Abstract
The basic structural unit of the signaling complex in bacterial chemotaxis consists of the chemotaxis kinase, CheA, a coupling protein, CheW, and chemoreceptors. These complexes play an important role in regulating the kinase activity of CheA and in turn controlling the rotational bias of the flagellar motor. Although individual 3D structures of CheA, CheW and chemoreceptors have been determined, yet the interaction between chemoreceptor and CheW is still unclear. We used Nuclear Magnetic Resonance (NMR) to characterize the interaction modes of chemoreceptor and CheW from Thermotoga maritima. We find that chemoreceptor binding surface is located near the highly conserved tip region of the N-terminal helix of the receptor, whereas the binding interface of CheW is placed between the beta-strand 8 of domain 1 and the beta-strands 1 and 3 of domain 2. The receptor-CheW complex shares a similar binding interface to that found in the “trimer of dimers” oligomer interface seen in the crystal structure of cytoplasmic domains of chemoreceptors from Escherichia coli. Based on the association constants inferred from fast exchange chemical shifts associated with receptor-CheW titrations, we estimate that CheW binds about 4 times tighter to its first binding site of the receptor dimer than to its second binding site. This apparent anticooperativity in binding may reflect the close proximity of the two CheW binding surfaces near the receptor tip or the further complicating the events at this highly conserved region of the receptor. This work describes the first direct observation of the interaction between chemoreceptor and CheW.
Keywords: chemotaxis, CheW, receptor, MCP, negative cooperativity
Introduction
The bacterial chemotaxis signaling system is one of the most well understood in biology. During chemotaxis, motile bacteria modulate their swimming behavior to direct their movement toward optimal environments by tracking temporal changes in chemical concentrations with high sensitivity and over a wide range of concentrations. The basic structural unit of the signaling complex in chemotaxis consists of chemotaxis kinase, CheA, coupling protein, CheW, and transmembrane receptors called methyl-accepting proteins, or MCPs. In the most completely understood receptor systems, reversible methylation and attractant ligand binding are opposing events that the signaling complexes use to regulate the autophosphorylation activity of CheA. This opposed modulation of kinase activity enables the cell to make comparison of the past and current environments and to control the activity of CheA in response to this comparison 1. Once autophosphorylated, CheA transfers its phosphoryl group to the small soluble protein CheY; then CheY~P diffuses to the flagellar motor, where it binds, and enhances the switching of the flagellar motor from the counter-clockwise to clockwise rotational state.
The best understood chemotaxis receptors consist of a periplasmic domain that interacts with the appropriate target ligand, a transmembrane region followed by a HAMP domain 1 that acts as a signal conversion module, and then by a large cytoplasmic domain. The cytoplasmic portion of the receptor interacts with CheA and CheW. The cytoplasmic region of the receptor consists of a long helical region that folds back on itself to form a long two-stranded anti-parallel coiled coil. Each monomer of the receptor is assembled into dimers to form a four stranded coiled coil that seems to be the basic structural unit of the cytoplasmic region of all chemotaxis receptors 2. To date, crystal structures of the receptor cytoplasmic domains from E. coli and T. maritima have been published. The cytoplasmic domains of all these receptors share the four stranded coiled coil structure and have a region of remarkable sequence similarity at the tip region where the individual chains fold back on themselves 2. In Figure 1, the crystal structure of a portion of the soluble receptor TM0014 is shown in a cartoon representation of the backbone (left) where the individual polypeptide chains can be seen. Residues 90 to 206 (TM001490–206) are shown in a space filling representation (right) and include the conserved tip region. This receptor region has been implicated by genetic and biochemical means as the site of interaction with CheA and CheW.
Figure 1.
Ribbon and worm diagrams of the structure of TM0014 (a) showing the two monomer chains (red and gray). (b) A representation of the short construct TM001490–206 in a space-filling view (red and gray) using the coordinates of TM0014. The ribbon and worm representations show those residues deleted in the shortened construct.
CheW is a coupling protein that plays an important role in the formation of the receptor – signaling complex. It contains two β sheet domains. Each domain consists of a five-stranded β –barrel that forms an internal hydrophobic core for protein – protein interaction. Its structure is quite similar to that of CheA P5 domain and it is known that they interact with each other.
