Abstract
Lentiviruses are capable of infecting many cells irrespective of their cycling status, stably inserting DNA copies of the viral RNA genomes into host chromosomes. This property has led to the development of lentiviral vectors for high-efficiency gene transfer to a wide variety of cell types, from slowly proliferating hematopoietic stem cells to terminally differentiated neurons. Regardless of their advantage over gammaretroviral vectors, which can only introduce transgenes into target cells that are actively dividing, lentiviral vectors are still susceptible to chromosomal position effects that result in transgene silencing or variegated expression. In this chapter, various genetic regulatory elements are described that can be incorporated within lentiviral vector backbones to minimize the influences of neighboring chromatin on single-copy transgene expression. The modifications include utilization of strong internal enhancer-promoter sequences, addition of scaffold/matrix attachment regions, and flanking the transcriptional unit with chromatin domain insulators. Protocols are provided to evaluate the performance as well as the relative biosafety of lentiviral vectors containing these elements.
Keywords: Lentiviral vectors, position effects, transgene silencing, insulators, enhancer blocking elements, scaffold/matrix attachment regions, insertional mutagenesis, genotoxicity
1. Introduction
Gammaretroviral vectors derived from Moloney murine leukemia virus (MLV) integrate into the host cell genome and therefore have been widely used for stable gene transfer (1). One of the main limitations of gammaretroviral vectors, however, is their dependence on nuclear membrane dissolution during mitosis for access to and integration into target cell chromosomes (2). By comparison, lentiviruses such as human immunodeficiency virus type 1 (HIV-1) have the capacity to infect many non-dividing as well as dividing cells (3,4). This property prompted the development of HIV-1-based lentiviral vectors that can efficiently transduce slowly proliferating cells such as hematopoietic stem cells as well as terminally differentiated cells such as neurons (1,5,6).
Another drawback of conventional MLV-based gammaretroviral vectors is their frequent silencing after transduction of certain cell types, particularly embryonic stem cells (ESCs) (7–9). Silencing elements have been identified in the MLV long terminal repeat (LTR) (10,11) and the primer-binding site (12–14), the removal of which has resulted in improved gammaretroviral vectors that exhibit an increased probability of transgene expression in ESCs and other primitive cells (1,15–18). Additionally, position effects due to chromatin structure at the vector insertion site contribute to gammaretroviral vector silencing shortly after integration, to expression variegation (in which variable expression is observed within the clonal progeny of a target cell), and to extinction of expression (in which expression is down-regulated with time or during differentiation) (19–22). Accumulating data suggest that lentiviral vectors are also susceptible to stem cell-associated and chromosomal position effects that result in variegated expression and silencing (23–29).
Progress in the field of gene expression and chromatin structure has led to the identification of genetic regulatory elements capable of counteracting position effects (reviewed in ref. 1): strong enhancer-promoters allow maintenance of active chromatin configurations even at unfavorable heterochromatin insertion sites; scaffold/matrix attachment regions (S/MARs) serve to organize chromatin into structural domains via anchorage to the nucleoskeleton; and chromatin domain insulators act as barrier elements that can physically block the influence of repressive chromatin structure. Most insulators also posses enhancer blocking activity distinct from their anti-silencer properties.
1.1. Strong Enhancer-Promoter Sequences
Silencing is facilitated by a large number of protein factors which in some cases can be antagonized by proteins such as transcriptional activators (30). For example, some strong promoters have been shown to counteract chromosomal position effects or withstand silencing induced during differentiation (31,32). Enhancers and promoters are believed to be foci for changes in chromatin structure although the mechanisms underlying these changes are incompletely understood (33–35). In cultured cells, it has been shown that enhancers could retard silencing of integrated reporter genes without significantly affecting the level at which they are expressed (36,37). These results suggest that enhancers function to disrupt or prevent the formation of repressive chromatin structures, thus permitting maintenance of gene expression.
Some of the commonly used strong promoters include the murine stem cell virus (MSCV) LTR (17), the human elongation factor 1α (EF1α) promoter (38,39) and the composite promoter (CAG) composed of the cytomegalovirus (CMV) immediate early region enhancer linked to chicken β-actin promoter sequences (40). The MSCV LTR is permissive for expression in murine and human hematopoietic stem/progenitor cells (HSPCs) with long-term hematopoietic repopulating potential (18,41–44). The EF1α promoter drives widespread expression in vivo (45) and has been identified as a strong promoter in primary human CD34+ HSPCs (46,47). The composite CAG promoter functions efficiently in murine ESCs and in transgenic mice (48,49). We previously developed a series of self-inactivating (SIN) HIV-1-based lentiviral vectors containing a variety of strong viral and cellular enhancer-promoters (43,50). Of those evaluated, the MSCV LTR, the EF1α promoter and the CAG promoter consistently mediated high and sustained transgene expression in the cell types studied.
We subsequently described an MSCV-based SIN gammaretroviral vector, MSinSB, which utilizes a modified version of the composite CAG promoter (32). When used to transduce murine ESCs, the MSinSB vector directed high transgene expression levels which were maintained during in vitro hematopoietic differentiation of the cells. This was in sharp contrast to the parental MSCV gammaretroviral vector which was tested in parallel and found to almost completely silence during the ESC differentiation process. Persistent high level expression of the MSinSB gammaretroviral vector was also demonstrated in murine bone marrow transplant recipients and following in vitro myelomonocytic differentiation of human CD34+ cord blood HSPCs.
1.2. Scaffold/Matrix Attachment Regions (S/MARs)
Eukaryotic cells have evolved specialized DNA elements known as boundary elements that mark the borders of adjacent chromatin domains and protect active regions from the repressive effects of nearby heterochromatin. One class of element, termed S/MARs, anchors chromatin to a skeleton of protein cross-ties called the nuclear scaffold (in metaphase) or nuclear matrix (in interphase) (51–57). It has been suggested that S/MARs function by forming chromatin loops, bringing enhancers and other distal regulatory elements in close proximity to their corresponding promoter sequences. Additionally, they may augment transgene expression by protecting the enhancer from DNA methylation (58), which may involve localized attraction of histone acetylation (59). In this regard, insertion of the S/MAR believed to define the upstream border of the human interferon-β locus (IFN-SAR) into gammaretroviral vectors has been shown to result in improved transgene expression (55,60–62).
We introduced the IFN-SAR into a series of HIV-1-based lentiviral vectors and analyzed reporter expression in transduced populations and individual clones of the human CD34+ HSPC line KG1a (50). After 4 months of continuous culture, significantly higher levels of transgene expression were observed from three different internal promoters in cells transduced with the modified vectors containing the IFN-SAR by comparison to the respective parental lentiviral vector backbones. Others have reported that inclusion of a S/MAR from the mouse immunoglobulin kappa light chain gene significantly increased the probability of transgene expression from SIN lentiviral vectors containing two different internal promoters (63).
