Abstract
Diastereomeric 8,5′-cyclopurine 2′-deoxynucleosides, containing a covalent bond between the deoxyribose and the purine base, represent an important class of DNA damage induced by ionizing radiation. The 8,5′-cyclo-2′-deoxyguanosine lesion (cdG) has been recently reported to be a strong block of replication and highly mutagenic in Escherichia coli. The 8,5′-cyclopurine-2′-deoxyriboses are suspected to play a role in the etiology of neurodegeneration in xeroderma pigmentosum patients. These lesions cannot be repaired by base excision repair, but they are substrates for nucleotide excision repair. The structure of an oligodeoxynucleotide duplex containing a site-specific S-cdG lesion placed opposite dC in the complementary strand was obtained by molecular dynamics calculations restrained by distance and dihedral angle restraints obtained from NMR spectroscopy. The S-cdG deoxyribose exhibited the O4′-exo (west) pseudorotation. Significant perturbations were observed for the β, γ, and χ torsion angles of the S-cdG nucleoside. Watson-Crick base pairing was conserved at the S-cdG•dC pair. However, the O4′-exo pseudorotation of the S-cdG deoxyribose perturbed the helical twist and base pair stacking at the lesion site and the 5′-neighbor dC•dG base pair. Thermodynamic destabilization of the duplex measured by UV melting experiments correlated with base stacking and structural perturbations involving the modified S-cdG•dC and 3′- neighbor dT•dA base pairs. These perturbations may be responsible for both the genotoxicity of this lesion and its ability to be recognized by nucleotide excision repair.
Introduction
Hydroxyl radicals cause a variety of damage in DNA, affecting the nucleobases1 or deoxyribose sugars,2 or both,3 as in the case of tandem 8,5′-cyclopurine 2′-deoxynucleoside lesions.4 At 2′-deoxyguanosines in DNA, hydrogen abstraction by a hydroxyl radical at the C5′ position of the deoxyribose followed by attack at the C8 carbon of guanine forms an N7-centered radical, which may be oxidized to produce diastereomeric 8,5′-cyclo-2′-deoxyguanosines (cdG).4–10 The corresponding 8,5′-cyclo-2′-deoxyadenosines (cdA) have also been characterized.3,4,7,9–15 For both cdG and cdA, the diastereomeric ratio at the C5′ position depends on experimental conditions and DNA conformation.5–7,13,16–18
The 8,5′-cyclopurine-2′-deoxynucleosides are believed to be important contributors to the genetic toxicology of oxidative stress and inflammation.4 They have been detected at the nucleotide level,5,11 in DNA,5,19–21 and cells in vitro,6 in human urine,18 and in vivo.21–23 The formation of 8,5′-cyclopurine-2′-deoxynucleosides might contribute to skin cancer risk in xeroderma pigmentosum complementation group C (XP-C) patients.24 They are also believed to play roles in Cockayne syndrome,21 breast and ovarian cancer,22 and familial Mediterranean fever.25
In Escherichia coli, S-cdG is a block to DNA replication, is highly mutagenic, and is refractory to repair.26 It induced 34% mutation upon induction of the SOS response. Most mutations were S-cdG → A mutations, though S-cdG → T mutations and deletions of the 5′-neighbor dC at low level also were observed.26 It has been reported in a preliminary study that S-cdG does not block primer elongation by Klenow DNA polymerases, and dATP is preferentially incorporated opposite the lesion.27
Computational studies predicted that the incorporation of the cdA stereoisomers into DNA would result in helical distortions at the lesion site.28–30 Both the R- and S-diastereomers of the 8,5′-cA ribonucleoside have been crystallized.31,32 Both exhibited the anti conformation about the glycosyl bond with χO4′-C1′-N9-C8 = 29.8° or 27.4°, respectively. The fused six-member ring C8-N9-C1′-O4′-C4′-C5′ adopted the half-chair conformation with the O4′ and C4′ out of plane. The deoxyribose adopted the O4′-exo (0T1) pseudorotation with P = ~289° and τm = ~48°. Molecular mechanics calculations predicted that the cdA diasetereomers maintain the O4′-exo pseudorotation when placed opposite dT in DNA.28 The NMR data and ab initio calculations suggest that incorporation of the S-cdA into di- or tri-nucleotides does not change the O4′-exo deoxyribose pseudorotation.33
Herein, we report the structure of the S-cdG•dC pair in 5′-d(GTGCXTGTTTGT)-3′•5′-d(ACAAACACGCAC)-3′, containing the DNA sequence of p53 codons 272–275, where X denotes the S-cdG (Scheme 1). The lesion is located in codon 273. The S-cdG remains stacked into the duplex and participates in Watson-Crick hydrogen bonding with the complementary dC. However, the S-cdG deoxyribose shifts to the O4′-exo pseudorotation with P = 280°. This alters the γ and δ backbone torsion angles. Additionally, the β and χ torsion angles are changed from those in B-DNA. The twist and base pair shift helicoidal parameters are perturbed at the C4•G21 and X5•C20 base pairs. The purine ring is anti about the glycosyl bond and the fused six-membered ring adopts the half-chair conformation with O4′ and C4′ out of plane.
Scheme 1.
Numbering scheme of the oligodeoxynucleotide duplex containing the (5′S)-8,5′-cyclo-2′-deoxyguanosine (S-cdG) 5′-nucleotide.
Results
Synthesis and Characterization of the S-cdG-Modfied Oligodeoxynucleotide
The S-cdG -modified oligodeoxynucleotide 5′-GTGCXTGTTTGT-3′, containing the sequence of p53 codons 272-275 in which the lesion was located in codon 273, was synthesized by a modification of the method reported by Romieu (Scheme S1 in the Supporting Information).34 The synthesis of N2-isobutyryl-5′-phenylthio-2′,5′-dideoxyguanosine gave 70% yield from N2-isobutyryl-2′-deoxyguanosine. The yield was improved to 91% when the exocyclic N2-dG amino group was protected with DMF. However, the DMF protection was unstable in the subsequent NaBH4 reduction step. Therefore, after cyclization and TBDMS protection of the 3′-hydroxyl group, it was replaced with an isobutyryl group. The modified oligodeoxynucleotide was synthesized using solid phase phosphoramidite chemistry and characterized by HPLC and MALDI-TOF mass spectrometry.
NMR Resonance Assignments
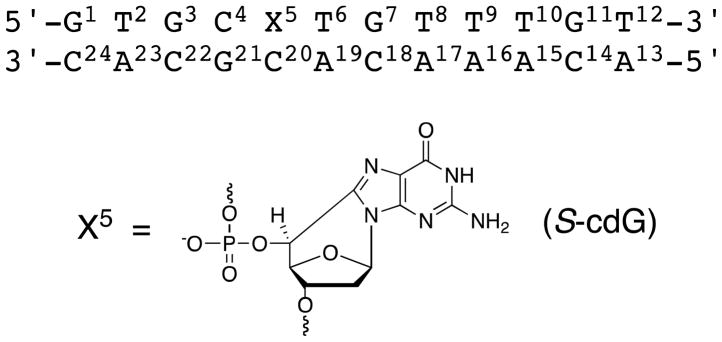
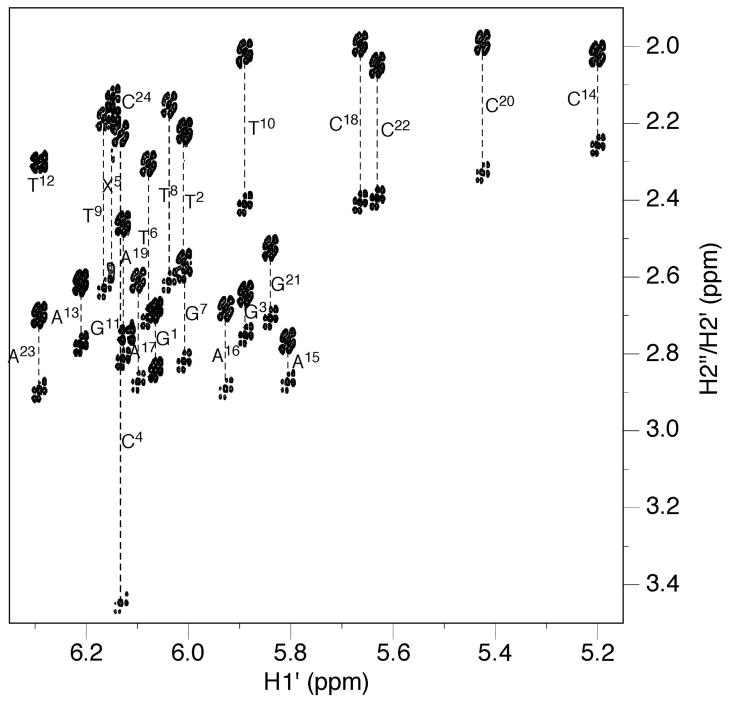
The non-exchangeable protons of the S-cdG-modified duplex were assigned based upon the sequential connectivity of the base proton H6 or H8 dipolar couplings with H1′ deoxyribose protons (Figure 1).35,36 For the modified strand, the NOE sequential connectivity was observed from G1 to C4. Since the S-cdG nucleotide lacked a proton at the C8 carbon, the sequential connectivity exhibited an interruption at X5. However, the X5 H1′ proton was identified at 6.14 ppm; it exhibited a weak X5 H1′→T6 H6 NOE, suggesting that the distance between these two protons was greater than in B-DNA. The sequential NOE connectivity resumed from T6 to T12. For the modified strand, all of the deoxyibose H1′ protons were observed within a narrow chemical shift window, between 5.8–6.3 ppm. The complete sequential NOE connectivity was observed for the complementary strand.