There have been several studies on the interaction of the chemoreceptor complexes with other chemotactic proteins; 9. Although the crystal structure of CheA containing the CheW and receptor coupling domains (P4P5) in T. maritima has been solved 4, the interaction between the chemoreceptors and CheW is still unclear. The 3D structures of CheW and TM0014 have been solved by NMR and X-ray crystallography respectively. The chemotaxis proteins from T. maritima are well-behaved at millimolar concentration and are stable at up to 90°C. This makes them well suited for structural analysis by solution nuclear magnetic resonance (NMR) methods. Here, we report the identification of a binding interface between chemoreceptor TM0014 and CheW from T. maritima using NMR methods.
Results
TM0014 is a soluble receptor without the transmembrane regions 10. Each monomer in the four-helix bundle dimer has 213 residues. The extended shape of this protein makes its effective size significantly larger than a globular protein of comparable molecular weight. Its rotational diffusion properties are unfavorable for detailed NMR studies of the backbone of the dimer. To facilitate high-resolution NMR study of the receptor backbone, we constructed a shorter fragment of the receptor, TM001490–206, that contains the helix bundle tip region of the full-length receptor, and a 5 His tag, and an additional tyrosine residue (used to determine protein concentration) at its N-terminus. As expected, the tyrosine mutant and TM001490–206 both showed the same chemical shift perturbation in the 1H-15N TROSY-HSQC spectra upon addition of CheW, confirming that the tyrosine mutant does not perturb the CheW-receptor interaction.
Sequential assignment of the receptor fragment
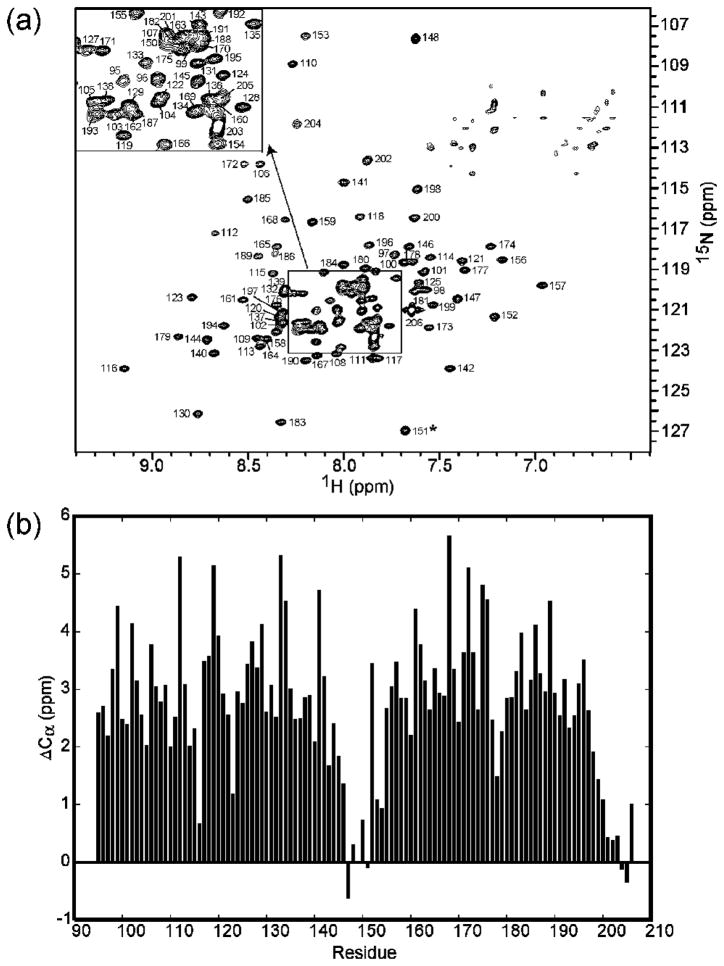
The backbone amide assignments of the receptor fragment TM001490–206 were completed using TROSY-based HNCACB, HNCA, HNCOCA, and HNCOCACB experiments (see Experimental Procedures). We used U-[2H,15N,13C]-labeled TM001490–206 for the backbone assignment. Figure 2a showed the assigned 1H-15N TROSY-HSQC spectrum of TM001490–206. The backbone amide resonances of TM001490–206 were well dispersed with mostly sharp resonances at 40°C. A total of 6 residues were unassigned because we were not able to detect their backbone amide resonances in the 3D spectra. These residues were Lys90, Ser91, Gly92, Thr93, and Asn 94 at the N-terminus, and Glu149 located right at the center of the tip region. The missing resonances of Lys90, Ser91, Gly92, Thr93, Asn 94, and Glu149 could be due to dynamic effects of conformational heterogeneity and/or solvent-exchange effects. Figure 2b shows a plot of chemical shift index10 as a function of residue number. As can be seen in the figure, the secondary structure of the fragment is largely helical with a distinct break at residues 146–151 corresponding to the position where the polypeptide chain folds back on itself to form the tip of the receptor.
