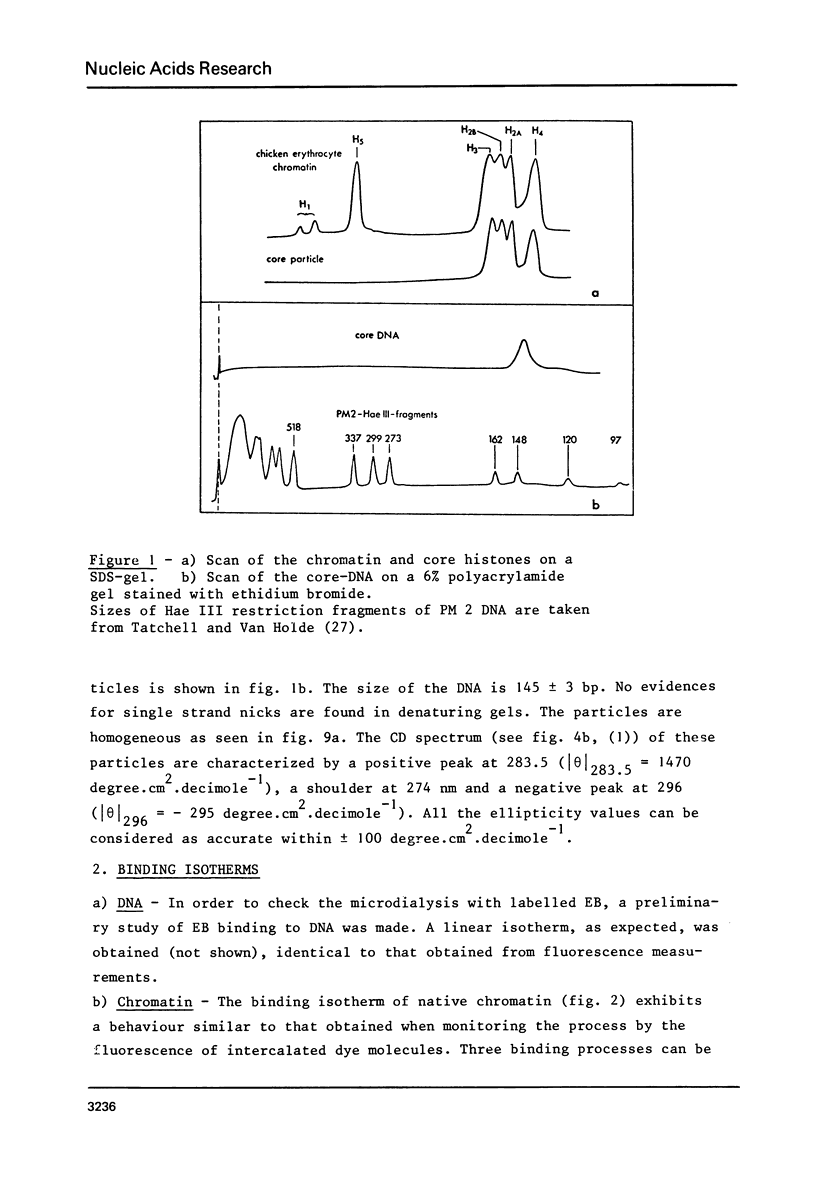
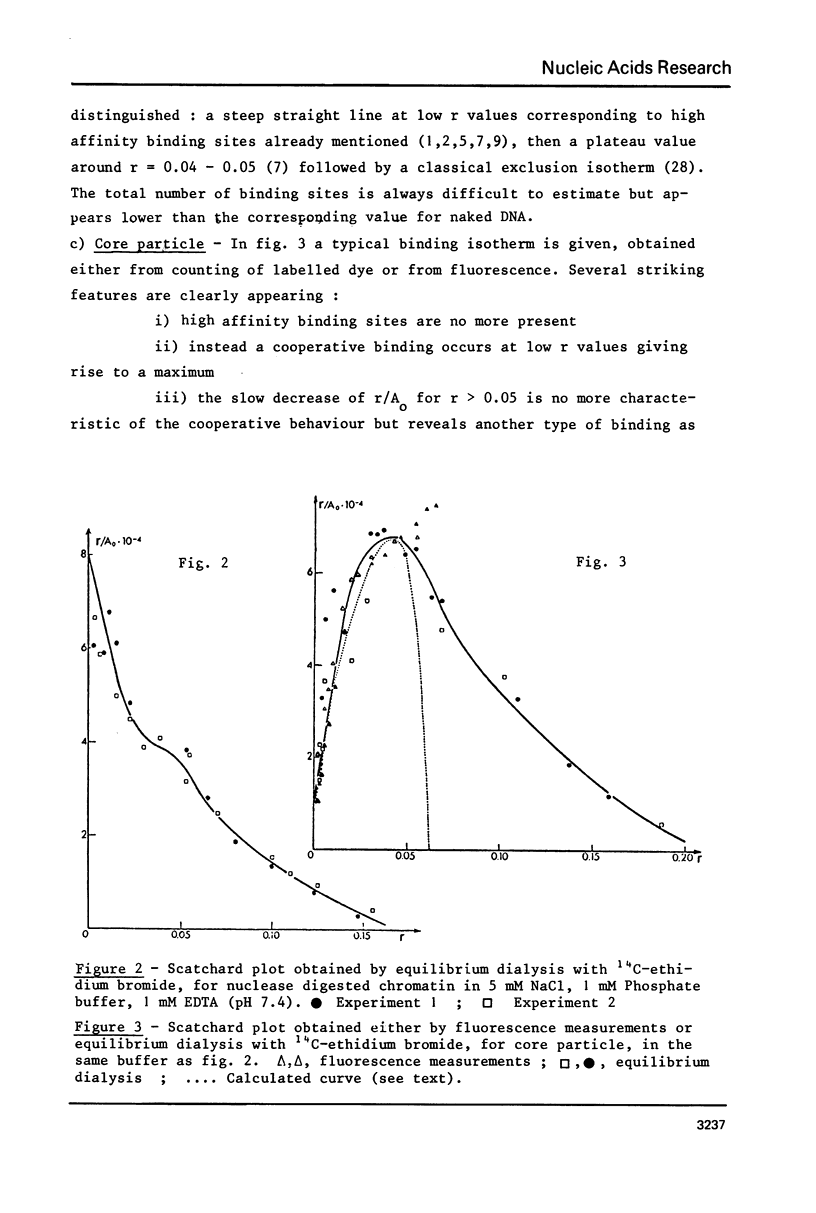
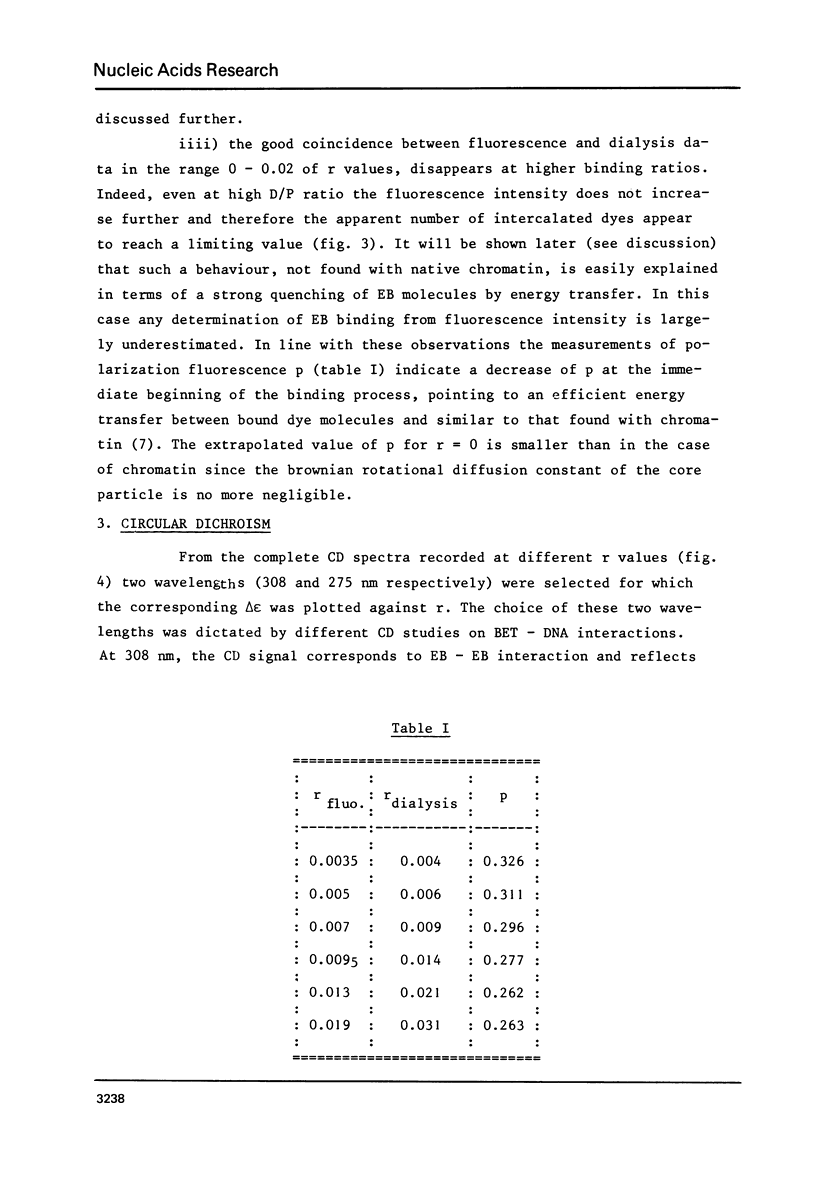
Abstract
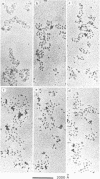
Ethidium bromide intercalation into DNA of nuclease digested erythrocyte chromatin and core particle, was followed at low ionic strength by fluorescence measurements, equilibrium dialysis using 14C labelled dye, circular dichroism and electron microscopy. High affinity binding sites in the chromatin are no more present in the core particle, i.e. when the linker is removed. In the case of core particle, a cooperative process occurs, accompanied by a partial stripping of the DNA from the core histone. Finally two populations of core particles can be detected by electron microscopy as far as their binding properties are concerned.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aktipis S., Kindelis A. Optical properties of the deoxyribonucleic acid-ethidium bromide complex. Effect of salt. Biochemistry. 1973 Mar 13;12(6):1213–1221. doi: 10.1021/bi00730a031. [DOI] [PubMed] [Google Scholar]
- Aktipis S., Martz W. W., Kindelis A. Thermal denaturation of the DNA-ethidium complex. Redistribution of the intercalated dye during melting. Biochemistry. 1975 Jan 28;14(2):326–331. doi: 10.1021/bi00673a019. [DOI] [PubMed] [Google Scholar]