Figure 1.
NOE connectivity of base H8/H6 protons with deoxyribose H1′ protons of the S-cdG containing duplex. A. Modified strand. B. Complementary strand.
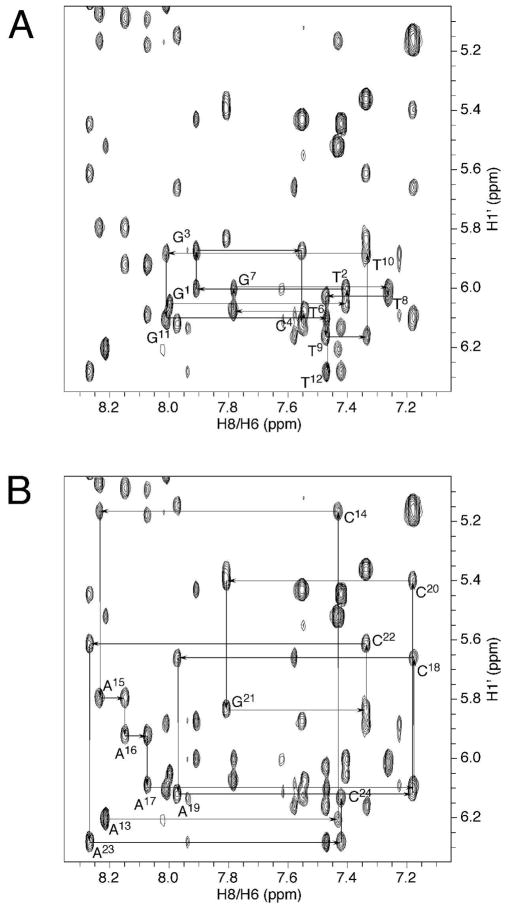
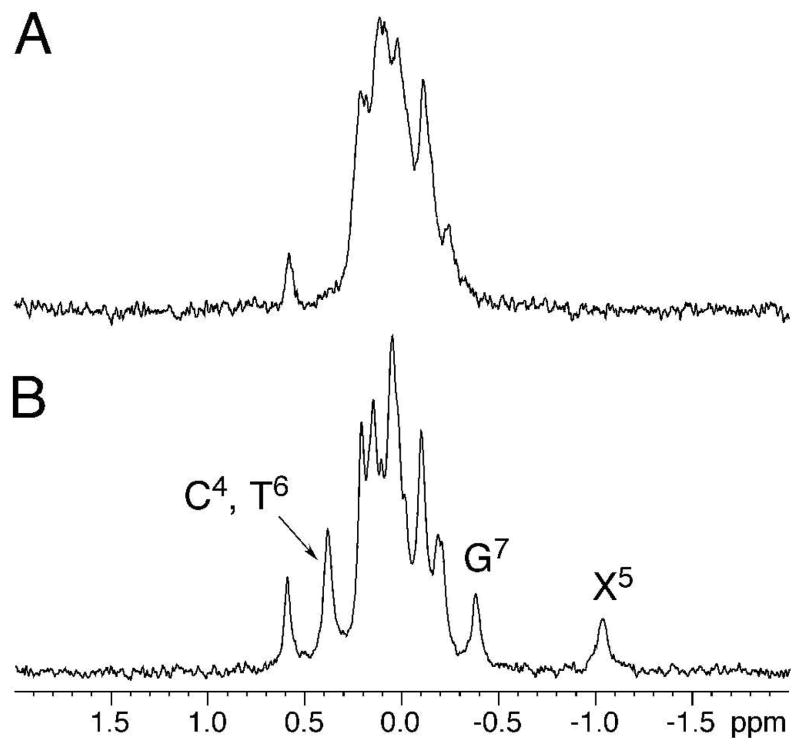
The assignments of X5 deoxyribose protons were made by analysis of scalar and dipolar couplings. Figure 2 displays a tile plot derived from a NOESY spectrum obtained at 60 ms mixing time. X5 H1′ exhibited strong dipolar couplings with H2′ and H2″; weak scalar couplings were also observed. H3′ exhibited strong dipolar couplings with H2′, H2″ and H4′, whereas the scalar couplings were unobservable. H4′ exhibited both scalar and dipolar couplings with the single H5′ proton. The geminal H2′ and H2″ protons were assigned from their NOEs to H1′ and H3′. H2′ exhibited a weaker NOE with H1′ than did H2″, whereas it exhibited a stronger NOE with H3′ than did H2″. In B-DNA, H2″ resonances are usually more downfield than H2′ resonances. However, the X5 H2′ resonance was observed at 2.55 ppm, whereas the H2″ resonance was observed at 2.27 ppm. For the remainder of the duplex, the H2′, H2″, H3′, and H4′ deoxyribose resonances were assigned unequivocally. The resonance assignments of the non-exchangeable DNA protons are tabulated in Table S1 of the Supporting Information.
Figure 2.
Tile plot derived from a NOESY spectrum obtained at a mixing time of 60 ms showing the assignment of S-cdG non-exchangeable protons.
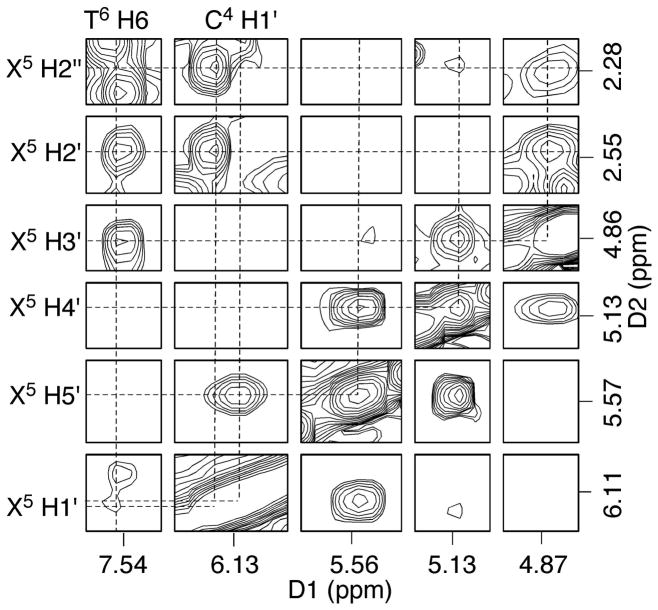
The resonances of the base imino protons were assigned based on sequential connectivity in NOESY spectra and the assignments were supported by NOEs to the amino protons of Watson-Crick base pairs (Figure 3).37 The NOE sequential connectivity was observed from G1→T2→G3→G21 to X5, and from G7→T8→T9→T10 to G11. At and adjacent to the lesion site, G21 N1H exhibited NOEs with C4 N4H1 and N4H2, and X5 N1H exhibited NOEs with the complementary C20 N4H1 and N4H2. At the 3′-neighbor base pair, the T6 N3H resonance was not observed, but A19 H2 exhibited NOEs to both X5 N1H and G7 N1H, suggesting A19 was still intercalated. Except for the terminal base pairs, the remaining NOE cross-peaks arising from Watson-Crick hydrogen bonding were observed.
Figure 3.
Assignment of the base imino and amino protons based on the NOE connectivity. NOE interactions of the imino protons with the opposite base arising from Watson-Crick base pairing are labeled as: (a) X5 N1H → C20 N4H2, (b) X5 N1H → A19 H2, (c) X5 N1H → C20 N4H1, (d) G7 N1H → C18 N4H2, (e) G7 N1H → A19 H2, (f) G7 N1H → A17 H2, (g) G7 N1H → C18 N4H1, (h) G11 N1H → C14 N4H2, (i) G11 N1H → C14 N4H1, (j) G3 N1H → C22 N4H2, (k) G3 N1H → C22 N4H1, (l) G21 N1H → C4 N4H2, (m) G21 N1H → C4 N4H1, (n) T2 N3H → A23 H2, (o) T10 N3H → A15 H2, (p) T9 N3H → A16 H2, and (q) T8 N3H → A17 H2.