Figure 2.
The assigned 1H-15N TROSY HSQC spectrum and the secondary structure analysis of TM001490–206. (a) 1H-15N correlation map of 15N-labeled TM001490–206 from T. maritima collected at 40°C and pH 7.4. The assignment of backbone amides is indicated with sequence numbers. Asterisks indicate aliasing of the peaks from outside the spectral window along 15N dimension. The central region enclosed by the square is expanded and represented as a small section in the upper left corner of the spectrum. (b) The differences between the observed Cα chemical shifts of residues 90–206 and their respective random-coil chemical shifts were plotted as a function of residue number. Continuous stretches of positive ΔCα are indicative of helices, while the small stretches with both positive and negative ΔCα values at residues 146–151 are suggestive of loops.
The receptor binding interface of CheW
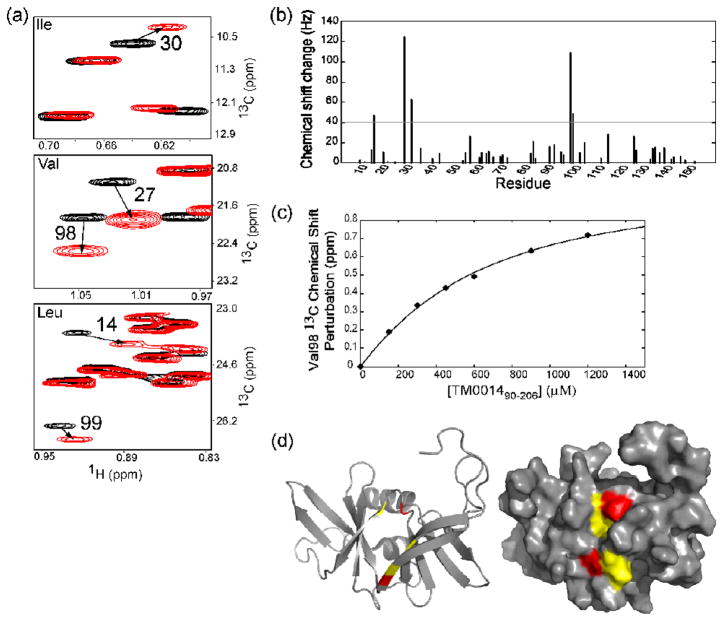
We performed chemical-shift-perturbation mapping with {[U-2H,12C], Ileγ1-[13CH3], Leu,Val-[13CH3]}-labeled12 but otherwise deuterated CheW by titrating it with protonated and deuterated TM001490–206 and TM0014. The CheW interface residues are identified by comparing the spectra collected with a protonated partner and a deuterated partner13. Figure 3a shows the Ile, Leu and Val methyl side chain spectrum of {[U-2H,12C], Ileγ1-[13CH3], Leu,Val-[13CH3]}-labeled CheW in the presence of an excess of deuterated TM001490–206. Comparing the isoleucine, valine, and leucine methyl group spectra of CheW alone (black) and CheW in the presence of TM001490–206 (red), respectively, we observed residue specific chemical shift changes in CheW upon binding TM001490–206. Essentially identical chemical shift changes were observed when we added an excess of intact TM0014 to the methyl labeled CheW (Supplement data S1), showing that the shorter receptor construct contains the same binding site for CheW as the intact receptor. The methyl resonances of CheW residues Val27 and Val98 broadened greatly when bound to protonated TM001490–206 but were sharper and could be observed at higher saturation with deuterated receptor. This difference in broadening between using the protonated and deuterated versions of TM001490–206, most likely reflects the proximity of the valine methyl groups of CheW to protons on the receptor13. The strongly distance dependent dipolar broadening is reduced when the protons of the receptor are replaced by deuterons.