1.3. Insulators
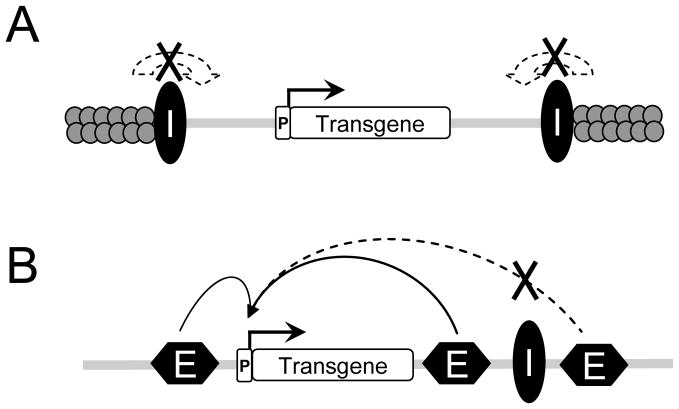
Chromatin domain insulators are DNA boundary elements capable of suppressing repressive position effects and in most cases also blocking the action of distant enhancers (64,65). The best studied vertebrate element is a 1.2-kilobase (kb) fragment encompassing a DNase I hypersensitive site at the 5′ end (5′HS4) of the chicken β-globin locus (66,67). Extensive characterization by Felsenfeld and colleagues has shown that the chicken β-globin 5′HS4 insulator can protect against chromosomal position effects and also provide enhancer blocking function (Fig. 5.1) (65–72). Almost all of the enhancer blocking activity of the 1.2-kb 5′HS4 insulator fragment can be conferred by a 42-bp sequence (DNase I-footprinted region II; FII) that is bound by the zinc finger protein CTCF (CCCTC-binding factor) (69,71). Consistent with this role, analyses of the genome-wide distribution of CTCF binding sites demonstrate a tendency of CTCF to reside in loci that separate chromosomal domains (73), perhaps facilitating the formation of chromatin loops (72,74). Recent studies have revealed that cohesin complexes, best known for sister chromatid cohesion, contribute to CTCF-dependent enhancer blocking (75,76). It is important to point out, however, that the contribution of CTCF to the anti-silencer properties associated with the barrier function of insulators remains unclear (71,76–78).
Fig. 5.1.
Schematic diagram illustrating the two main and separate properties of insulators. (A) An insulator element (I) can protect against chromosomal position effects by blocking the advancement of adjacent repressive chromatin. (B) Many insulators also provide enhancer blocking function by preventing the activity of an enhancer (E), but only when placed between the enhancer and a promoter (P).
In an attempt to eliminate chromosomal position effects and transgene silencing, we and others have used insulators to flank gammaretroviral and lentiviral vectors. Some improvements were obtained when a single copy of the 1.2-kb fragment of the chicken β-globin 5′HS4 insulator was added (50,77,79–81). However, in our experience, the titers of 5′HS4 insulator-containing vectors were generally reduced (50). Moreover, Felsenfeld and colleagues reported superior protection in cell culture transfection and Drosophila transgenesis experiments when the transgene was flanked by two direct copies of the 1.2-kb 5′HS4 fragment (66). In further work by this group, the chicken β-globin 5′HS4 insulator was mapped to a 5′ 250-bp fragment (encompassing the FII region containing the CTCF binding site), two copies of which were shown to provide complete insulator function (70). Therefore, as one approach toward alleviating ESC differentiation-associated gammaretroviral extinction, we inserted two copies of the 250-bp 5′HS4 core in tandem into the U3 region of the 3′ LTR of a MSCV-based gammaretroviral vector (32). However, analysis of proviral DNA from stably transduced cells indicated that the direct repeat configuration was highly unstable, with one copy of the 5′HS4 insulator core being deleted at high frequency during replication. More recently, a 400-bp fragment consisting of the 5′HS4 insulator 250-bp core plus 150 bp of 3′ flanking sequences has been reported to protect gammaretroviral vector and lentiviral vector transgene expression to the same degree as the full-length 1.2-kb fragment (82).
Given the ability of the chicken β-globin 5′HS4 insulator and the IFN-SAR to individually provide some protection against position effects to gammaretroviral vectors, and in view of their complementary properties and structural features—i.e., the core element of the 5′HS4 insulator is GC-rich with a high density of cytidine-guanosine (CpG) dinucleotides (reminiscent of a CpG island), whereas S/MARs are AT-rich sequences (57)—plus the fact that lentiviral vectors can readily accommodate large DNA inserts (83), we were interested in evaluating the utility of combining these elements (50). Our findings in the human CD34+ HSPC line KG1a and in primary human CD34+ cord blood cells demonstrated that position-effect protection of lentiviral vector-mediated transgene expression provided by combinatorial association of the 5′HS4 insulator and the IFN-SAR was superior to that conferred by either element alone. The lentiviral vector backbone utilizing the 5′HS4 insulator/IFN-SAR combination has also been successfully used to confer stable long-term transgene expression in human ESCs and their differentiated progeny (84,85).
Kwaks et al. identified a number of novel human genetic elements that exhibit enhancer blocking activity in a screen of genomic DNA fragments that protected a reporter gene from repression by the chromatin-associated repressor proteins, heterochromatin protein 1 or the Polycomb group protein HPC2 (86). One of these so-called “anti-repressor” elements, element 40, has subsequently been combined with the IFN-SAR and shown to minimize variegated expression in lentiviral transgenic mice (87,88).
Insulation of the transgene from the host cell genome has the reciprocal benefit of protecting the host cell genome against insertional mutagenesis caused by integrating vectors (i.e., reducing their genotoxic potential) (89–91). We recently reported the development of a 77-bp element, FII/BEAD-A (FB), which combines the minimal enhancer blocking components of the chicken β-globin 5′HS4 insulator (a modified 42-bp FII region containing the CTCF binding site) and a homologous CTCF binding site-containing region from the human T-cell receptor α/δ BEAD-1 insulator (92). With a new flow cytometry-based assay, we showed that the FB element was as effective in enhancer blocking activity as the prototypical 1.2-kb 5′HS4 insulator fragment. Notably, using an in vitro insertional mutagenesis assay involving primary murine HSPCs (93), we found that both SIN gammaretroviral and lentiviral vectors containing the FB element exhibited greatly reduced transforming potential—to background levels under the experimental conditions used—compared to their unshielded counterparts.
This chapter describes strategies to incorporate strong enhancer-promoter sequences, the IFN-SAR, and the chicken β-globin 5′HS4 insulator into lentiviral vectors, and provides protocols to evaluate insulator performance as well as the relative biosafety of the modified vectors.
2. Materials
2.1. Barrier Function Assay
2.1.1. Design and Construction of Reporter Plasmids for Barrier Function Assay
Plasmids: The reporter plasmid, pRL-TKGFP-CMVE, has been described previously (92). The plasmid and the corresponding plasmid map can be obtained from the authors upon request. Starting material plasmids: Herpes simplex virus thymidine kinase (TK) promoter plasmid (e.g., pRL-TK; Promega Corp., Madison, WI, USA); green fluorescent protein (GFP) gene plasmid (e.g., Clontech Laboratories, Inc., Mountain View, CA, USA, pAcGFP1-N1; Invitrogen Corp., Carlsbad, CA, USA, pcDNA™6.2/N-EmGFP-GW/TOPO®; Stratagene Corp., La Jolla, CA, USA, phrGFP II-N); CMV immediate early region enhancer plasmid (e.g., Clontech Laboratories, pCMV-DsRed-Express); chicken β-globin 5′HS4 insulator plasmid (e.g., pJC13-1 (66)).