Deoxyribose Coupling Constants
Figure 4 displays the expansion of an ECOSY spectrum38 in the region of deoxyribose H1′ correlations with H2′ and H2″. The 3JH1′-H2′ and 3JH1′-H2″ coupling constants were measured from the multiplicities of the cross peaks. The 3JH1′-H2′ and 3JH1′-H2″ values for X5 were 2.6 and 7.0 Hz, respectively. Consistently, the H1′-H2′ cross peak was weak. The 3JH4′-H5′ was 5.4 Hz, whereas the 3JH3′-H4′ was not measureable. Except for the terminal nucleotides, the 3JH1′-H2′ for all other nucleotides were 8–10 Hz and the 3JH1′-H2″ were 5–7 Hz, suggesting that the deoxyriboses adopted C1′-exo or C2′-endo conformations. The 3J coupling constants for the deoxyribose protons are tabulated in Table S2 of the Supporting Information.
Figure 4.
An expansion of the ECOSY spectrum used for the measurement of 3JH1′-H2′ and 3JH1′-H2″ coupling constants. Except for X5, G11, and T12, all H2″ protons exhibited greater chemical shifts than H2′ protons. The geminal H2′ and H2″ protons of G11 and T12 were not resolved, and X5 H2″ was upfield from H2′.
Phosphodiester Backbone Conformation
The 31P resonances were assigned from a 31P-H3′ HMBC spectrum (Figure S2 in the Supporting Information). Except for X5, each phosphodiester exhibited a heteronuclear coupling with H3′ of the 5′-neighbor. Figure 5 displays the 31P NMR of the S-cdG containing duplex compared with the corresponding unmodified duplex. At the modified nucleotide the 31P resonance shifted upfield, indicating a backbone perturbation at the modified base. The other 31P resonances were clustered within a modest chemical shift range, centered in the spectral region characteristic of B DNA.
Figure 5.
31P NMR of the S-cdG containing duplex compared with the corresponding unmodified duplex. A. Unmodified duplex. B. S-cdG containing duplex.
Chemical Shift Perturbations
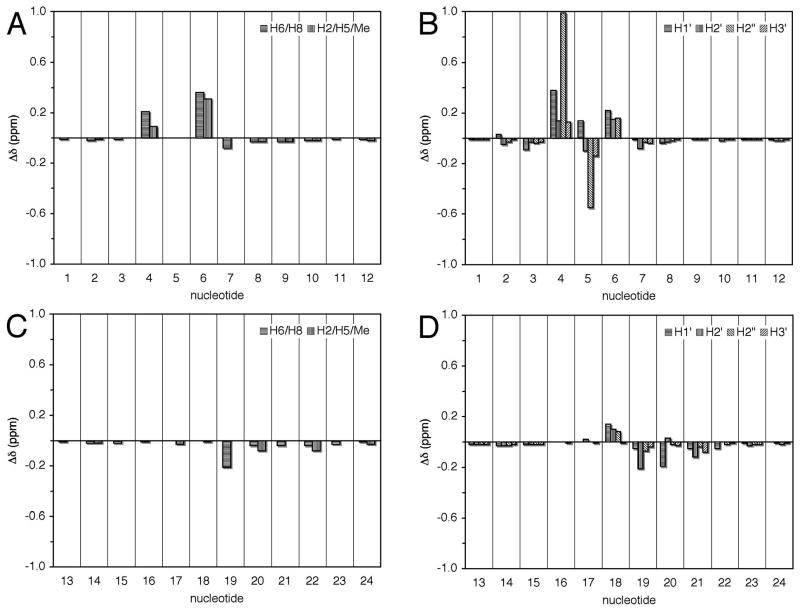
Chemical shifts of the non-exchangeable protons between the S-cdG containing duplex and the corresponding unmodified duplex were compared (Figure 6). Significant changes were observed at X5 and the 5′- and 3′-neighboring nucleotides of the modified strand. C4 H6, H1′ and H2″ shifted downfield by 0.21, 0.38, and 0.99 ppm, respectively; X5 H2″ shifted upfield by 0.55 ppm; and T6 H6, CH3, and H1′ shifted downfield by 0.36, 0.31, and 0.22 ppm, respectively. In contrast, the chemical shift perturbations for the complementary strand were small, with the exception of A19 H2′, which shifted upfield by 0.21 ppm.
Figure 6.
Proton chemical shift perturbations of the S-cdG containing duplex compared with the unmodified duplex. A. Base protons of the S-cdG-modified strand. B. Deoxyribose protons of the S-cdG-modified strand. C. Base protons of the complementary strand. D. Deoxyribose protons of the complementary strand.
Thermal Stability of the S-cdG Modified Duplex
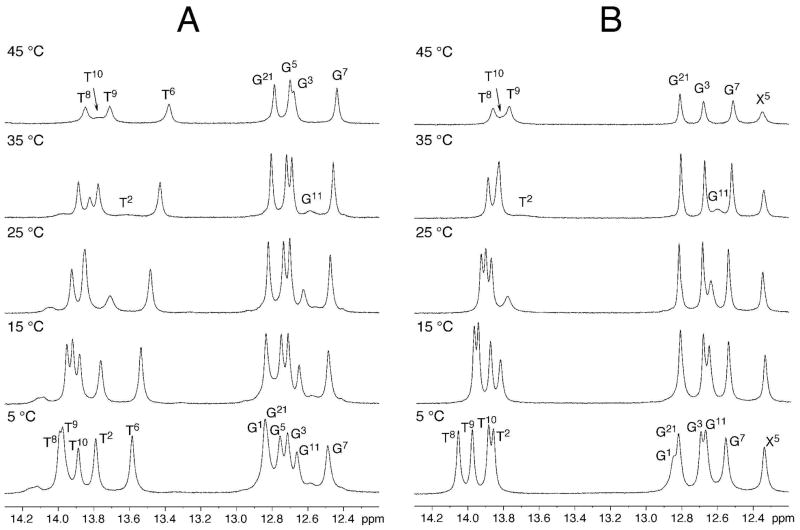
The thermal melting of the modified duplex containing the S-cdG was monitored using UV spectroscopy in 100 mM NaCl at pH 7.0. It exhibited a melting temperature (Tm) of 46 ± 1 °C, as compared to the unmodified DNA that exhibited a Tm: of 55 °C. (Figure S1 in the Supporting Information). Thus, the incorporation of S-cdG reduced the Tm by 9 °C. Figure 7 displays 1H NMR of the S-cdG containing duplex and the corresponding unmodified duplex at different temperatures. In the modified duplex, the X5 imino resonance exhibited significantly more line broadening at 45 °C than the corresponding G5 imino resonance of the unmodified duplex. For the modified duplex, the T6 N3H resonance was not observed at 5 °C, suggesting that the S-cdG nucleotide also significantly perturbed the 3′-flanking T6•A19 base pair.
Figure 7.
1H NMR of the S-cdG containing duplex compared with the corresponding unmodified duplex at different temperatures. A. Unmodified duplex. B. S-cdG containing duplex.
Structural Refinement
A total of 426 distance restraints, including 274 intra-nucleotide and 152 inter-nucleotide restraints were calculated from the intensities of NOE cross-peaks using MARDIGRAS (Table S3 in the Supporting Information).39 A total of 29 NOEs involving the S-cdG protons were used as restraints. A total of 45 empirical distance restraints arising from Watson-Crick base pairing interactions were used, as were 165 empirical torsion angle restraints that were applied to refine the non-terminal nucleotides. These were justified based upon the NMR data, which suggested that structural perturbations for the duplex were localized at and adjacent to the lesion site. No base pair distance restraints were used for the T6•A19 base pair, and no torsion angle restraints were used for the C4•G21, X5•C20 and T6•A19 base pairs. The restraints used for the structure refinement are summarized in Table 1.
Table 1.
rMD Restraints and Statistical Analysis of rMD Converged Structures of the S-cdG Containing Duplex.
| Total restraints for rMD calculation | 636 |
|
| |
| Experimental NOE distance restraints | 426 |
| Intranucleotide NOE restraints | 274 |
| Internucleotide NOE restraints | 152 |
| NOEs of S-cdG | 29 |
| Empirical basepairing restraints | 45 |
| Empirical torsion angle restraints | 165 |
| Backbone torsion angles restraints | 95 |
| Deoxyribose torsion angles restraints | 70 |
|
| |
| Structure Statisticsa
| |
| NMR R-factor (R1x) (×10−2) | 5.75 |
| Intranucleotide NOEs | 4.66 |
| Internucleotide NOEs | 7.97 |
| RMSD deviation of refined structures | 0.55 |
Mixing time used to calculate R1x was 250ms. , where (a0) and (ac) are the intensities of observed (non-zero) and calculated NOE cross peaks, respectively.