Figure 3.
The interaction between the coupling protein CheW and the soluble receptor fragment TM001490–206. (a) A superimposition of 1H-13C HMQC spectra of ILV methyl groups of CheW (black) and CheW in the presence of TM001490–206 (red). Residues with significant chemical shifts are numbered with the arrows indicating the direction of the shift. (b) Measured chemical shift perturbations of CheW by the presence of receptor as a function of residue number represented as combined chemical shift (ΔH2+ΔN2)1/2 in hertz. (c) Changes in chemical shift of V98 of CheW by TM001490–206. The calculated binding curve is shown with a best-fit dissociation constant of 300 μM. (d) TM001490–206 binds to a hydrophobic surface of CheW, formed by β-strands 1, 2, 3, and 8 and the loop bridging β-strands 2 and 3. The residues the showing the largest chemical shift changes are shown in red (residues 27 and 98) and the residues with moderate shifts are shown in yellow (residues 14, 30 and 99).
Resonances from other neighboring residues, including Leu14, Ile30 and Leu99, also shifted moderately in the presence of TM001490–206. The largest chemical shift changes were localized in one hydrophobic patch on CheW, as illustrated in the surface representation of CheW shown in Figure 3d. The residues of CheW with the largest shifts, and the residues with more moderate shifts, were colored red and yellow, respectively. As can be seen, significant changes in chemical shift were observed in residues located in β-strands 1–3 and 8 of CheW. We assayed the binding affinity of CheW in the presence of TM001490–206 by plotting the chemical shift perturbation of a given resonance from its free position by different concentrations of TM001490–206. Several peaks shifted progressively as the receptor fragment was added, showing that the binding was in the fast exchange regime. The titration curve for Val98 was shown in Figure 3c as a plot of the observed chemical shift of the methyl resonance of Val98 as a function of TM001490–206 concentration. These data were fitted to a hyperbolic binding isotherm, and the dissociation constant was determined to be ~ 300 uM.
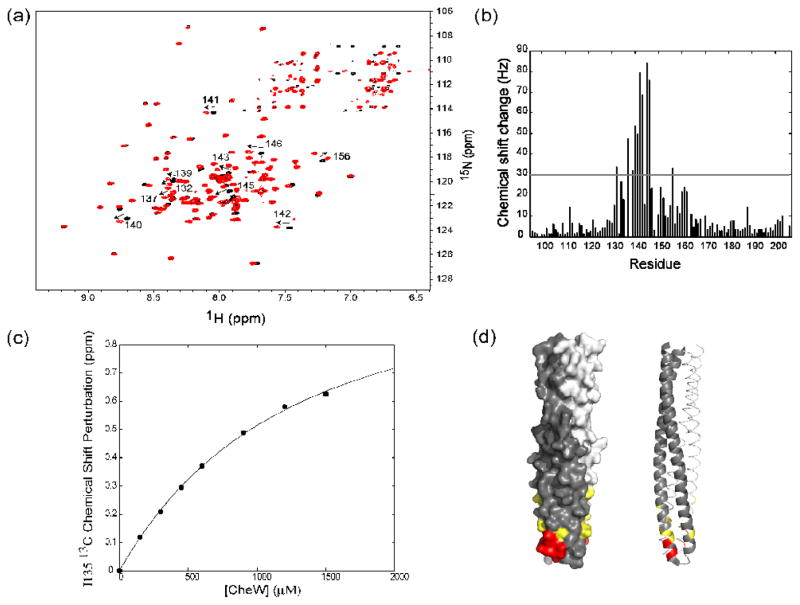
The CheW binding interface ot TM001490–206
The binding surface of TM001490–206 in contact with CheW was observed, using a series of 1H-15N TROSY-HSQC spectra of TM001490–206 taken over the course of a titration with CheW. Figure 4a showed the superimposition of two 1H-15N HSQC spectra of TM001490–206 (black) and TM001490–206-CheW complex (red). Significant chemical shift changes were observed in TM001490–206 due to the binding of CheW. The backbone amide resonances of residues 132, 137, 139, 140, 141, 142, 143, 145, 146 and 156 of TM001490–206 shifted significantly when associated with CheW. It should be noted that the backbone amide of residue 135 did not shift significantly but its methyl resonance did shift (not shown). Figure 4b showed the observed chemical shift perturbation for each assigned residue of TM001490–206 as a plot of the observed chemical shift change as a function of residue number. Significant chemical shift changes were localized in one area of TM001490–206. Figure 4d showed the residues, with observed combined chemical shift changes above 30 Hz, colored red on the crystal structure of TM0014. These residues formed a well-defined hydrophobic patch at the tip region of TM0014 and were exposed to the solvent where CheW can interact. We estimated the strength of the CheW-receptor interaction by plotting the observed chemical shift of the methyl resonance peak of Ile135 as a function of CheW concentration in Figure 4c. The curve suggested a dissociation constant of ~1.2 mM.