Restriction and modifying enzymes.
DH5α competent bacteria (Invitrogen).
Luria-Bertani media (LB): LB + 50 μg/ml Ampicillin (LB-AMP); AMP-agar plates.
QIAquick Gel Extraction Kit; QIAGEN Plasmid Maxi Kit (Qiagen, Valencia, CA, USA).
2.1.2. Testing of Reporter Plasmids for Barrier Function
Cell line: K562 human erythroleukemia cells (American Type Culture Collection, Manassas, VA, USA, cat. no. CCL-243).
K562 culture medium: Iscove’s modified Dulbecco’s medium (IMDM; Mediatech, Herndon, VA, USA) plus 10% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, Utah, USA), L-glutamine (2 mM), penicillin (50 IU/ml) and streptomycin (50 μg/ml).
Phosphate-buffered saline (PBS).
Electroporation cuvettes.
Electroporator.
Analytical flow cytometer (such as a FACSCalibur instrument, BD Biosciences, San Jose, CA, USA).
Fluorescence-activated cell sorter (such as a FACSAria instrument, BD Biosciences).
2.2. Enhancer Blocking Assay
2.2.1. Design and Construction of Reporter Plasmids for Enhancer Blocking Assay
Plasmids: The reporter plasmid, pRL-TKGFP-CMVE-TKRFP, has been described previously (92). The plasmid and the corresponding plasmid map can be obtained from the authors upon request. Starting material plasmids: pRL-TKGFP-CMVE and herpes simplex virus TK promoter plasmid described in Subheading 2.1.1; red fluorescent protein (RFP) gene plasmid (e.g., pDsRedT4-N1 plasmid (94)).
Restriction and modifying enzymes.
DH5α competent bacteria (Invitrogen).
QIAquick Gel Extraction Kit, QIAGEN Plasmid Maxi Kit (Qiagen).
2.2.2. Testing of Reporter Plasmids for Enhancer Blocking Activity
Cell lines: 293T/17 (293T) human embryonic kidney cell line (American Type Culture Collection, cat. no. CRL-11268) (see Note 1); K562 cells (American Type Culture Collection, cat. no. CCL-243).
293T culture medium: Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech) supplemented with 4.5 g/l glucose, 2 mM L-glutamine, 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% heat-inactivated FBS. Store at 4°C and warm up to 37°C before use.
K562 culture medium prepared as described in Subheading 2.1.2.
2.5 M CaCl2: Dissolve 183.7 g CaCl2 dihydrate (tissue culture grade) in deionized, distilled water. Bring the volume up to 500 ml and filter-sterilize using a 0.22-μm nitrocellulose filter. Stable at −20°C.
2× N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES)-buffered saline (2× HBS): 50 mM HEPES (Sigma-Aldrich Corp., St. Louis, MO, USA), 280 mM NaCl, 1.5 mM Na2HPO4. Titrate to pH 7.05 with 5 N NaOH. Filter sterilize using a 0.22-μm nitrocellulose filter. Store as single use aliquots at −20°C.
PBS.
Electroporation cuvettes.
Electroporator.
Analytical flow cytometer.
2.3. Lentiviral Vector Construction
2.3.1. Strong Enhancer-Promoter Sequences
Plasmids: Lentiviral vector plasmid containing reporter gene, e.g., GFP (see Note 2); MSCV LTR promoter plasmid (17); human EF1α promoter plasmid (e.g., pEF-BOS (39)); CAG promoter plasmid (e.g., pBacMam-2; Novagen, Inc., Madison, WI, USA).
Restriction and modifying enzymes.
DH5(competent bacteria (Invitrogen).
QIAquick Gel Extraction Kit; QIAGEN Plasmid Maxi Kit (Qiagen).
2.3.2. S/MAR Element
2.3.3. 5′HS4 Insulator
2.4. Lentiviral Vector Function
2.4.1. Transgene Expression in Cell Lines
Replication-defective lentiviral vector particles (95): produce and titer as described in other Chapters of this book.
Cell line: K562 cells.
K562 culture medium prepared as described in Subheading 2.1.2.
Polybrene stock solution (Sigma-Aldrich Corp.; hexadimethrine bromide, cat. no. H9268). Prepare stock of 6 mg/ml (1000 ×) in sterile deionized, distilled water; aliquot and store at −20°C.
Analytical flow cytometer.
2.4.2. Stability of the Inserted Elements
Genomic DNA Extraction Kit (Sigma-Aldrich Corp.).
PCR primers (Sigma-Aldrich Corp.), 0.05 μmole scale.
Taq DNA polymerase, dNTP mix (Promega Corp.).
PCR thermocycler.
2.4.3. Transgene Expression Following Differentiation of Transduced Human HSPCs
Replication-defective lentiviral vector particles (95): Produce and titer as described in other Chapters of this book.
Human CD34+ cord blood cells (StemCell Technologies, Vancouver, BC, Canada).
Human HSPC culture medium: IMDM containing 10% FBS, 100 μM β-mercaptoethanol (Sigma-Aldrich Corp.) plus human stem cell factor (SCF; 100 ng/mL), human Flt-3 ligand (100 ng/mL) and human thrombopoietin (20 ng/mL).
Recombinant fibronectin fragment (RetroNectin; Takara Mirus Bio, Madison, WI, USA).
Protamine sulfate solution (1000×; 4 mg/ml) (Sigma-Aldrich Corp.). Prepare stock of 4 mg/ml (1000 ×) in sterile deionized, distilled water; aliquot and store at −20°C.
Human HSPC differentiation medium: IMDM medium containing 10% heat-inactivated FBS, L-glutamine (2 mM), and penicillin (50 IU/mL), plus human interleukin-3 (IL-3; 20 ng/mL), human interleukin-6 (IL-6; 20 ng/mL) and human granulocyte-macrophage colony-stimulating factor (20 ng/mL).
Antibodies: anti-CD34 and anti-CD14 monoclonal antibodies conjugated to allophycocyanin (BD Biosciences).
Analytical flow cytometer.
2.4.4. Transgene Expression Following ESC Differentiation
Replication-defective lentiviral vector particles (95): Produce and titer as described in other Chapters of this book.
ESC line adapted for growth on gelatin-coated culture dishes (e.g., CCE (16); StemCell Technologies, cat. no. 00300).
ESC culture medium: Grow undifferentiated CCE ESCs on 0.1% gelatin-coated dishes in DMEM with 4.5 g/l glucose and 4 mM L-glutamine plus 150 μM β-mercaptoethanol, 15% FBS, 10 ng/mL murine leukemia inhibitory factor (LIF; StemCell Technologies).
ESC embryoid body differentiation medium: IMDM containing 1% methylcellulose, 2 mM L-glutamine, 150 μM β-mercaptoethanol, 15% FBS, and 40 ng/ml murine SCF (StemCell Technologies).
ESC hematopoietic differentiation medium: IMDM with 1% methylcellulose, 2 mM L-glutamine, 1% bovine serum albumin, 10 μg/ml insulin, 200 μg/ml transferrin, 150 μM β-mercaptoethanol, 15% FBS, 150 ng/ml SCF, 30 ng/ml murine IL-3, 30 ng/ml murine IL-6, and 3 U/ml erythropoietin (StemCell Technologies).