The rMD calculations for the S-cdG containing duplex were performed from initial A- and B-form starting structures. Ten final structures, five each for A- and B-DNA starting structures, with lowest energies, were obtained. All structures converged as indicated by pairwise RMSD comparisons (Table 1). The accuracies of the emergent structures were evaluated by comparison of theoretical NOE intensities calculated by CORMA40 for the refined structure to the experimental NOE intensities to yield sixth root residuals (R1x).41 The overall residuals, as well as the residuals for intra- or inter-nucleotide NOEs, were consistently less than 0.1 (Table 1). R1x values for each nucleotide were less than 0.15 (Figure S3 in the Supporting Information). Thus, the refined structures provided accurate depictions of the NOE data.
Structure of the S-cdG-Containing Duplex
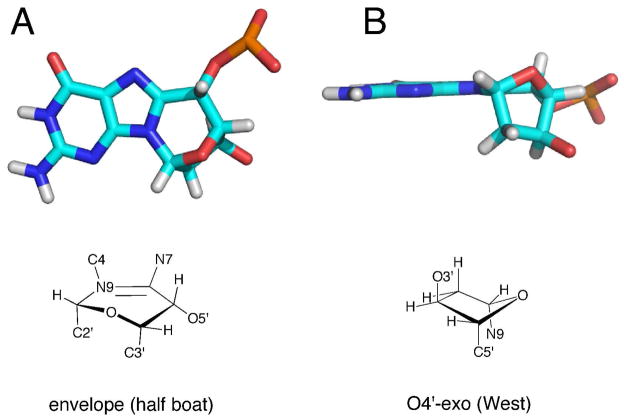
The significant perturbations involved the modified strand. Figure 8 shows an expanded view at the lesion site. The S-cdG nucleotide was in the O4′-exo, “west” pseudorotation (Figure 9B), with P = 280.2° and τm = 47.6°. The heavy atoms N9, O3′, and C5′ were axial about the deoxyribose ring. With the exception of the terminal nucleotides, all other deoxyribose pseudorotations were either C1′-exo or C2′-endo. Figure 9A displays the six-membered ring C8-N9-C1′-O4′-C4′-C5′ conformation. It adopted the envelope (half boat) conformation. Helicoidal analysis of the backbone torsion angles showed that at the lesion site, the β (P-O5′-C5′-C4′) angle shifted from the characteristic ~180° to −87°. The γ (O5′-C5′-C4′-C3′) angle shifted from ~ 50° to −67°. Modest perturbations of the δ (C5′-C4′-C3′-O3′) and ζ (C3′-O3′-P-O5′) torsion angles were also observed from ~ 120° to +149° and from ~ −90° to −59°, respectively. There was also a modest change for the glycosyl torsion angle χ from ~ −120° to −157°. C4 H2″ was proximate to the X5 purine ring. In contrast, X5 H2″ was farther from the X5 purine ring compared to the H2″ protons in B DNA.
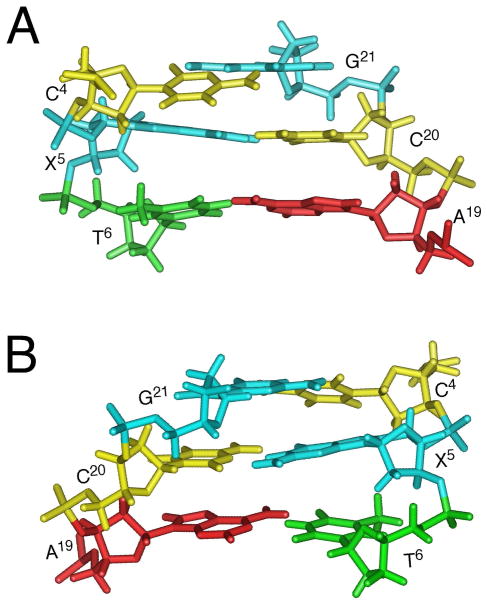
Figure 8.
Expanded views of the refined structure of the S-cdG containing duplex at the lesion site. A. View from the major groove. B. View from minor groove.
Figure 9.
Ring conformations of the S-cdG in the refined structure. A. Six-member ring C8-N9-C1′-O4′-C4′-C5′. B. 2′-deoxyribose.
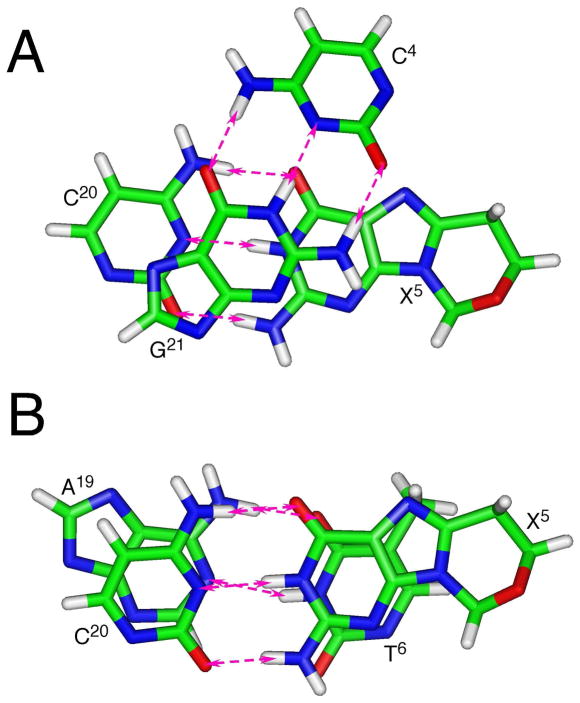
Figure 10 shows the base stacking and base pairing at the lesion site. The 5′-neighbor C4•G21 base pair exhibited a shift of −1.0 Å resulting in the displacement of C4 toward the major groove. At the C4→X5 step, an increased twist of 49° with respect to the X5•C20 base pair was evident. In contrast, the helix was underwound at the X5→T6 step. Additionally, the 3′-neighbor base pair T6•A19 exhibited a greater than normal base pair opening of −11.3°.
Figure 10.
Base pairing and base stacking of the refined structure of the S-cdG containing duplex at the lesion site. The pink arrows indicate anticipated hydrogen bonding interactions. A. the C4•G21 and X5•C20 base pairs. B. The X5•C20 and T6•A19 base pairs.
Molecular Dynamics Calculations in Explicit Solvent
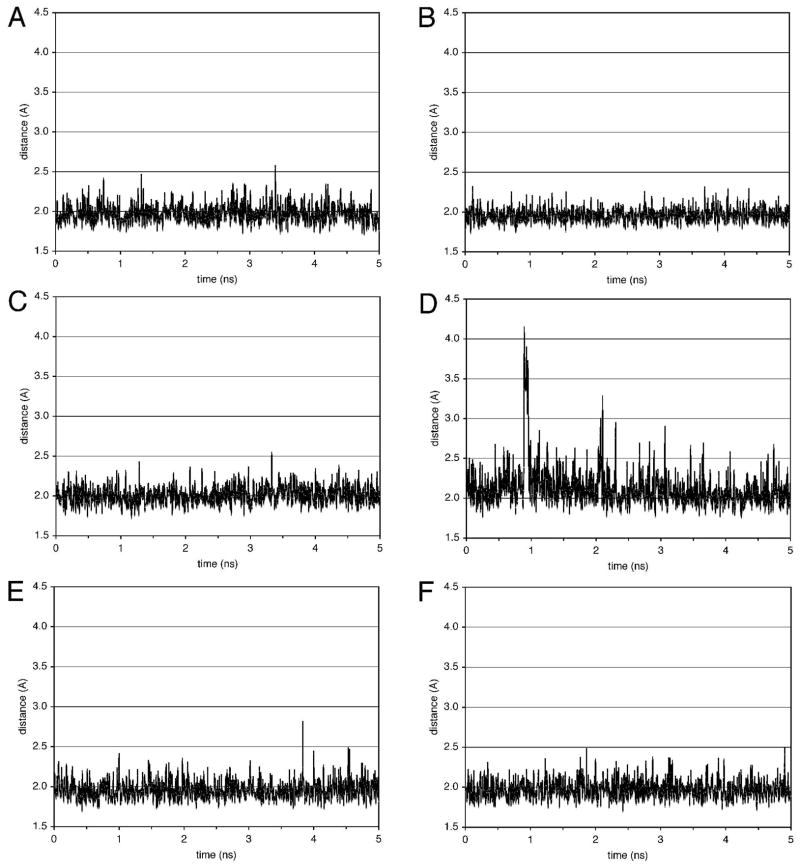
A molecular dynamics simulation was carried out in explicit water at constant pressure at 300 K, for 5 ns. The distances of the atoms involving in the Watson-Crick hydrogen bonding were measured in the trajectories. Figure 11 shows the distances of guanine N1H → cytosine N3 and the thymine N3H → adenine N1 of some base pairs observed in the trajectories. During this simulation, no changes in the monitored distances were observed for the G•C and C•G base pairs including the damaged X5•C20 base pair. In contrast, at the T6•A19 base pair, an opening occurred at ~0.9 ns, as indicated by the distances of T6 O2 → A19 H2, T6 N3H → A19 N1, and T6 O4 → A19 N6H1 jumping from ~ 3.5 Å to ~ 5.5 Å, ~ 2.0 Å to ~ 3.5 Å, and from ~ 1.8 Å to ~ 2.2 Å, respectively. Other non-terminus T•A base pairs exhibited no remarkable changes.