Figure 4.
The interaction of TM001490–206 with the coupling protein CheW. (a) A superimposition of 1H-15N TROSY HSQC spectra of TM001490–206 (black) and TM001490–206 in the presence of CheW (red). Large chemical-shift changes (indicated by arrows), are seen in the tip region of the receptor (residues 132, 137, 139, 140, 141, 142, 143, 145, 146, and 156). (b) Measured combined chemical shifts changes in the presence of CheW plotted as a function of residue number for TM001490–206. (c) Changes in chemical shift of TM001490–206 as a function of CheW concentration. The calculated binding curve is shown for residue 135 with best-fit dissociation constant of 1200 μM. (d) Mapping of the residues of TM001490–206 perturbed upon binding of CheW on the structure TM001490–206. Residues with chemical shifts larger than 50 Hz are shown in red; those with shifts between 30 and 50 Hz are shown in yellow.
As shown from the data presented in Figure 3, the NMR based titration of labeled CheW by unlabeled receptor fragment suggests an affinity about four fold stronger than is observed when labeled receptor is titrated with unlabeled CheW. This discrepancy is well outside of experimental error and reflects the stoichiometry and nature CheW-receptor interaction. When CheW is titrated by excess receptor fragment, the binding is predominated by one CheW bound per receptor dimer. However, when receptor fragment is titrated by excess CheW, the binding now reflects a combination of both one and two CheW molecules bound per receptor dimer. If each monomer of the receptor dimer bound CheW identically and independently of whether a CheW was bound at the other monomer, both titrations should show the same apparent affinity. However, if the binding of the first CheW results in a lower affinity for the binding of the second, it would require a higher CheW concentration to fully saturate the receptor dimer than the concentration of receptor needed to saturate CheW. This is exactly what is observed.
Discussion
Our results suggest that TM001490–206 retains the native structure of intact TM0014. Static light scattering data (not shown) confirm the dimer nature of the shortened version of TM0014 as expected. The chemical shift index results demonstrate that the secondary structure of TM001490–206 consists of two helices with a distinct break at residues 146–151, in agreement with the crystal structure of TM0014. When we superimpose the isoleucine methyl side chain spectrum of TM0014 with TM001490–206 (Supplement data S2), TM001490–206 shows a subset of the resonances observed for TM0014. This suggests that TM0014 and TM001490–206 share a common structure. The TM0014 construct was shortened further to form TM0014107–191. TM0014107–191 has a very different isoleucine methyl chemical shift spectrum when compared to the other two fragments (Supplement data S2). In addition, TM0014107–191 is not as stable as TM001490–206 and TM0014 and TM0014107–191 tends to precipitate at high concentration. This suggests that the length of the receptor plays an important role in maintaining the conformation of the tip region as well as providing structural stability.
In this study, we have identified the interaction interfaces of TM001490–206 and CheW complex in solution by chemical shift perturbation techniques. The receptor interaction surface of CheW consists of residues located in the solvent-exposed patches of β strands 1, 3, and 8 as summarized in Figure 3d. The presence of TM0014 or TM001490–206 cause similar methyl chemical shift changes in the HMQC spectra of CheW (see supplemental data). This confirms that the binding of TM001490–206, and CheW has not been affected by shortening the TM0014 fragment. Previous studies from genetic suppressor screens and biochemical assays suggested residues of E. coli CheW to be important for interaction with the receptors. These homologous residues are also involved in the interaction between CheW and receptor in T. maritima, consistent with the high degree of structural similarity, although their amino acid sequences were not very similar 3,14.