Monoclonal antibodies: anti-CD41 and anti-CD45 conjugated to R-phycoerythrin (BD Biosciences).
Analytical flow cytometer.
2.4.5. Murine HSPC Immortalization Assay
Female C57BL/6 mice (6- to 8-week old; The Jackson Laboratory, Bar Harbor, ME, USA; cat. no. 000664) used as bone marrow donors. All procedures involving mice must follow the guidelines set forth in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and be approved by an Institutional Animal Care and Use Committee.
5-fluorouracil (5-FU; Sigma-Aldrich Corp., cat. no. F6627). Store the stock at room temperature, avoiding light. Prepare a fresh working solution of 15 mg/ml in PBS immediately before use.
Erythrocyte lysing solution: 154 mM NH4Cl 10 mM NaHCO3, and 0.082 mM sodium ethylenediaminetetraacetic acid (EDTA), pH 7.3. Commercial lysing solutions are also available. Store at room temperature.
Murine HSPC culture medium: IMDM supplemented with 4.5 g/l glucose, 2 mM L-glutamine, 50 IU/ml penicillin, 50 μg/ml streptomycin, 15% heat-inactivated FBS, 100 ng/ml murine SCF, 30 ng/ml murine IL-3 and 10 ng/ml murine IL-6. Store at 4°C and warm up to 37°C before use.
Recombinant fibronectin fragment (RetroNectin; Takara Mirus Bio).
Polybrene stock solution (1000×; 8 mg/ml) prepared as described in Subheading 2.1.3. Aliquot and store at −20°C.
3. Methods
3.1. Barrier Function Assay
As discussed in Subheading 1.3, one of the defining properties of an insulator is to function as a barrier to prevent adjacent heterochromatin from advancing into and silencing gene expression. The barrier function assay will determine if the test element can reliably shield a stably integrated transgene from chromosomal position effects and allow it to be expressed in a copy number-dependent manner (Fig. 5.2A). In the absence of an insulator, transgene expression tends to be copy number-independent and is frequently extinguished in cells over time; when shielded with an insulator, transgene expression should be stable with a good correlation between vector copy number and expression levels. Felsenfeld and colleagues established an assay to test the barrier activity of the 1.2-kb fragment of the chicken β-globin 5′HS4 insulator (68). A reporter expressing a portion (Tac subunit) of the human IL-2 receptor driven by an erythroid-specific promoter and enhancer was constructed and integrated into a chicken erythroid cell line. In the absence of the insulator, extended culture of the transfected cells often led to loss of transgene expression, reflecting integration site position effects. When the same reporter plasmid was flanked on each side by two direct copies of the 1.2-kb 5′HS4 insulator and used to transfect the cells, stable transgene expression was obtained even after extended period of time in culture. The following is a modified version of the above barrier function assay that takes advantage of the intrinsic fluorescence of the GFP reporter gene in order to facilitate generation of single-copy clones and analyze transgene expression levels by fluorescence-activated cell sorting and flow cytometry.
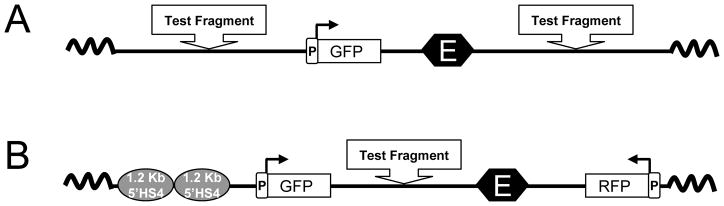
Fig. 5.2.
Schematic diagram showing the reporter plasmid pRL-TKGFP-CMVE used in the flow cytometry-based barrier function assay (A) and the reporter plasmid pRL-TKGFP-CMVE-TKRFP used in the flow cytometry-based enhancer blocking assay (B). Abbreviations: 1.2 kb 5′HS4, 1.2-kb fragment of the chicken β-globin 5′HS4 insulator; GFP, enhanced GFP gene; RFP, DsRed.T4 gene; E, human CMV immediate early region enhancer; P, herpes simplex virus TK promoter.
3.1.1. Design and Construction of Reporter Plasmids for Barrier Function Assay
The reporter plasmid pRL-TKGFP-CMVE (Fig. 5.2A) can be used to determine the barrier function activity of any test fragments with insulator properties.
Subclone one or more copies of the test element with potential insulating activity upstream of the TK-GFP expression cassette and an equivalent number of copies of the test element downstream of the CMV enhancer (see Note 3).
Construct a positive control insulated reporter plasmid by removing two direct copies of the full-length 1.2-kb chicken β-globin 5′HS4 insulator from plasmid pJC13-1 and inserting them upstream of the TK-GFP expression cassette and downstream of the CMV enhancer.
3.1.2. Testing of Reporter Plasmids for Barrier Function
Culture the K562 cells in K562 culture medium. Maintain cells at 37°C in a humidified incubator containing 5% CO2.
Electroporate 1 × 107 K562 cells (at 300 V and 950 μF) with 1 μg of each linearized reporter plasmids in 0.5 ml PBS. After 10 days of culture perform single cell sorting of the GFP+ cells using a fluorescence-activated cell sorter.
Grow up individual clones, extract genomic DNA and identify clones with single-copy integrations by Southern blot analysis. Digest the genomic DNA with enzymes that cut only once within the reporter plasmid, hybridize Southern blots with a GFP-specific probe, and pick clones that exhibit a single band.
Perform flow cytometric analysis on single-copy clones immediately and at one month intervals for up to 3 months of continuous culture. Without any insulators, many clones will express little or no GFP. Also, many of the clones that are initially GFP+ will gradually extinguish expression over time. Expect most or all of the single-copy clones to express and maintain GFP expression in the presence of full insulator activity (see Note 4).
3.2. Enhancer Blocking Assay
The enhancer blocking assay is used to measure the ability of a test element to shield a promoter from an enhancer when it is placed between the enhancer and a promoter (66,96) (Fig. 5.1B). Chung et al. developed an enhancer blocking assay to test the ability of the 1.2-kb chicken β-globin 5′HS4 insulator and shield a reporter expressing a neomycin (neo) resistance gene from the action of a strong enhancer (66). An erythroid-specific enhancer and promoter were used to drive neo gene expression in stably transfected human K562 erythroleukemia cells and geneticin-resistant colonies were counted. Using this assay, it was shown that the 1.2-kb 5′HS4 fragment was effective in reducing the number of drug-resistant colonies, which was an indication of the strength of enhancer blocking activity.
We have recently developed a flow cytometry-based assay (92) by constructing a plasmid in which enhancer blocking activity is measured by down-modulation of expression of a GFP reporter gene when a test fragment is inserted between a herpes simplex virus TK promoter-driven GFP expression cassette and a downstream human CMV enhancer (Fig. 5.2B). To control for variations in transfection efficiency, GFP expression levels are compared to those of an analogous CMV enhancer-TK promoter-driven RFP reporter gene expression cassette contained in the same construct. To allow use of circular plasmids in transient transfection experiments and eliminate influences of flanking chromatin structure on GFP expression in stable transfection experiments with linearized plasmids, two copies of the full-length 1.2-kb 5′HS4 insulator were introduced upstream of the GFP expression cassette. The various control and test constructs are transiently transfected into human 293T cells or stably transfected into human K562 erythroleukemia cells. Forty-eight hours following transient transfection of 293T cells and 2–4 weeks after stable transfection of K562 cells, GFP fluorescence can be measured by flow cytometric analysis and normalized with respect to RFP fluorescence.