Figure 11.
Distances of guanine N1H → cytosine N3 and the thymine N3H → adenine N1 of some base pairs in the trajectories of the molecular dynamics simulations conducted in explicit solvent at 300 K. A. T2•A23 base pair. B. C4•G21 base pair. C. X5•C20 base pair. D. T6•A19 base pair. E. T8•A17 base pair. F. T9•A16 base pair.
Discussion
Interest in the 8,5′-cyclopurine-2′-deoxynucleoside lesions has been piqued by evidence that in mammalian cells 8,5′-cyclo-2′-deoxyadenosine (cdA) diastereomers3,4,7,9–15 are repaired by nucleotide excision repair (NER),42,43 an idea that was suggested earlier,5,6 and not by base excision repair. Also, bacterial DNA N-glycosylases endo III and FpG do not excise S-cdG from DNA, suggesting that like cdA, it also is a substrate for NER.27 Although the repair of S-cdG by the human NER system remains to be determined, Jasti et al.26 demonstrated that in DNA the S-cdG lesion was incised by the UvrABC nuclease of E. coli. The covalent bond between C8 of guanine and C5′ of the deoxyribose in the 8,5′-cyclopurines locks the modified nucleotide in the anti conformation. This is believed to hinder the flipping of the purine ring from the duplex, which is consistent with the observation that the 8,5′-cyclopurine-2′-deoxynucleosides are not repaired by BER.42,43 If not repaired, the S-cdG lesion is mutagenic. In SOS-induced E. coli, a mutation frequency of 34% was observed. Most mutations were S-cdG→A mutations, though S-cdG→T mutation and a deletion of the 5′-neighbor C also was observed.26 Hence, it was of interest to determine the structure of S-cdG in DNA.
Structure of S-cdG in DNA
The present study reveals that S-cdG remains stacked into the duplex and participates in Watson-Crick hydrogen bonding with the complementary dC. However, the S-cdG deoxyribose shifts to the O4′-exo pseudorotation, as opposed to the “south” pseudorotation (C2′-endo) observed in B-DNA, or the “north” pseudorotation (C3′-endo) in A-DNA.44,45 This corroborates computational studies on 8,5′-cyclopurine-2′-deoxynucleosides.28 Crystal structures of the cA ribonucleoside also exhibited the O4′-exo pseudorotation,31,32 and an NMR and DFT study of di- and tri-deoxynucleotides containing S-cdA indicated the O4′-exo deoxyribose.33 The O4′-exo pseudorotation introduces significant helicoidal perturbation into the modified strand of DNA. This involves changes in the S-cdG phosphodiester backbone torsion angles β (P-O5′-C5′-C4′), γ (O5′-C5′-C4′-C3′), δ (C5′-C4′-C3′-O3′), and ζ (C3′-O3′-P-O5′) from ~ 180° to −87°, from ~ 50° to −67°, from ~ 120° to 149°, and from ~ −90° to −59°, respectively. These changes perturb the helicoidal twist and base pair shift parameters at the C4•G21 and X5•C20 base pairs from ~ 30° to 49° and from ~ 0 Å to −1.0 Å, respectively. These changes are consistent with the upfield shift of the 31P resonance at S-cdG. These conclusions also are consistent with computational studies, which predict that the O4′-exo pseudorotation of the cdA deoxyribose should alter the helical twist parameter for the modified cdA•dT base pair as compared to the flanking base pairs.28 In addition, the modified cdA•dT base pair exhibited an altered base pair shift parameter. The altered ζ backbone torsion angle of S-cdG (−59°) results in the greater than normal base opening of −11.3° for the 3′-neigbhor T6•A19 base pair. (Figure S7 in the Supporting Information). Additionally, the glycosyl torsion angle χ (O4′-C1′-N9-C2) torsion angle is modified from ~ −120° to −157°. This places the six-member ring C8-N9-C1′-O4′-C4′-C5′ into the half-boat conformation. The bond between X5 C8 and C5′ pulls X5 H4′ and H5′ closer to the purine ring as compared to the H4′, and H5″ protons in B-DNA. This is consistent with the downfield chemical shifts of both X5 H4′ and H5′. In contrast, X5 H2″ is farther from the X5 purine ring compared to the H2″ protons in B-DNA, consistent with its upfield shift compared to that in the unmodified duplex.
Thermodynamic Considerations
Energetically, the O4′-exo pseudorotation is disfavored due to the axial orientation of all substituent heavy atoms.28 The helical perturbation of the modified strand associated with the unusual O4′-exo deoxyribose at the lesion site is consistent with the 9 °C decrease in the Tm of the modified duplex as compared to the unmodified control. The destabilization likely involves structural perturbations observed for the modified X5•C20 and 3′-neighbor T6•A19 base pairs, and accompanying base stacking perturbations. Indeed, the X5 imino resonance exhibits increased line broadening at 45 °C as compared to the G5 imino resonance of the unmodified duplex (Figure 7). Exchange-mediated line broadening of DNA imino protons is normally associated with the formation of an open state of the base pair in which the imino proton is freed from its hydrogen bond and is accessible to the base that catalyzes the proton exchange,46–48, 49,50 but S-cdG is locked in the anti conformation about the glycosyl bond and incapable of flipping out of the duplex. It seems possible that if the complementary nucleotide C20 nucleotide flips out, this might facilitate proton exchange by allowing water to enter the duplex to access the X5 imino proton, but more detailed studies of the exchange kinetics of the X5 and neighboring imino protons are warranted.49,50 For the modified duplex, the T6 N3H resonance is not observed, suggesting increased exchange with solvent for the imino proton of the 3′-flanking T6•A19 base pair (Figure 7). This may be a consequence of altered ζ backbone torsion angle of S-cdG, which results in the opening of the 3′-neighbor base pair. While the MD simulations occur on a different timescale than the NMR experiments, in the MD simulations, transient opening of the T6•A19 base pair is predicted (Figure 11). In contrast, the thermal melting experiments (Figure 7) suggest that the 5′-neighbor C4•G21 base pair is more stable with respect to imino proton exchange.
Structure-Activity Relationships
(a) DNA Repair
In human global genome NER, the XPC/HR23B complex51–55 is believed to be involved in damage recognition. The XPA protein is also essential for NER. Yang et al.56 reported that it exists as a homodimer either in the free state or as a complex with RPA. For example, it binds to mismatched bubble substrates, including the C8-dG adducts of AF, AAF, and 1-nitropyrene, and the T[6,4]T photoproducts.57 XPA is proposed to be involved in the verification of DNA damage.54,55 It may also recruit repair factors and stabilize repair intermediates since it binds more efficiently to undamaged ds-ssDNA junctions with ssDNA branches,57 intermediate structures found in NER.
The destabilization of the S-cdG modified duplex and the perturbation of the X5•C20 and T6•A19 base pairs is likely relevant with respect to NER. Thermal destabilization of the duplex is believed to modulate recognition of a diverse group of damages by XPC.54,58–61 From studies of the yeast XPC orthologue Rad4 bound to DNA containing a cyclobutane pyrimidine dimer, Min and Pavletich62 concluded that Rad4 may exploit the destabilization of two base pairs. Interestingly, the 5R-thymine glycol lesion, another substrate for NER, also destabilizes two base pairs in DNA.63 The perturbation of the X5•C20 and T6•A19 base pairs in the S-cdG modified duplex may facilitate extrusion of both C20 and A19 (but not X5) out of the helix, enabling XPC/HR23B to recognize S-cdG prior to recruiting XPA.
(b) Error-Prone Replication Bypass
The bond between C8 of guanine and C5′ of 2′-deoxyribose locks the N-glycosyl torsion angle of S-cdG in the anti domain. Therefore, during translesion synthesis, an incoming dCTP can form a Watson–Crick base pair, whereas an incoming dTTP might form a wobble pair. The insertion of both dATP and dTTP were noted in pol V-dependent TLS by Jasti et al.26 Significantly, they noted the genotoxicity of the S-cdG lesion, which implied that DNA polymerases have difficulty in bypassing this locked nucleotide. They speculated that accommodation of the S-cdG lesion within the active site of the polymerase likely involves rotational adjustments of the nucleoside around the glycosyl bond. 26 Thus, future structural studies of template•primers containing the S-cdG lesion complexed with error-prone polymerase will be of interest.