The CheW interaction surface of TM001490–206 has been identified and consists mainly of residues near or at the tip region, on the N-terminal side of the tip of the receptor (N-terminal helix). A few residues near the hairpin loop on the C-terminal side (C-terminal helix) of the receptor showed small chemical shift changes when CheW was added. This suggests that the residues on the N-terminal side play major role whereas the residues on the C-terminal side play a more minor role in the receptor and CheW interaction. Lui and Parkinson 7 determined a few suppressor mutation sites on the receptors, including two residues located near our proposed binding sites. Mehan et al. 9 used chemical modifications to map regions of the E. coli receptor Tar that interfered with normal regulation of CheA and found a sub-population of sites near the receptor tip that inhibited activation of CheA in the presence of CheW. These are in similar locations to those we have identified in TM001490–206 A recent study by Mowery et al.15 showed that all R366 mutants of Tsr, which are equivalent to R146 of TM001490–206, either impaired or destroyed Tsr function, possibly by disrupting the interaction of the receptor and CheW and in turn prevented the formation of the signaling complexes. This evidence seems to agree well with the chemical shift perturbation studies.
Currently there are two predominant structural models for receptor-receptor interaction in an extended signaling network; the trimer of dimers model, based on crystal packing of the E. coli serine receptor cytoplasmic domain 16, and the hedge row of dimers model based on the crystal packing of a T. maritima receptor cytoplasmic domain 4. The trimer of dimers model has been generally supported by electron tomography 17,18, genetic studies 19, and biochemical analysis 20,21,22. Recently, there has been a report about the universal architecture of chemoreceptor array among many bacteria, including T. maritima 18, that favors the hexagonal symmetry, the trimers of dimers model. In addition, a series of recent studies from the Hazelbauer laboratory suggests that trimers of receptor dimers are critical for receptor function 23,24,25 in the E. coli system. The preponderance of current evidence supports the trimer of dimers view of receptor organization.
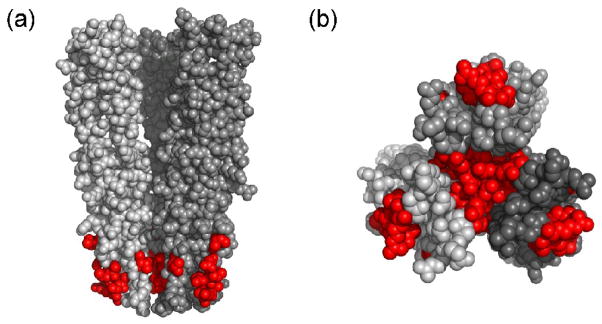
Analysis from the Parkinson laboratory 19 suggests that there are 11 principal trimer contact residues that are highly conserved among chemotactic bacteria. Somewhat surprisingly, 6 of those 11 trimer contact residues are also involved in the receptor and CheW interaction based on our NMR results. These residues were mapped onto the 3D structure of the cytoplasmic domain of Tsr in the trimer of dimer arrangement. In Figure 5, one can see that these residues are also involved in the receptor interaction. This implies that the trimer interface shares similar contact regions with the receptor and CheW. Earlier evidence by Studdert and Parkinson supported this idea. They demonstrated that the presence of CheW and CheA prevented the exchange between members with recently made receptor molecules and stabilized the trimer formation 21. One explanation could be that the principal trimer contact is blocked by CheW binding to receptors and the formation of trimers of dimers as well as the interaction between the receptor and CheW are a competitive process. Recently, Cardozo et al. showed that over-expression of CheW in E. coli cells, resulted in disruption of receptor arrays 26, supporting the notion that CheW binding and trimer formation are competitive processes.
Figure 5.
Mapping of contact residues of TM001490–206 that are involved in the CheW interaction (red) onto the 3D crystal structure of E. coli trimer of dimer Tsr receptor. (a) Space-filling side view of cytoplasmic domain of Tsr residues 335–446 in the trimer of dimer arrangement. Residues 377, 381, 384, 385, 388, 398 of Tsr correspond to residues 135, 139, 142,143, 146, and 156 of TM0014. (b) A bottom view of the space-filling model of the trimer of dimers.