3.2.1. Design and Construction of Reporter Plasmids for Enhancer Blocking Assay
The reporter plasmid pRL-TKGFP-CMVE-TKRFP (Fig. 5.2B) can be used to determine the enhancer blocking activity of any test fragments with insulator properties.
Subclone one or more copies of the test element with potential enhancer blocking activity between the TK-GFP expression cassette and the CMV enhancer at a unique ClaI restriction enzyme site.
Construct a reporter plasmid to control for spacing between the TK promoter of the TK-GFP expression cassette and the CMV enhancer by inserting a fragment of bacteriophage λ DNA equivalent in size to the test fragment at the unique ClaI restriction enzyme site.
For comparison, construct a positive control insulated reporter plasmid by inserting one or two copies of the full-length 1.2-kb chicken β-globin 5′HS4 insulator between the TK-GFP expression cassette and the CMV enhancer at the unique ClaI restriction enzyme site.
3.2.2. Testing of Reporter Plasmids for Enhancer Blocking Activity
Culture the 293T and the K562 cells in their corresponding culture media. Maintain cells at 37°C in a humidified incubator containing 5% CO2.
Transiently transfect 293T cells with plasmid DNA (20 μg) by calcium phosphate coprecipitation. Forty-eight hours following the transient transfection of 293T cells, measure GFP fluorescence by flow cytometric analysis and normalize with respect to RFP expression levels.
Electroporate 1 × 107 K562 cells (at 300V and 950μF) using 20 μg of each linearized reporter plasmids in 0.5 ml PBS. After 10 days of culture, perform single cell sorting of the GFP+ cells using a fluorescence-activated cell sorter. Two to four weeks after stable transfection of K562 cells, measure GFP fluorescence by flow cytometric analysis and normalize with respect to RFP fluorescence intensity. Obtain an indication of the strength of the enhancer blocking activity of the test fragment based on the relative decrease in GFP expression.
3.3. Lentiviral Vector Construction
3.3.1 Strong Enhancer-Promoter Sequences
SIN HIV-1-based lentiviral transfer vectors utilizing constitutively active viral and cellular promoters to drive expression of the GFP reporter gene have been described previously (43,50). Examples of promoters are given below that could be utilized as internal promoters in any SIN lentiviral vector (see Note 5).
MSCV LTR promoter: a 302-bp NheI–SmaI fragment of the U3 region and part of the R region of the MSCV LTR (nucleotides − 266 to +30 relative to the U3/R boundary) can be obtained from the MSCV gammaretroviral vector (17).
Human EF1α gene promoter: a 1.2-kb HindIII-EcoRI fragment, which includes 203 bp of 5′ flanking region, 33 bp of the first exon, 943 bp of the first intron and 10 bp of the second exon (located 20 bp upstream of the EF1α ATG initiation codon), can be obtained from the plasmid pEF-BOS (39).
CAG promoter: a 1.8-kb SphI-SmaI fragment, which contains 365 bp of the CMV enhancer, 277 bp of the chicken β-actin promoter and a 908-bp hybrid intron with a 3′ splice site from the rabbit β-globin gene, can be excised from the pBacMam-2 plasmid.
Subclone different promoter fragments into the lentiviral vector upstream of the transgene or a selectable marker.
Prepare a high quality plasmid DNA for transient transfection and generation of vector particles.
3.3.2. S/MAR Element
One of the best characterized S/MAR elements is a 2.2-kb fragment (element E) from the upstream border of the human interferon-β (IFN-β) gene, of which an 800-bp core (IFN-SAR) contributes most of the S/MAR activity (52,55,56).
Obtain the IFN-SAR as an 800-bp fragment from plasmid pCL and clone in reverse orientation into the 3′ untranslated region of the transgene, immediately upstream of the 3′ LTR.
Prepare a high quality plasmid DNA for transient transfection and generation of vector particles.
3.3.3 5′HS4 Insulator
The 5′HS4-containing vectors can be constructed by inserting the 1.2-kb fragment of pJC13-1 plasmid containing the chicken β-globin 5′HS4 insulator in direct orientation into the deleted U3 region of the 3′ LTR.
Prepare a high quality plasmid DNA for transient transfection and generation of vector particles.
3.4. Lentiviral Vector Function
A convenient strategy to flank the lentiviral transgene expression cassette with a boundary element is to subclone the fragment of interest into the U3 region of the 3′ LTR of the vector, which is transferred to the 5′ LTR during viral replication. A potential limitation of this approach (e.g., in the case of an element the size of the 1.2-kb chicken β-globin 5′HS4 insulator) is a decrease in vector titer, perhaps due to topological or size constraints (50). Because full insulator activity may require multiple copies of the element in question (66), another potential cause for concern in addition to size constraints is that direct repeats are frequently unstable during viral replication (32). Besides a flanking configuration (55,57), a single copy of a S/MAR element (the IFN-SAR) has been shown to provide beneficial effects (50,55,60–62), so this might be an option in certain instances. Other elements have been described that have been reported to help counteract insertion site-associated chromosomal position effects when used as a single copy in the context of a lentiviral vector, such as locus control regions (LCRs) and ubiquitously acting chromatin opening elements (UCOEs) (97,98). However, their generally large size (>2 kb) could also pose size constraints as far as titer is concerned (83). Therefore, when incorporating any boundary or novel genetic regulatory elements into lentiviral vectors, especially multiple copies, it is absolutely essential to confirm the fidelity of vector sequence transmission while concurrently assessing vector performance.
3.4.1. Transgene Expression in Cell Lines
To evaluate the activity of S/MAR and insulator elements in the context of lentiviral vectors, produce replication-defective vector particles and measure titers. Compare the titers between the parental vector and the vectors containing the various elements by flow cytometric analysis of GFP fluorescence.
To assess vector performance, transduce K562 cells using low multiplicities of infection (MOI) and compare the levels of GFP transgene expression achieved by the vectors containing the various elements to those achieved with the parental vector by flow cytometric analysis of GFP fluorescence. Pooled populations or clones containing single-copy integrations (obtained by fluorescence-activated cell sorting and Southern blot analysis) can be followed initially and at one month intervals for up to 3 months of continuous culture.
3.4.2. Stability of the Inserted Elements
To document proper transmission to the 5′ LTR and stability of insulators incorporated into the 3′ LTR, extract genomic DNA from vector-transduced cells.
Use primer pairs designed to specifically PCR-amplify the 5′ and 3′ LTR sequences separately and analyze for the presence of DNA bands having the expected sizes (see Note 6).
Sequence the DNA fragments to further confirm the structurally intact presence of the insulator following transmission from the 3′ LTR to the 5′ LTR.
3.4.3. Transgene Expression Following Differentiation of Transduced Human HSPCs
The effects of S/MAR and insulator elements on transgene expression can also be studied in primary human myelomonocytic cells following transduction and in vitro differentiation of human CD34+ HSPCs.