Conclusions
The structure of S-cdG has been determined when placed opposite dC in DNA. The S-cdG•dC and the flanking base pairs maintain Watson-Crick hydrogen bonding. However, S-cdG exhibits the O4′-exo deoxyribose pseudorotation in DNA. This introduces significant helicoidal and base stacking perturbations into the duplex. The imino proton of the 3′-neighbor T•A base pair undergoes increased exchange with solvent, whereas the 5′-neighbor C•G base pair is only moderately influenced. Collectively, these structural and thermodynamic perturbations may be important in modulating the recognition of the S-cdG lesion during nucleotide excision repair.
Experimental Section
Synthesis
(a) N2-((Dimethylamino) methylene)-2′-deoxyguanosine (1)
To a suspension of 2′-deoxyguanosine (10 g, 35.06 mmol) in dry methanol (100 mL), N,N-dimethylformamide dimethyl acetal (18.7mL, 140.24 mmol) was added dropwise with vigorous stirring. The mixture was stirred at room temperature under argon for 72 h. The solid product was isolated by filtration, washed with cold methanol and dried. The product was isolated as a white solid in quantitative yield.
(b) N2-DMF-5′-phenylthio-2′,5′-dideoxyguanosine (2)
N2-DMF-2′-deoxyguanosine 1 (1 g, 3.74 mmol) and diphenyl disulfide (1.63g, 7.48 mmol) were dissolved in 15 mL dry DMF under argon, and PBu3 (1.85mL, 7.48mmol) was slowly added dropwise and the mixture was stirred at room temperature for 6 h. The reaction was monitored by thin layer chromatography (TLC) (90/10 CH2Cl2/MeOH, v/v). The reaction was quenched with 10 mL water and evaporated to a glassy syrupy residue. It was purified by silica gel column chromatography with a step gradient of methanol (0–7%) in DCM as the mobile phase. The product was isolated as white foam (1.42g, yield of 91 %)
(c) N2-DMF-5′,8-cyclo-2′,5′-dideoxyguanosine (3)
Previously crushed N2-DMF-5′,8-cyclo-2′,5′-dideoxyguanosine 2 (1.4g, 3.38 mmol) and triethyl phosphate were added to argon-purged 2 L quartz reactor, dissolved in 1 L dry acetonitrile via sonication. This solution was degassed by bubbling argon for 40 min. The reactor was sealed under argon atmosphere and irradiated at 254 nm UV light for 20 h. The reaction was monitored using TLC (85/15 CHCl3/MeOH, v/v). The solution was evaporated to dryness and the resulting brownish yellow solid was purified by silica gel column chromatography with a step gradient of methanol (0–10%) in CHCl3. The product was isolated as a light yellowish white solid (0.53 g, yield of 52%).
(d) N2-DMF-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′,5′-dideoxyguanosine (4)
N2-DMF-5′,8-cyclo-2′,5′-dideoxyguanosine 3 (1.4g, 4.6 mmol) and imidazole (1.27g, 18.7 mmol) were dried and dissolved in 20 mL dry DMF. TBDMS-Cl (1.39g, 9.2 mmol) was added to this solution while stirring under nitrogen atmosphere. The reaction mixture was stirred at room temperature for 20 h and monitored by TLC (93/7 CHCl3/MeOH, v/v). The solvent was dried under nitrogen and the resulting semi solid was purified on a silica gel column chromatography with a step gradient of methanol (0–3%) in chloroform. The product was isolated as a white solid (1.35g, yield of 70 %).
(e). N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′,5′-dideoxyguanosine (5)
N2-DMF-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′,5′-dideoxyguanosine 4 (1.1g, 2.63 mmol) was dissolved in a mixture of 50 mL methanol and 10 mL 29% aq. ammonia, and stirred overnight at room temperature. The solvents were removed under reduced pressure. The resulting white powder was co-evaporated with 5 mL dry pyridine 3 times. This white solid was dissolved in 12 mL dry pyridine, a few crystals of DMAP was added to it and isobutyryl chloride (0.56mL, 5.26 mmol) was added to it dropwise under nitrogen atmosphere. This reaction mixture was stirred at room temperature for 8 h and monitored by TLC (93/7 CHCl3/MeOH, v/v). The solvent was dried under reduced pressure and the resulting yellow solid was purified by silica gel column chromatography with a step gradient of methanol (0–2%) in DCM. The product was isolated as a white solid (1.0 gm, yield of 88 %).
(f). (5′S)-N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′-deoxyguanosine (7)
N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′,5′-dideoxyguanosine 5 (1.0 g, 2.3 mmol) was dissolved in 250 mL dry 1,4 dioxane, SeO2 (1.28 g, 11.5 mmol) and the mixture was refluxed for 24 h. The reaction was monitored by TLC (93/7 CHCl3/MeOH, v/v). The hot solution was passed through a celite pad and washed with 20 mL 10% methanol in chloroform. The filtrate was dried under reduced pressure to produce brownish white powder of N2-isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′-oxo-5′,8-cyclo-2′-deoxyguanosine. This product was added to 50 mL methanol and NaBH4 (0.174g, 4.6 mmol) was added to it in 3 portions. This reaction mixture was stirred at room temperature for 1 hr and the reaction was monitored by TLC (93/7 CHCl3/MeOH, v/v). The excess borohydride was neutralized by addition of 1N HCl dropwise to the solution. The solution was passed through a celite pad and evaporated to dryness. The resulting yellow solid was purified by silica gel column chromatography with a step gradient of methanol (0–5%) in chloroform. The product was isolated as a white solid (0.37 g, yield of 36%).
(g). (5′S)-N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5′,8-cyclo-2′-deoxyguanosine (8)
(5′S)-N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′,8-cyclo-2′-deoxyguanosine 6 (0.4 g, 0.89 mmol) was dissolved in 3 mL dry pyridine and evaporated to dryness. This process was repeated twice. The residual white solid was dissolved in 10 mL dry pyridine and DMT-Cl (0.92 g, 2.7 mmol) and a few crystals of DMAP were added to it. The mixture was heated at 80°C and stirred under nitrogen atmosphere for 8 h. It was monitored by TLC (94/5/1 CHCl3/MeOH/NEt3, v/v). The solution was cooled to ~5°C on an ice bath and quenched with methanol. The solvents were removed under reduced pressure and the resulting yellow solid was purified by silica gel column chromatography with a step gradient of methanol (0–1%) in chloroform containing 1% TEA. The product was isolated as a white solid (0.39 g, yield of 58 %).
(h). (5′S)-N2-Isobutyryl-5′-O-(4,4′-dimethoxytrityl)-5′,8-cyclo-2′-deoxyguanosine
(5′S)-N2-Isobutyryl-3′-O-(tert-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5′,8-cyclo-2′-deoxyguanosine 7 (0.3 g, 0.40 mmol) was dissolved in 15 mL of dry THF and a solution of 1M TBAF in THF (.8 mL, 0.8 mmol) was added. The reaction was stirred under nitrogen atmosphere for 5 h and monitored by TLC (92/7/1 CHCl3/MeOH/NEt3, v/v). The solvents were removed under reduced pressure and the resulting yellow solid was purified by silica gel column chromatography with a step gradient of methanol (0–2%) in chloroform containing 1% TEA. The product was isolated as a white solid (0.24 g, yield of 95%).
(i). (5′S)-5′,8-Cyclo-2′-deoxyguanosine Phosphoramidite Derivative
(5′S)-N2-Isobutyryl-5′-O-(4,4′-dimethoxytrityl)-5′,8-cyclo-2′-deoxyguanosine (from the previous reaction) (0.092 mg, 0.14 mmol) was dissolved in dry dichloromethane and evaporated to dryness. This process was repeated twice. The solid was dissolved in 5 mL of dry dichloromethane and kept under argon. Diisopropylethylamine (51 μl, 0.29 mmol) was added to it, then 2-Cyanoethyl N,N-diisopropylchlorophosphoramidite (34 μl, 0.15 mmol) was added to the stirring solution dropwise. The reaction was checked by TLC (95/4/1 CHCl3/MeOH/NEt3, v/v). After 1 hr the solution was cooled to ~5°C with an ice bath, 51 μl of DIEA and 0.2 mL of methanol was added to it. The solvents were removed under reduced pressure and the resulting light yellow semi solid was purified twice on a silica gel column chromatography with a step gradient of methanol (0–1%) in chloroform containing 1% TEA. The product was isolated as a white solid (0.084 g, yield of 70%).