The remarkable sequence conservation observed at the tip of chemotaxis receptors in virtually all motile bacteria is not seen in CheW. CheW homologs show multiple substitutions throughout the sequence, although these changes are generally conservative. This suggests that the receptor tips have a more complex biochemical role, involving receptor array assembly as well as simple binding interactions with CheW and CheA. Previous studies have revealed that most mutants at the tip of the E. coli serine receptor are defective in their ability to mediate chemotactic responses, but it has not been easy to assign one simple biochemical defect to these mutations 19.
The apparent negative cooperativity we observe in CheW binding suggests that there might be steric hindrance caused by CheW binding to the first binding site of the receptor, which makes it difficult to fit another CheW on the second binding site of the receptor at the relatively small receptor tip. It is also possible that when the first CheW binds, it might produce structural changes to the second binding site that result in lowered affinity. Alternatively, since the P5 domain of CheA is structurally homologous to CheW 2,5, one of the CheW binding sites on the receptor might be the true binding surface for CheW, while the weaker binding we see could represent the binding surface for the CheA P5 domain on the receptor. The relatively low affinity of the first binding is in contrast to that seen in the E. coli system, where CheW appears to bind much more strongly to the chemotaxis receptors 27. This affinity difference is reversed when considering the CheW – CheA interaction. Thermotoga CheW binds much more strongly to its CheA than does the E. coli CheW to its CheA. It remains unclear what the physiological consequences might be of these differences in affinity and what role the apparent negative cooperativity we observe may have on the signaling properties of the receptor-CheA-CheW complex.
Experimental Procedures
Protein expression and purification
Sample Preparation
The TM0014 construct was received from Brian Crane’s laboratory. PCR methods were used to generate a DNA fragment encoding TM0014 from Lys90 to Thr206 to create a TM001490–206 construct. The DNA encoding TM001490–206 was subcloned into the vector pET28a (Novagen), and the N-terminal histidine of the his tag was mutated to tyrosine using QuickChange mutagenesis (Stratagene). TM001490–206 proteins with an N-terminal His5 tags were transformed and over expressed in E. coli strain BL21(RIL DE3) (Novagen). During the log phase of bacterial growth, isopropyl β-D-thiogalactopyranoside (IPTG) was added at 1mM concentration. U-[15N,13C,2H] - labeled TM001490–206 were grown in minimal media which consisted of 1 mM magnesium sulfate, 0.1 mM calcium chloride, 0.5 μg/ml of thiamine, and 100 μg/ml of ampicillin, with the addition of 1g/L of 15NH4Cl as the main nitrogen source, and 2g/L U[13C,2H]-glucose or 2g/L of U-[12C,2H]-glucose (CIL, Andover, MA) as the main carbon source. {[U-2H,12C], Ileγ1-[13CH3]}-labeled TM001490-206 and TM0014 and {[U-2H,12C], Ileγ1-[13CH3], Leu,Valδ-[13CH3]}-labeled CheW were prepared using the method previously described by Kay laboratory 12. His5-tagged TM001490–206 was purified using nickel affinity column (Ni-NTA Agarose, QIAGEN) and size exclusion chromatography. CheW was over expressed and purified as previously described 3. Purified proteins were dialyzed in 50mM Na2HPO4 and 1mM EDTA at pH 7.4. All NMR samples contained 0.02% sodium azide and 10% D2O.
NMR data collection and processing
NMR data were collected at 40°C on a Varian 600Mhz or a Bruker 800Mhz spectrometer, each using a 1H/13C/15N/2H cryogenically cooled probe equipped with a Z pulsed-field gradient. Sequential assignments of TM001490–206 were accomplished with a U-[15N,13C,2H] sample of TM001490–206, using TROSY b ased 2D Heteronuclear Single Quantum Coherence (HSQC) and TROSY based 3D triple resonance HNCACB 28,29,30, HN(CO)CACB, HNCA 28,29,31,32,33 and HN(CO)CA experiments 34. All titration experiments were performed. In the first titration, 300 μM {[U-2H,12C], Ileγ1-[13CH3]}-labeled TM001490–206 was titrated with 2H-labeled CheW to a concentration between 0 and 1500 μM. At each titration point a Heteronuclear Multiple Quantum Coherence (HMQC) spectrum was collected at 40C. The same titration was repeated with 300 μM {[U-2H,12C], Ileγ1-[13CH3], Leu,Valδ-[13CH3]}-labeled CheW mixed with varying concentrations of [U-2H]-labeled TM0014107–191 between 0 and 1500 μM. The titration analysis was done assuming fast exchange using one to one binding of CheW to TM001490–206 monomer units to form the CheW - TM001490–206 complex. 1H-13C HMQC spectra were collected at CheW: TM001490–206 molar ratios of 0, 0.50, 0.75, 1.0, 2.0, 3.0, 4.0, and 5.0 when methyl TM001490–206 spectra were observed. A similar titration was performed to obtain methyl CheW spectra with TM001490–206:CheW molar ratios of 0, 0.50, 0.75, 1.0, 2.0, 3.0, and 4.0. Binding constants were estimated by fitting the observed population-weighted displacement of the resonance peaks from free to bound states during the titration.