Obtain human CD34+ cord blood cells and culture in 12-well tissue culture plates coated with recombinant fibronectin fragment (2 μg/cm2) at a density of 1 × 106 cells per well. Prestimulate the cells for 48 hours in human HSPC culture medium.
Transduce the cells for 24 hours with lentiviral vector particles (2 × 106 transducing units [TU]/mL; MOI, 2) in the presence of protamine sulfate (4 μg/mL). Add fresh medium and culture the cells for an additional 72 hours.
To evaluate and compare the levels of GFP transgene expression in CD14+ monocytes derived from transduced human CD34+ cord blood cells, culture the cells for 7 weeks in human HSPC differentiation medium.
Maintain all cultures at 37°C in a humidified atmosphere containing 5% CO2. At weekly intervals, stain cells with anti-CD14 monoclonal antibodies conjugated to allophycocyanin and measure GFP fluorescence in CD14+ cells by flow cytometric analysis (see Note 7).
3.4.4. Transgene Expression Following ESC Differentiation
Most gammaretroviral vectors experience severe down-regulation of transgene expression upon differentiation of ESCs into more mature cells in vitro (32). As noted in the Introduction, when assessed at the single vector copy per cell level, transgenes delivered by lentiviral vectors have also been found to be susceptible to variegated expression and silencing in ESCs and their differentiated progeny (23,26,29). Consequently, it is important to verify efficacy of lentiviral vector-directed transgene expression in differentiating ESC systems prior to gene function studies. The example provided involves differentiation of ESCs into hematopoietic cells.
Prepare lentiviral vector particles and transduce monolayers of CCE ESCs cells in the presence of polybrene at 8 μg/ml.
Determine stability of transgene expression by measuring GFP fluorescence by flow cytometric analysis after 10 days and 1 month of continuous culture.
Use a two-step method for in vitro hematopoietic differentiation of CCE ESCs (16). The primary differentiation of ESCs into embryoid bodies can be induced by removal of LIF and plating a single cell suspension of cells in ESC embryoid body differentiation medium.
Perform hematopoietic differentiation of embryoid bodies by plating single cell suspensions of day 9 embryoid bodies in ESC hematopoietic differentiation medium.
Stain differentiated cells with anti-CD41 and anti-CD45 monoclonal antibodies and measure GFP fluorescence of CD41+ and/or CD45+ cells by flow cytometric analysis.
3.4.5. Murine HSPC Immortalization Assay
Dissolve 5-FU in sterile PBS (15 mg/ml) immediately before use and intravenously inject into mice (150 mg/kg body weight) using a 27-G needle attached to a 1-ml syringe.
Harvest HSPC-enriched bone marrow cells 4 days after the 5-FU injection. Flush the hind limbs with PBS containing 2% FBS using a 21-G needle attached to a 5-ml syringe (see Note 8).
Lyse erythrocytes by incubating total bone marrow cells in erythrocyte lysis solution for 10 min at room temperature followed by centrifugation at 375 × g for 5 min.
Coat 35-mm suspension culture plates with 2 μg/cm2 recombinant fibronectin fragment. Transfer the nucleated cells to plates at a density of 5 × 105 cells/ml and culture for 48 hours in murine HSPC culture medium at 37°C in a humidified atmosphere containing 5% CO2.
Transduce the cells for 3 consecutive days (4 hours each day) by incubation with vector conditioned medium in the presence of 8 μg/ml polybrene supplemented with 100 ng/ml murine SCF, 30 ng/ml murine IL-3 and 10 ng/ml murine IL-6.
Culture the transduced cells in murine HSPC culture medium minus murine SCF and murine IL-6. Passage cells every 3 days for 2 weeks. After bulk culture, plate 100 cells into each well of a 96-well plate and continue culturing them in HSPC culture medium minus murine SCF and murine IL-6. Two weeks later, examine the plates and determine the replating frequency based on the number of wells that contain proliferating cell populations.
Expand proliferating cell populations to establish immortalized cell lines.
Extract genomic DNA from the immortalized cells and subject to Southern blot analysis to determine vector copy number. Perform integration site analysis [e.g., by linear amplification-mediated polymerase chain reaction (LAM-PCR)].
Acknowledgments
This work was supported in part by National Institutes of Health grants R01HL65519 and R01HL66305, and by an Elaine H. Snyder Cancer Research Award and a King Fahd Endowed Professorship (to R.G.H.) from The George Washington University.
Footnotes
The 293T/17 cell line is a clone of the 293T (293tsA1609neo) human embryonic kidney cell line (99), which was selected for high transfectability and capability of producing high-titer vector stocks. The 293T cell line is a derivative of 293 cells into which the simian virus 40 (SV40) large T antigen gene was inserted. 293 cells express the adenovirus serotype 5 E1A 12S and 13S gene products, which strongly transactivate transcription from expression vectors containing the human CMV enhancer-promoter (100). Expression of the SV40 large T antigen by 293T cells may stimulate extrachromosomal replication of plasmids containing the SV40 origin of replication during transient transfection.
Besides academic sources, lentiviral vector backbones and packaging systems are available commercially (e.g., Clontech Laboratories, Inc., Mountain View, CA, USA; Invitrogen Corp., Carlsbad, CA, USA; Stratagene Corp., La Jolla, CA, USA).
Common recombinant DNA techniques are used for plasmid DNA preparations, restriction enzyme digestions and subclonings. Detailed protocols for each technique can be obtained from commercial sources, as well as from various standard molecular biology manuals. Accordingly, these techniques have not been described here.
When GFP is used as the reporter gene (101), it is useful to wait ~5 days before analyzing the transduced cells by flow cytometry. This will minimize the contribution of false positive signals because of pseudotransduction, which is the direct transfer of reporter protein present in the vector supernatants or incorporated into the vector particles to the target cells; this is particularly problematic for vesicular stomatitis virus-G-protein pseudotyped vectors (102,103). Note that it has also been shown that transgenes can be efficiently transiently expressed from unintegrated lentiviral vectors during this timeframe (104).
Both the EF1α and the CAG promoters contain introns for augmented transgene expression (38,40). Consequently, they are relatively large in size (1.2 and 1.8 kb, respectively). Size constraints and the fact that introns in gammatroviruses are removed during the viral life cycle (105) have precluded routine use of these promoters in gammaretroviral vectors. Lentiviral vectors based on HIV-1 are capable of transporting intron-containing sequences to target cells through a process that involves the binding of the viral Rev protein to a cis-acting RNA sequence known as the Rev-responsive element (106).
The presence of shorter or longer PCR products than expected indicates deletions or rearrangements of the inserted elements.
At the 7-week time point in the experiment, ~30% to 50% of the nonadherent cells should be CD14+ monocytes. Detailed protocols for staining of hematopoietic cells and detection of cell surface antigens by immunofluorescence flow cytometric analysis may be found on the websites of the monoclonal antibody manufacturers.
It should be possible to obtain 3–4 × 106 bone marrow cells from each 5-FU-treated mouse. The bone marrow cells can be used directly or further enriched for more primitive HSPCs using various magnetic- or fluorescence-activated cell sorting procedures (107). When culturing murine HSPCs, keep the cell density at 0.5 × 106 cells/ml.