Oligodeoxynucleotides
The 5′-d(GTGCGTGTTTGT)-3′ and 5′-d(ACAAACACGCAC)-3′ were synthesized and purified by anion-exchange chromatography by the Midland Certified Reagent Co. (Midland, TX). The dodecamer containing the S-cdG 5′-d(GTGCXTGTTTGT)-3′, where X represents the S-cdG, was synthesized, purified, and characterized using a slightly amended procedure of the synthesis reported by Romieu et al.34 The purity of the modified oligodeoxynucleotide was assessed by HPLC and mass spectrometry. Oligodeoxynucleotides were desalted by chromatography on Sephadex G-25. The 5′-d(GTGCGTGTTTGT)-3′ or 5′-d(GTGCXTGTTTGT)-3′ were annealed with the complementary strand 5′-d(ACAAACACGCAC)-3′ in buffer containing 10 mM NaH2PO4, 100 mM NaCl, and 50μM Na2EDTA (pH 7.0), respectively. The resulting duplexes were heated to 95 °C for 10 min, and cooled to room temperature. They were purified by DNA Grade hydroxylapatite chromatography using a gradient from 10 to 200 mM NaH2PO4 in 100 mM NaCl, 50 μM Na2EDTA (pH 7.0), and desalted using Sephadex G-25.
Melting Temperature
Melting temperatures of the DNA duplexes were measured in 10 mM NaH2PO4, 100 mM NaCl, 50 μM EDTA (pH 7.0) by UV/vis spectroscopy at 260 nm. The strand concentration was 10 μM. The thermal scan proceeded from 10 to 80 °C with an interval of 1 °C. The melting temperatures were calculated by differentiating the absorbance profiles.
NMR
Samples were at 1.0 mM strand concentration. Samples for the non-exchangeable protons were dissolved in 500 μL in 10 mM NaH2PO4, 100 mM NaCl, 50 μM Na2EDTA (pH 7.0). They were exchanged with D2O and suspended in 280 μL 99.996% D2O. The pH was adjusted with dilute DCl or NaOD. Experiments were performed at 800 MHz. COSY and NOESY spectra were recorded with 512 real data in the t1 dimension and 2048 real data in the t2 dimension. NOESY spectra were zero-filled during processing to create a matrix of 1024 × 1024 real points. NOESY experiments used TPPI quadrature detection64 and mixing times of 60, 150, 200 and 250 ms. The relaxation delay was 1.5 s. The TOCSY mixing time was 80 ms. The temperature was 25 °C. Chemical shifts were referenced to water. Data were processed using TOPSPIN65 and analyzed with the program SPARKY.66 The ECOSY data were recorded with 1024 real data in the t1 dimension and 4096 real data in the t2 dimension.38 The spectrum was zero-filled during process to create a matrix of 2048 × 16384 to increase digital resolution. The temperature was 30 °C. Samples for the observation of exchangeable protons were dissolved in 500 μL of 10 mM NaH2PO4, 100 mM NaCl, 50 μM EDTA, (pH 7.0) containing 9:1 H2O:D2O (v/v) (pH 7.0). Experiments were performed at 500 MHz. The temperature was 5 °C. The Watergate sequence was used for water suppression.67 The mixing time was 250 ms. The 31P-H1 experiments were carried out at the 1H frequency of 600 MHz. 31P -H3′ 3J couplings were applied to determine the phosphodiester backbone conformation.31P chemical shifts were referenced using indirect shift ratios.69
Distance and Dihedral Angle Restraints
Footprints were drawn around NOE crosspeaks obtained at a mixing time of 250 ms. Their intensities were determined by volume integrations. These were combined as necessary with intensities generated from complete relaxation matrix analysis of a starting structure to generate a hybrid NOE intensity matrix.41,70 The program MARDIGRAS39,40,71 iteratively refined the hybrid intensity matrix and optimized agreement between calculated and experimental NOE intensities. The RANDMARDI algorithm39 carried out iterations, randomizing peak volumes within limits specified by the input noise level.71 Calculations were initiated using isotropic correlation times of 2, 3, and 4 ns. Analysis of these data yielded experimental distance restraints used in rMD calculations (Table S3 in the Supporting Information), and the corresponding standard deviations for the distance restraints.
The deoxyribose pseudorotational angles (P) were estimated by examining the 3JHH of sugar protons.72 The data were fit to curves relating the coupling constants to the pseudorotation (P), sugar pucker amplitude (φ), and the percentage S-type conformation. The pseudorotation and amplitude ranges were converted to the five dihedral angles ν0 to ν4. Coupling constants measured from 1H-31P HMBC spectra were applied73,74 to the Karplus relationship75 to determine the backbone dihedral angle ε (C4′-C3′-O3′-P), related to the H3′-C3′-O3′-P angle by a 120° shift. The ζ (C3′-O3′-P-O5′) backbone angles were calculated from the correlation between ε and ζ in B-DNA.68 Empirical restraints preserved Watson-Crick hydrogen bonding and prevented propeller twisting between base pairs, except for A6•T19 base pair. Except for the modified, the flanking, and the terminal base pairs, other backbone torsion angle restraints were using empirical data derived from B-DNA.44
Molecular Dynamics Calculations
Restrained molecular dynamics (rMD) calculations for the modified oligodeoxynucleotide duplexes utilized a simulated annealing approach.76 The partial charges on the cdG nucleotide (Figure S5 in the Supporting Information) were obtained from density function theory (DFT) calculations using a neutral total charge, utilizing the B3LYP/6-31G* basis set and the program GAUSSIAN.77 To obtain the starting structures used for rMD calculations, the cdG-modified duplex was energy minimized using 200 iterations with the conjugate gradients algorithm. The rMD calculations were conducted with AMBER78 and the parm99 force field. The generalized Born (GB) model79 with parameters developed by Tsui and Case80 was used for implicit water simulation. The program CORMA was utilized to calculate the NOE intensities from the structures emergent from rMD calculations.
Molecular dynamics simulations in explicit water were performed using the AMBER force field. The average structure converged from the simulated annealing rMD calculations was used as the starting structure. This was placed in an 8.0 Å cubic TIP3P water box in each direction.81 The necessary Na+ ions were added to neutralize the duplex. The system was subjected to 1000 iterations of potential energy minimization using steepest descents. The solvent was brought to thermal equilibrium by a MD simulation at constant volume for 10,000 iterations with an integrator time of 1 fs, at 300 K. After equilibration of the system at 300 K, MD calculations were performed at constant pressure for 5 ns with an integrator time of 1 fs. Bond lengths involving hydrogens were fixed with the SHAKE algorithm.82 The particle mesh Ewald (PME) method was used to approximate non-bonded interactions.83,84 The cutoff radius for non-bonded interactions was 8.0 Å. The PTRAJ program from the AMBER package was used to analyze the MD trajectories. Helicoidal analyses were carried out with the programs CURVES85 and 3DNA.86
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA-55678 (M.P.S.) and ES-013324 (A.K.B.).
Footnotes
Supporting Information Available. The supporting information includes the complete reference (77); Scheme S1, synthesis of the phosphoramidite of S-cdG; Tables S1, 1H chemical shifts of the non-exchangeable protons of the S-cdG modified duplex; S2, 3J coupling constants and deoxyribose pseudorotation of the S-cdG modified duplex; S3, NOE distance restraints for the S-cdG modified duplex, and Figures S1, 31P-H3′ HMBC spectrum of the S-cdG modified duplex; S2, nucleotide-by-nucleotide sixth root residuals for the average refined structure of the S-cdG modified duplex; S3, backbone torsion angles of the refined structure of the S-cdG-contiaining duplex; S4, base pairing and base stacking helicoidal parameters of the S-cdG-containing duplex; and S5, calculated electrostatic potentials for the S-cdG nucleotide. This material is available free charge via the internet at http://pubs.acs.org.
