Abstract
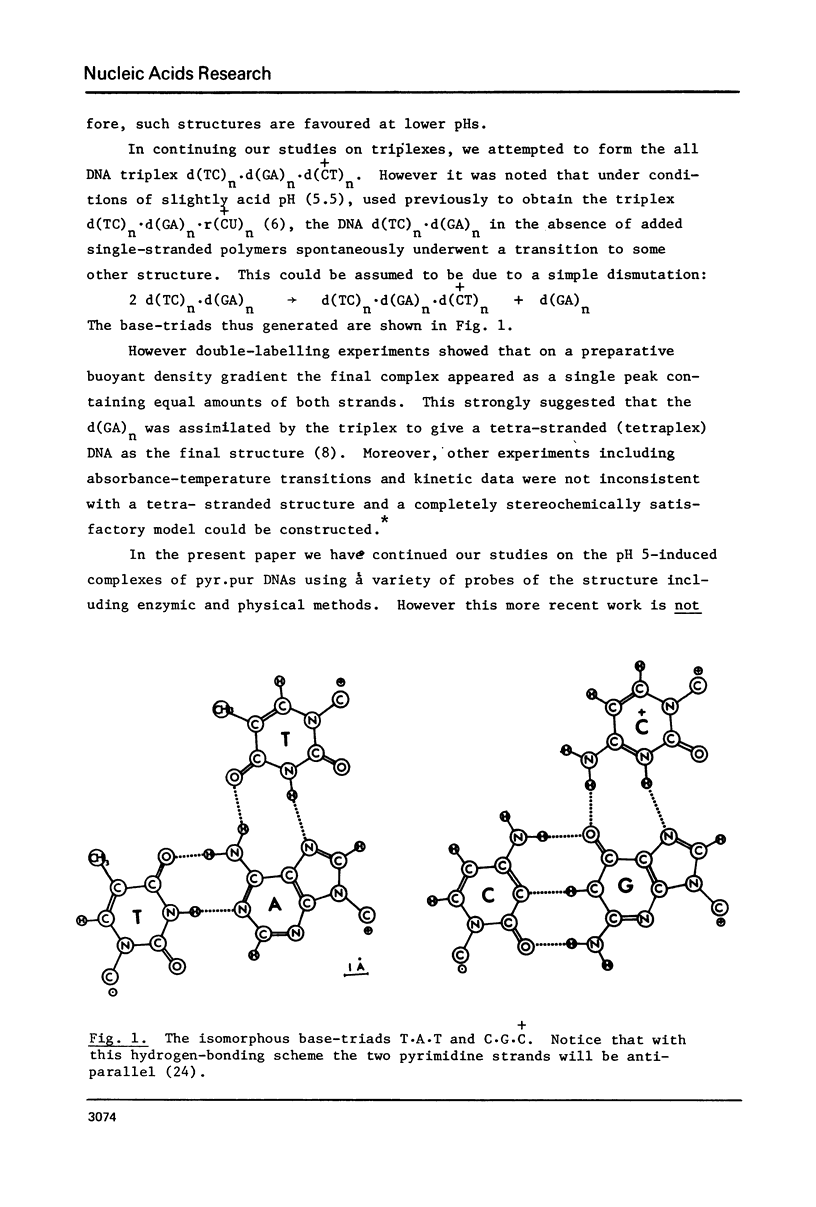
(Pyrimidine)n . (purine)n DNAs of repeating sequences form a distinctive complex on lowering the pH below 6. Previously this complex was thought to be tetra-stranded. The present work is inconsistent with this view, and four lines of evidence show that the complex consists of a triplex together with a poly d(purine) possessing secondary structure. Formula: (see text). (a) S1 nuclease digestion leads to degradation of 50% of the poly d(purine) content of the pH 5-induced complex. (b) Buoyant density studies demonstrate that there is no interaction between the triplex and added free poly d(purine) and also that the complex formed from duplex DNA contained poly d(purine) which is free to form a triplex on addition of an appropriate poly d(pyrimidine) in the correct stoichiometry. (c) The hyperchromic shifts of the triplex and poly d(purine), upon melting, are mutually independent. (d) The circular dichroism spectrum of the complex is simply the weighted average of a triplex together with a free poly d(purine). The triplexes have tm's approximately 20 degrees higher than the corresponding duplexes under comparable conditions and they are extremely resistant to various deoxyribonucleases; properties which may prove useful for their isolation from natural sources.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- BERNARDI G., GRIFFE M. STUDIES ON ACID DEOXYRIBONUCLEASE. II. ISOLATION AND CHARACTERIZATION OF SPLEEN-ACID DEOXYRIBONUCLEASE. Biochemistry. 1964 Oct;3:1419–1426. doi: 10.1021/bi00898a005. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C. Spacing of polypyrimidine regions in mouse DNA as determined by poly(adenylate, guanylate) binding. J Mol Biol. 1978 Jun 5;121(4):541–559. doi: 10.1016/0022-2836(78)90399-6. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Zimm B. H. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J Mol Biol. 1965 Jul;12(3):525–536. doi: 10.1016/s0022-2836(65)80310-2. [DOI] [PubMed] [Google Scholar]