References
- 1.Hawley RG. Progress toward vector design for hematopoietic stem cell gene therapy. Curr Gene Ther. 2001;1:1–17. doi: 10.2174/1566523013348904. [DOI] [PubMed] [Google Scholar]
- 2.Miller DG, Adam MA, Miller AD. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–42. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–6. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–10. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 6.Ramezani A, Hawley RG. Overview of the HIV-1 lentiviral vector system. Curr Protoc Mol Biol. 2002;16.21:1–15. doi: 10.1002/0471142727.mb1621s60. [DOI] [PubMed] [Google Scholar]
- 7.Speers WC, Gautsch JW, Dixon FJ. Silent infection of murine embryonal carcinoma cells by Moloney murine leukemia virus. Virology. 1980;105:241–4. doi: 10.1016/0042-6822(80)90171-3. [DOI] [PubMed] [Google Scholar]
- 8.Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–8. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 9.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–13. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 10.Hilberg F, Stocking C, Ostertag W, Grez M. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1987;84:5232–6. doi: 10.1073/pnas.84.15.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiher H, Barklis E, Ostertag W, Jaenisch R. Two distinct sequence elements mediate retroviral gene expression in embryonal carcinoma cells. J Virol. 1987;61:2742–6. doi: 10.1128/jvi.61.9.2742-2746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–9. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 13.Petersen R, Kempler G, Barklis E. A stem cell specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11:1214–21. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–6. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller G, Wall C, Fong AZC, Hawley TS, Hawley RG. Overexpression of HOX11 leads to the immortalization of embryonic precursors with both primitive and definitive hematopoietic potential. Blood. 1998;92:877–87. [PubMed] [Google Scholar]
- 17.Hawley RG, Lieu FHL, Fong AZC, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–8. [PubMed] [Google Scholar]
- 18.Hawley RG, Hawley TS, Fong AZC, Quinto C, Collins M, Leonard JP, et al. Thrombopoietic potential and serial repopulating ability of murine hematopoietic stem cells constitutively expressing interleukin-11. Proc Natl Acad Sci USA. 1996;93:10297–302. doi: 10.1073/pnas.93.19.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992;2:907–12. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 20.Rivella S, Sadelain M. Genetic treatment of severe hemoglobinopathies: the combat against transgene variegation and transgene silencing. Semin Hematol. 1998;35:112–25. [PubMed] [Google Scholar]
- 21.Emery DW, Stamatoyannopoulos G. Stem cell gene therapy for the β-chain hemoglobinopathies. Problems and progress. Ann N Y Acad Sci. 1999;872:94–107. doi: 10.1111/j.1749-6632.1999.tb08456.x. [DOI] [PubMed] [Google Scholar]
- 22.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 23.Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, et al. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–94. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–38. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawliuk R, Westerman KA, Fabry ME, Payen E, Tighe R, Bouhassira EE, et al. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–71. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 26.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 27.Persons DA, Hargrove PW, Allay ER, Hanawa H, Nienhuis AW. The degree of phenotypic correction of murine β-thalassemia intermedia following lentiviral-mediated transfer of a human γ-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–83. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- 28.Yao S, Sukonnik T, Kean T, Bharadwaj RR, Pasceri P, Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–6. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 30.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–72. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 31.Aparicio OM, Gottschling DE. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–46. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 32.Ramezani A, Hawley TS, Hawley RG. Stable gammaretroviral vector expression during embryonic stem cell-derived in vitro hematopoietic development. Mol Ther. 2006;14:245–54. doi: 10.1016/j.ymthe.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrester WC, Thompson C, Elder JT, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–63. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenuwein T, Forrester WC, Qiu RG, Grosschedl R. The immunoglobulin mu enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993;7:2016–32. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 35.Pikaart M, Feng J, Villeponteau B. The polyomavirus enhancer activates chromatin accessibility on integration into the HPRT gene. Mol Cell Biol. 1992;12:5785–92. doi: 10.1128/mcb.12.12.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters MC, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, et al. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–95. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 37.Francastel C, Walters MC, Groudine M, Martin DI. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–69. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 38.Kim DW, Uetsuki T, Kaziro Y, Yamaguchi N, Sugano S. Use of the human elongation factor 1α promoter as a versatile and efficient expression system. Gene. 1990;91:217–23. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucl Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 41.Cheng L, Du C, Lavau C, Chen S, Tong J, Chen BP, et al. Sustained gene expression in retrovirally transduced, engrafting human hematopoietic stem cells and their lympho-myeloid progeny. Blood. 1998;92:83–92. [PubMed] [Google Scholar]
- 42.Dorrell C, Gan OI, Pereira DS, Hawley RG, Dick JE. Expansion of human cord blood CD34+CD38− cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID repopulating cell (SRC) frequency: dissociation of SRC phenotype and function. Blood. 2000;95:102–10. [PubMed] [Google Scholar]
- 43.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–69. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 44.Gao Z, Golob J, Tanavde VM, Civin CI, Hawley RG, Cheng L. High levels of transgene expression following transduction of long-term NOD/SCID-repopulating human cells with a modified lentiviral vector. Stem Cells. 2001;19:247–59. doi: 10.1634/stemcells.19-3-247. [DOI] [PubMed] [Google Scholar]
- 45.Taboit-Dameron F, Malassagne B, Viglietta C, Puissant C, Leroux-Coyau M, Chereau C, et al. Association of the 5′HS4 sequence of the chicken β-globin locus control region with human EF1α gene promoter induces ubiquitous and high expression of human CD55 and CD59 cDNAs in transgenic rabbits. Transgenic Res. 1999;8:223–35. doi: 10.1023/a:1008919925303. [DOI] [PubMed] [Google Scholar]
- 46.Chang L-J, Urlacher V, Iwakuma T, Cui Y, Zucali J. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 1999;6:715–28. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- 47.Ye Z-Q, Qui P, Burkholder JK, Turner J, Culp J, Roberts T, et al. Cytokine transgene expression and promoter usage in primary CD34+ cells using particle-mediated gene delivery. Hum Gene Ther. 1998;9:2197–205. doi: 10.1089/hum.1998.9.15-2197. [DOI] [PubMed] [Google Scholar]
- 48.Araki K, Imaizumi T, Okuyama K, Oike Y, Yamamura K. Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. J Biochem. 1997;122:977–82. doi: 10.1093/oxfordjournals.jbchem.a021860. [DOI] [PubMed] [Google Scholar]
- 49.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Let. 1997;407:313–9. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 50.Ramezani A, Hawley TS, Hawley RG. Performance- and safety-enhanced lentiviral vectors containing the human interferon-β scaffold attachment region and the chicken β-globin insulator. Blood. 2003;101:4717–24. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- 51.Bode J, Maass K. Chromatin domain surrounding the human interferon-β gene as defined by scaffold-attached regions. Biochemistry. 1988;27:4706–11. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- 52.Mielke C, Kohwi Y, Kohwi-Shigematsu T, Bode J. Hierarchical binding of DNA fragments derived from scaffold-attached regions: correlation of properties in vitro and function in vivo. Biochemistry. 1990;29:7475–85. doi: 10.1021/bi00484a017. [DOI] [PubMed] [Google Scholar]