Supplementary Material
Acknowledgments
We thank Brian Crane and Abiola Pollard (Cornell University) for the TM0014 construct. Robert Levenson provided discussion and technical advice. This work was support by NIH grant GM59544 to FWD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A. 2007;104:2885–90. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–5. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 4.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–7. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 5.Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002;277:22251–9. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- 6.Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interaction surfaces on CheW. J Biol Chem. 2002;277:23596–603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 7.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–51. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–7. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehan RS, White NC, Falke JJ. Mapping out regions on the surface of the aspartate receptor that are essential for kinase activation. Biochemistry. 2003;42:2952–9. doi: 10.1021/bi027127g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–44. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–51. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 12.Tugarinov V, Kay LE. An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR. 2004;28:165–72. doi: 10.1023/B:JNMR.0000013824.93994.1f. [DOI] [PubMed] [Google Scholar]
- 13.Hamel DJ, Dahlquist FW. The contact interface of a 120 kD CheA-CheW complex by methyl TROSY interaction spectroscopy. J Am Chem Soc. 2005;127:9676–7. doi: 10.1021/ja052517m. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Hu Y, Fu W, Xia B, Jin C. Solution structure of the bacterial chemotaxis adaptor protein CheW from Escherichia coli. Biochem Biophys Res Commun. 2007;360:863–7. doi: 10.1016/j.bbrc.2007.06.146. [DOI] [PubMed] [Google Scholar]
- 15.Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:8065–74. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–92. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A. 2007;104:3777–81. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–6. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–5. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci U S A. 2005;102:15623–8. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–22. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005;8:116–21. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–5. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Khursigara CM, Subramaniam S, Hazelbauer GL. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol Microbiol. 2011;79:677–85. doi: 10.1111/j.1365-2958.2010.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Method Enzymol. 2007;423:317–35. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 26.Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol Microbiol. 2010;75:1171–81. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–82. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 28.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. [13C]-constant-time [15N,1H]-TROSY-HNCA for sequential assignments of large proteins. J Biolmol NMR. 1999;14:85–8. doi: 10.1023/a:1008346931993. [DOI] [PubMed] [Google Scholar]
- 29.Loria JP, Rance M, Palmer AG., 3rd Transverse-relaxation-optimized (TROSY) gradient-enhanced triple-resonance NMR spectroscopy. J Magn Reson. 1999;141:180–4. doi: 10.1006/jmre.1999.1891. [DOI] [PubMed] [Google Scholar]
- 30.Meissner A, Sorensen OW. Sequential HNCACB and CBCANH protein NMR pulse sequences. J Magn Reson. 2001;151:328–31. doi: 10.1006/jmre.2001.2374. [DOI] [PubMed] [Google Scholar]
- 31.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–90. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pervushin K, Gallius V, Ritter C. Improved TROSY-HNCA experiment with suppression of conformational exchange induced relaxation. J Biomol NMR. 2001;21:161–6. doi: 10.1023/a:1012484027195. [DOI] [PubMed] [Google Scholar]
- 33.Salzmann M, Wider G, Pervushin K, Wuthrich K. Improved sensitivity and coherence selection for [15N,1H]-TROSY elements in triple resonance experiments. Journal of Biomolecular NMR. 1999;15:181–4. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- 34.Yang D, Kay LE. Improved lineshape and sensitivity in the HNCO-family of triple resonance experiments. J Biomol NMR. 1999;14:273–276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.