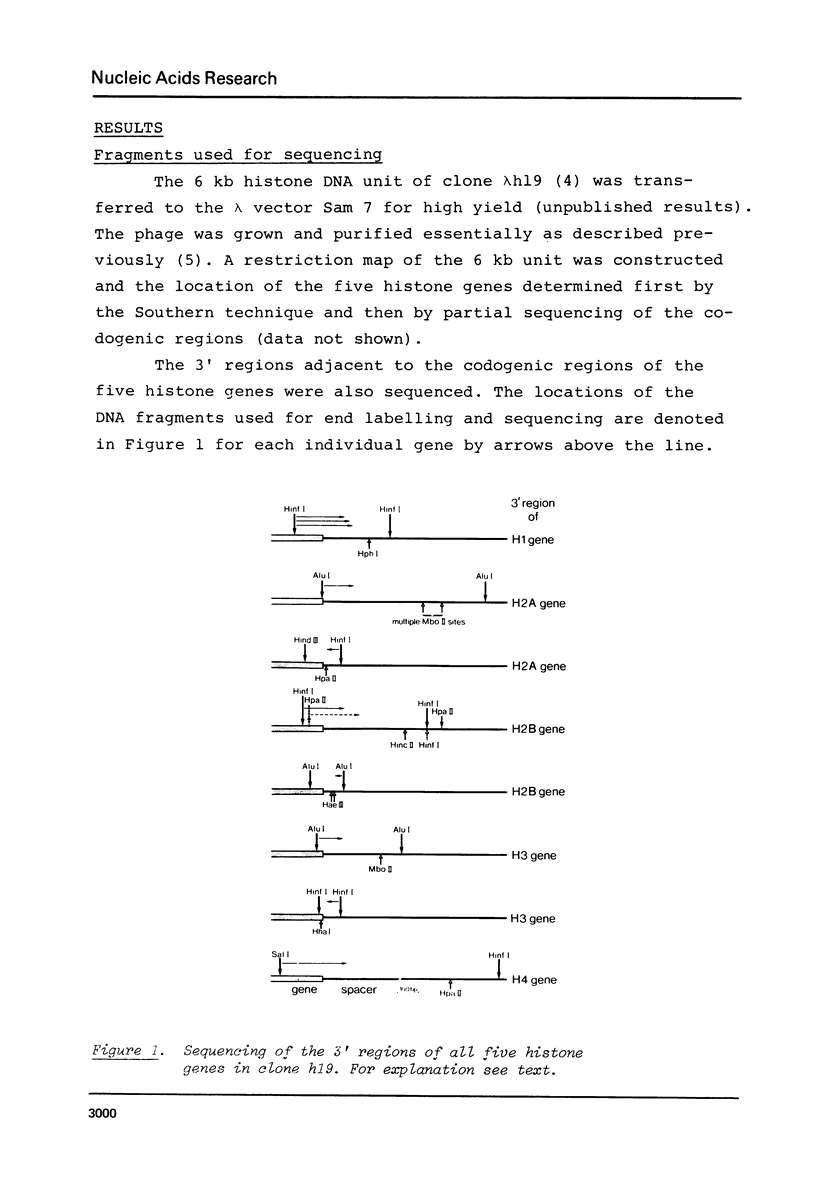








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnstiel M. L., Schaffner W., Smith H. O. DNA sequences coding for the H2B histone of Psammechinus miliaris. Nature. 1977 Apr 14;266(5603):603–607. doi: 10.1038/266603a0. [DOI] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Brandt W. F., Strickland W. N., Strickland M., Carlisle L., Woods D., von Holt C. A histone programme during the life cycle of the sea urchin. Eur J Biochem. 1979 Feb 15;94(1):1–10. doi: 10.1111/j.1432-1033.1979.tb12864.x. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Smith H. O., Schaffner W., Gross K. W., Birnstiel M. L. Integration of eukaryotic genes for 5S RNA and histone proteins into a phage lambda receptor. Nucleic Acids Res. 1976 Oct;3(10):2617–2632. doi: 10.1093/nar/3.10.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Brown D. D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978 Dec;15(4):1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- Kunkel N. S., Hemminki K., Weinberg E. S. Size of histone gene transcripts in different embryonic stages of the sea urchin, Strongylocentrotus purpuratus. Biochemistry. 1978 Jun 27;17(13):2591–2598. doi: 10.1021/bi00606a021. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Levy S., Childs G., Kedes L. Sea urchin nuclei use RNA polymerase II to transcribe discrete histone RNAs larger than messengers. Cell. 1978 Sep;15(1):151–162. doi: 10.1016/0092-8674(78)90091-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rich A., Schimmel P. R. Structural organization of complexes of transfer RNAs with aminoacyl transfer RNA synthetases. Nucleic Acids Res. 1977;4(5):1649–1665. doi: 10.1093/nar/4.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Dunn J. J. A nucleotide sequence from a ribonuclease III processing site in bacteriophage T7 RNA. Proc Natl Acad Sci U S A. 1977 Mar;74(3):822–826. doi: 10.1073/pnas.74.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Scherer G. E., Walkinshaw M. D., Arnott S. A computer aided oligonucleotide analysis provides a model sequence for RNA polymerase-promoter recognition in E.coli. Nucleic Acids Res. 1978 Oct;5(10):3759–3773. doi: 10.1093/nar/5.10.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wienand U., Schwarz Z., Feix G. Electrophoretic elution of nucleic acids from gels adapted for subsequent biological tests. Application for analysis of mRNAs from maize endosperm. FEBS Lett. 1979 Feb 15;98(2):319–323. doi: 10.1016/0014-5793(79)80208-2. [DOI] [PubMed] [Google Scholar]
- Zimmermann R. A., Mackie G. A., Muto A., Garrett R. A., Ungewickell E., Ehresmann C., Stiegler P., Ebel J. P., Fellner P. Location and characteristics of ribosomal protein binding sites in the 16S RNA of Escherichia coli. Nucleic Acids Res. 1975 Feb;2(2):279–302. doi: 10.1093/nar/2.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]