- 53.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, et al. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–7. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 54.Boulikas T. Nature of DNA sequences at the attachment regions of genes to nuclear matrix. J Cell Biochem. 1993;52:14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- 55.Schubeler D, Mielke C, Maass K, Bode J. Scaffold/mattrix-attached regions act upon transcription in a context-dependent manner. Biochemistry. 1996;35:11160–9. doi: 10.1021/bi960930o. [DOI] [PubMed] [Google Scholar]
- 56.Benham C, Kohwi-Shigematsu T, Bode J. Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J Mol Biol. 1997;274:181–96. doi: 10.1006/jmbi.1997.1385. [DOI] [PubMed] [Google Scholar]
- 57.Goetze S, Baer A, Winkelmann S, Nehlsen K, Seibler J, Maass K, et al. Performance of genomic bordering elements at predefined genomic loci. Mol Cell Biol. 2005;25:2260–72. doi: 10.1128/MCB.25.6.2260-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forrester WC, Fernandez LA, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–14. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez LA, Winkler M, Grosschedl R. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol Cell Biol. 2001;21:196–208. doi: 10.1128/MCB.21.1.196-208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agarwal M, Austin TW, Morel F, Chen J, Bohnlein E, Plavec I. Scaffold attachment region-mediated enhancement of retroviral vector expression in primary T cells. J Virol. 1998;72:3720–8. doi: 10.1128/jvi.72.5.3720-3728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Auten J, Agarwal M, Chen J, Sutton R, Plavec I. Effect of scaffold attachment region on transgene expression in retrovirus vector-transduced primary T cells and macrophages. Hum Gene Ther. 1999;10:1389–99. doi: 10.1089/10430349950018058. [DOI] [PubMed] [Google Scholar]
- 62.Dang Q, Auten J, Plavec I. Human β interferon scaffold attachment region inhibits de novo methylation and confers long-term, copy number-dependent expression to a retroviral vector. J Virol. 2000;74:2671–8. doi: 10.1128/jvi.74.6.2671-2678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park F, Kay MA. Modified HIV-1 based lentiviral vectors have an effect on viral transduction efficiency and gene expression in vitro and. in vivo Mol Ther. 2001;4:164–73. doi: 10.1006/mthe.2001.0450. [DOI] [PubMed] [Google Scholar]
- 64.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–14. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 67.Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–80. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–62. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 70.Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, et al. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA. 2002;99 (Suppl 4):16433–7. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, et al. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–8. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 73.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, et al. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 76.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 77.Yao S, Osborne CS, Bharadwaj RR, Pasceri P, Sukonnik T, Pannell D, et al. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–23. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 79.Emery DW, Yannaki E, Tubb J, Stamatoyannopoulos G. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc Natl Acad Sci USA. 2000;97:9150–5. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rivella S, Callegari JA, May C, Tan CW, Sadelain M. The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J Virol. 2000;74:4679–87. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yannaki E, Tubb J, Aker M, Stamatoyannopoulos G, Emery DW. Topological constraints governing the use of the chicken HS4 chromatin insulator in oncoretrovirus vectors. Mol Ther. 2002;5:589–98. doi: 10.1006/mthe.2002.0582. [DOI] [PubMed] [Google Scholar]
- 82.Aker M, Tubb J, Groth AC, Bukovsky AA, Bell AC, Felsenfeld G, et al. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum Gene Ther. 2007;18:333–43. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- 83.Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 84.Ma Y, Ramezani A, Lewis R, Hawley RG, Thomson JA. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 2003;21:111–7. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 85.Vieyra DS, Goodell MA. Pluripotentiality and conditional transgene regulation in human embryonic stem cells expressing insulated tetracycline-ON transactivator. Stem Cells. 2007;25:2559–66. doi: 10.1634/stemcells.2007-0248. [DOI] [PubMed] [Google Scholar]
- 86.Kwaks TH, Barnett P, Hemrika W, Siersma T, Sewalt RG, Satijn DP, et al. Identification of anti-repressor elements that confer high and stable protein production in mammalian cells. Nat Biotechnol. 2003;21:553–8. doi: 10.1038/nbt814. [DOI] [PubMed] [Google Scholar]
- 87.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet. 2006;38:479–83. doi: 10.1038/ng1766. [DOI] [PubMed] [Google Scholar]
- 88.Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci USA. 2008;105:13895–900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baum C, Fehse B. Mutagenesis by retroviral transgene insertion: risk assessment and potential alternatives. Curr Opin Mol Ther. 2003;5:458–62. [PubMed] [Google Scholar]
- 90.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–49. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 91.Ramezani A, Hawley TS, Hawley RG. Reducing the genotoxic potential of retroviral vectors. Methods Mol Biol. 2008;434:183–203. doi: 10.1007/978-1-60327-248-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramezani A, Hawley TS, Hawley RG. Combinatorial incorporation of enhancer blocking components of the chicken β-globin 5′HS4 and human T-cell receptor α/δ BEAD-1 insulators in self-inactivating retroviral vectors reduces their genotoxic potential. Stem Cells. 2008;26:3257–66. doi: 10.1634/stemcells.2008-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Modlich U, Bohne J, Schmidt M, Von KC, Knoss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–53. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–7. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 95.Ramezani A, Hawley RG. Generation of HIV-1-based lentiviral vector particles. Curr Protoc Mol Biol. 2002;16.22:1–15. doi: 10.1002/0471142727.mb1622s60. [DOI] [PubMed] [Google Scholar]
- 96.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–6. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 97.Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J, et al. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–57. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buzina A, Lo MY, Moffett A, Hotta A, Fussner E, Bharadwaj RR, et al. β-globin LCR and intron elements cooperate and direct spatial reorganization for gene therapy. PLoS Genet. 2008;4:e1000051. doi: 10.1371/journal.pgen.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–87. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gorman CM, Gies D, McCray G, Huang M. The human cytomegalovirus major immediate early promoter can be trans-activated by adenovirus early proteins. Virology. 1989;171:377–85. doi: 10.1016/0042-6822(89)90605-3. [DOI] [PubMed] [Google Scholar]
- 101.Hawley TS, Herbert DJ, Eaker SS, Hawley RG. Multiparameter flow cytometry of fluorescent protein reporters. Methods Mol Biol. 2004;263:219–38. doi: 10.1385/1-59259-773-4:219. [DOI] [PubMed] [Google Scholar]
- 102.Gallardo HF, Tan C, Ory D, Sadelain M. Recombinant retroviruses pseudotyped with the vesicular stomatitis virus G glycoprotein mediate both stable gene transfer and pseudotransduction in human peripheral blood lymphocytes. Blood. 1997;90:952–7. [PubMed] [Google Scholar]
- 103.Liu ML, Winther BL, Kay MA. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J Virol. 1996;70:2497–502. doi: 10.1128/jvi.70.4.2497-2502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, et al. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–32. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 105.Shimotohno K, Temin HM. Loss of intervening sequences in genomic mouse α-globin DNA inserted in an infectious retrovirus vector. Nature. 1982;299:265–8. doi: 10.1038/299265a0. [DOI] [PubMed] [Google Scholar]
- 106.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–7. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 107.Hawley RG, Ramezani A, Hawley TS. Hematopoietic stem cells. Methods Enzymol. 2006;419:149–79. doi: 10.1016/S0076-6879(06)19007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]