References
- 1.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Faseb J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 2.Dedon PC. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 3.Keck K. Z Naturforsch B. 1968;23:1034–1043. [PubMed] [Google Scholar]
- 4.Jaruga P, Dizdaroglu M. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Dizdaroglu M. Biochem J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Dirksen ML, Jiang HX, Robbins JH. Biochem J. 1987;241:929–932. doi: 10.1042/bj2410929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dirksen ML, Blakely WF, Holwitt E, Dizdaroglu M. Int J Radiat Biol. 1988;54:195–204. doi: 10.1080/09553008814551631. [DOI] [PubMed] [Google Scholar]
- 8.Chatgilialoglu C, Bazzanini R, Jimenez LB, Miranda MA. Chem Res Toxicol. 2007;20:1820–1824. doi: 10.1021/tx700282x. [DOI] [PubMed] [Google Scholar]
- 9.Boussicault F, Kaloudis P, Caminal C, Mulazzani QG, Chatgilialoglu C. J Am Chem Soc. 2008;130:8377–8385. doi: 10.1021/ja800763j. [DOI] [PubMed] [Google Scholar]
- 10.Chatgilialoglu C, Ferreri C, Terzidis MA. Chem Soc Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 11.Mariaggi N, Cadet J, Teoule R. Tetrahedron. 1976;32:2385–2387. [Google Scholar]
- 12.Raleigh JA, Kremers W, Whitehouse R. Radiat Res. 1976;65:414–422. [PubMed] [Google Scholar]
- 13.Fuciarelli AF, Shum FY, Raleigh JA. Biochem Biophys Res Commun. 1986;134:883–887. doi: 10.1016/s0006-291x(86)80502-2. [DOI] [PubMed] [Google Scholar]
- 14.Chatgilialoglu C, Guerra M, Mulazzani QG. J Am Chem Soc. 2003;125:3839–3848. doi: 10.1021/ja029374d. [DOI] [PubMed] [Google Scholar]
- 15.Navacchia ML, Chatgilialoglu C, Montevecchi PC. J Org Chem. 2006;71:4445–4452. doi: 10.1021/jo060197z. [DOI] [PubMed] [Google Scholar]
- 16.Jaruga P, Theruvathu J, Dizdaroglu M, Brooks PJ. Nucleic Acids Res. 2004;32:e87. doi: 10.1093/nar/gnh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belmadoui N, Boussicault F, Guerra M, Ravanat JL, Chatgilialoglu C, Cadet J. Org Biomol Chem. 2010;8:3211–3219. doi: 10.1039/c004531d. [DOI] [PubMed] [Google Scholar]
- 18.Jaruga P, Dizdaroglu M. Biochem Biophys Res Commun. 2010;397:48–52. doi: 10.1016/j.bbrc.2010.05.050. [DOI] [PubMed] [Google Scholar]
- 19.Dizdaroglu M, Jaruga P, Rodriguez H. Free Radic Biol Med. 2001;30:774–784. doi: 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 20.Jaruga P, Birincioglu M, Rodriguez H, Dizdaroglu M. Biochemistry. 2002;41:3703–3711. doi: 10.1021/bi016004d. [DOI] [PubMed] [Google Scholar]
- 21.Kirkali G, de Souza-Pinto NC, Jaruga P, Bohr VA, Dizdaroglu M. DNA Repair (Amst) 2009;8:274–278. doi: 10.1016/j.dnarep.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez H, Jaruga P, Leber D, Nyaga SG, Evans MK, Dizdaroglu M. Biochemistry. 2007;46:2488–2496. doi: 10.1021/bi062022p. [DOI] [PubMed] [Google Scholar]
- 23.Jaruga P, Xiao Y, Nelson BC, Dizdaroglu M. Biochem Biophys Res Commun. 2009;386:656–660. doi: 10.1016/j.bbrc.2009.06.107. [DOI] [PubMed] [Google Scholar]
- 24.Brooks PJ. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkali G, Tunca M, Genc S, Jaruga P, Dizdaroglu M. Free Radic Biol Med. 2008;44:386–393. doi: 10.1016/j.freeradbiomed.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Jasti VP, Das RS, Hilton BA, Weerasooriya S, Zou Y, Basu AK. Biochemistry. 2011;50:3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasparutto D, Bourdat AG, D’Ham C, Duarte V, Romieu A, Cadet J. Biochimie. 2000;82:19–24. doi: 10.1016/s0300-9084(00)00347-3. [DOI] [PubMed] [Google Scholar]
- 28.Miaskiewicz K, Miller JH, Fuciarelli AF. Nucleic Acids Res. 1995;23:515–521. doi: 10.1093/nar/23.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karwowski BT. J Molec Struct Theochem. 2009;915:73–78. [Google Scholar]
- 30.Karwowski BT. Org Biomol Chem. 2010;8:1603–1609. doi: 10.1039/b920373g. [DOI] [PubMed] [Google Scholar]
- 31.Birnbaum GI, Cygler M, Dudycz L, Stolarski R, Shugar D. Biochemistry. 1981;20:3294–3301. doi: 10.1021/bi00514a048. [DOI] [PubMed] [Google Scholar]
- 32.Haromy TP, Raleigh J, Sundaralingam M. Biochemistry. 1980;19:1718–1722. doi: 10.1021/bi00549a031. [DOI] [PubMed] [Google Scholar]
- 33.Karwowski BT. Tetrahedron-Asymmetr. 2008;19:2390–2395. [Google Scholar]
- 34.Romieu A, Gasparutto D, Cadet J. Chem Res Toxicol. 1999;12:412–421. doi: 10.1021/tx9802668. [DOI] [PubMed] [Google Scholar]
- 35.Reid BR. Q Rev Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 36.Patel DJ, Shapiro L, Hare D. Q Rev Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 37.Boelens R, Scheek RM, Dijkstra K, Kaptein R. J Magn Reson. 1985;62:378–386. [Google Scholar]
- 38.Griesinger C, Sorensen OW, Ernst RR. J Am Chem Soc. 1985;107:6394–6396. [Google Scholar]
- 39.Borgias BA, James TL. J Magn Reson. 1990;87:475–487. [Google Scholar]
- 40.Borgias BA, James TL. Methods Enzymol. 1989;176:169–183. doi: 10.1016/0076-6879(89)76011-0. [DOI] [PubMed] [Google Scholar]
- 41.James TL. Curr Opin Struct Biol. 1991;1:1042–1053. [Google Scholar]
- 42.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 44.Arnott S, Hukins DWL. Biochem Biophys Res Comm. 1972;47:1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 45.Saenger W. Principles of Nucleic Acid Structure. Springer; New York: 1984. [Google Scholar]
- 46.Teitelbaum H, Englander SW. J Mol Biol. 1975;92:79–92. doi: 10.1016/0022-2836(75)90092-3. [DOI] [PubMed] [Google Scholar]
- 47.Mandal C, Kallenbach NR, Englander SW. J Mol Biol. 1979;135:391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- 48.Englander SW, Kallenbach NR. Q Rev Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 49.Leroy JL, Kochoyan M, Huynh-Dinh T, Gueron M. J Mol Biol. 1988;200:223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 50.Folta-Stogniew E, Russu IM. Biochemistry. 1996;35:8439–8449. doi: 10.1021/bi952932z. [DOI] [PubMed] [Google Scholar]
- 51.Legerski R, Peterson C. Nature. 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- 52.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, Hoeijmakers JHJ, Hanaoka F. Embo J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 54.Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 55.Riedl T, Hanaoka F, Egly JM. Embo J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang ZG, Liu Y, Mao LY, Zhang JT, Zou Y. Biochemistry. 2002;41:13012–13020. doi: 10.1021/bi026064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Roginskaya M, Colis LC, Basu AK, Shell SM, Liu Y, Musich PR, Harris CM, Harris TM, Zou Y. Biochemistry. 2006;45:15921–15930. doi: 10.1021/bi061626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunz D, Hess MT, Naegeli H. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 59.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 60.Dip R, Camenisch U, Naegeli H. DNA Repair (Amst) 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Buterin T, Meyer C, Giese B, Naegeli H. Chem Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Min JH, Pavletich NP. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 63.Brown KL, Roginskaya M, Zou Y, Altamirano A, Basu AK, Stone MP. Nucleic Acids Res. 2010;38:428–440. doi: 10.1093/nar/gkp844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marion D, Wuthrich K. Biochem Biophys Res Comm. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 65.Bruker Biospin, Inc. 3.1. Billerica, MA: 2010. [Google Scholar]
- 66.Goddard TD, Kneller DG. University of California. San Francisco: 2006. [Google Scholar]
- 67.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 68.Gorenstein DG. Meth Enzymol. 1992;211:254–286. doi: 10.1016/0076-6879(92)11016-c. [DOI] [PubMed] [Google Scholar]
- 69.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. J Biomol NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 70.Keepers JW, James TL. J Magn Reson. 1984;57:404–426. [Google Scholar]
- 71.Liu H, Spielmann HP, Ulyanov NB, Wemmer DE, James TL. J Biomol NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 72.Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR. Biochemistry. 1993;32:4207–4215. doi: 10.1021/bi00067a007. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Zuiderweg ERP, Glick GD. J Am Chem Soc. 1995;117:2981–2991. [Google Scholar]
- 74.Geen H, Freeman R. J Magn Reson. 1991;93:93–141. [Google Scholar]
- 75.Lankhorst PP, Haasnoot AG, Erkelens C, Altona C. Nucleic Acids Res. 1984;12:5419–5428. doi: 10.1093/nar/12.13.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirkpatrick S, Gelatt CD, Jr, Vecchi MP. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 77.Frisch MJ, et al. Gaussian, Inc. Wallingford, CT: 2004. [Google Scholar]
- 78.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bashford D, Case DA. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 80.Tsui V, Case DA. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 81.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 82.Ryckaert J-P, Ciccotti G, Berendsen HJC. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 83.Darden T, York D, Pedersen L. J Chem Phys. 1993;12:10089–10092. [Google Scholar]
- 84.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;19:8577–8593. [Google Scholar]
- 85.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 86.Lu XJ, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.