- Denniston-Thompson K., Moore D. D., Kruger K. E., Furth M. E., Blattner F. R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Morgan A. R., Ratliff R. L. A comparison of the circular dichroism spectra of synthetic DNA sequences of the homopurine . homopyrimidine and mixed purine- pyrimidine types. Nucleic Acids Res. 1978 Oct;5(10):3679–3695. doi: 10.1093/nar/5.10.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Harwood S. J., Wells R. D. Micrococcus luteus deoxyribonucleic acid polymerase. Studies on the initiation of deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1970 Nov 10;245(21):5625–5634. [PubMed] [Google Scholar]
- Johnson D., Morgan A. R. Unique structures formed by pyrimidine-purine DNAs which may be four-stranded. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1637–1641. doi: 10.1073/pnas.75.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Morgan A. R., Coulter M. B., Flintoff W. F., Paetkau V. H. Enzymatic synthesis of deoxyribonucleic acids with repeating sequences. A new repeating trinucleotide deoxyribonucleic acid, d(T-C-C)n-d(G-G-A)n. Biochemistry. 1974 Apr 9;13(8):1596–1603. doi: 10.1021/bi00705a007. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Pulleyblank D. E. Native and denatured DNA, cross-linked and palindromic DNA and circular covalently-closed DNA analysed by a sensitive fluorometric procedure. Biochem Biophys Res Commun. 1974 Nov 27;61(2):396–403. doi: 10.1016/0006-291x(74)90970-x. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Murray N. L., Morgan A. R. Enzymatic and physical studies on the triplex dTn.dAn.rUn. Can J Biochem. 1973 Apr;51(4):436–449. doi: 10.1139/o73-051. [DOI] [PubMed] [Google Scholar]
- Price P. A., Stein W. H., Moore S. Effect of divalent cations on the reduction and re-formation of the disulfide bonds of deoxyribonuclease. J Biol Chem. 1969 Feb 10;244(3):929–932. [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Sederoff R., Lowenstein L. Polypyrimidine segments in Drosophila melanogaster DNA: II. Chromosome location and nucleotide sequence. Cell. 1975 Jun;5(2):183–194. doi: 10.1016/0092-8674(75)90026-4. [DOI] [PubMed] [Google Scholar]
- Shapiro A. L., Viñuela E., Maizel J. V., Jr Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem Biophys Res Commun. 1967 Sep 7;28(5):815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- Subramanian K. N., Dhar R., Weissman S. M. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. III. Construction of the total sequence of EcoRII-G fragment of SV40 DNA. J Biol Chem. 1977 Jan 10;252(1):355–367. [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]