- Angerer L. M., Georghiou S., Moudrianakis E. N. Studies on the structure of deoxyribonucleoproteins. Spectroscopic characterization of the ethidium bromide binding sites. Biochemistry. 1974 Mar 12;13(6):1075–1082. doi: 10.1021/bi00703a003. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Butour J. L., Delain E., Coulaud D., Le Pecq J. B., Barbet J., Roques B. P. Measurement of the expected DNA lengthening caused by mono-and bisintercalating drugs using electron microscopy. Biopolymers. 1978 Apr;17(4):872–886. doi: 10.1002/bip.1978.360170406. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Dalgleish D. G., Peacocke A. R. The circular dichroism in the ultraviolet of aminoacridines and ethidium bromide bound to DNA. Biopolymers. 1971 Oct;10(10):1853–1863. doi: 10.1002/bip.360101008. [DOI] [PubMed] [Google Scholar]
- Doenecke D. Binding of ethidium bromide to fractionated chromatin. Exp Cell Res. 1976 Jul;100(2):223–227. doi: 10.1016/0014-4827(76)90141-5. [DOI] [PubMed] [Google Scholar]
- Doenecke D. Ethidium bromide (EB) binding to nucleosomal DNA. Effects on DNA cleavage patterns. Exp Cell Res. 1977 Oct 15;109(2):309–315. doi: 10.1016/0014-4827(77)90010-6. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Fenske H., Eichhorn I., Böttger M., Lindigkeit R. Evidence of altered histone interactions, as investigated by removal of histones, in chromatin isolated from rat liver nuclei by a conventional method. Nucleic Acids Res. 1975 Oct;2(10):1975–1985. doi: 10.1093/nar/2.10.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Kovacs A. M., Champagne M., Daune M. Comparison between histones FV and F2a2 of chicken erythrocyte. II. Interaction with homologous DNA. Biochim Biophys Acta. 1975 Jun 2;395(1):16–27. doi: 10.1016/0005-2787(75)90229-4. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- LaRue H., Pallotta D. A study of the interaction between ethidium bromide and rye chromatin: comparison with calf thymus chromatin. Nucleic Acids Res. 1976 Sep;3(9):2193–2206. doi: 10.1093/nar/3.9.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Chan D. C., Piette L. H. Conformational state of DNA in chromatin subunits. Circular dichroism, melting, and ethidium bromide binding analysis. Nucleic Acids Res. 1976 Nov;3(11):2879–2893. doi: 10.1093/nar/3.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. J., Daune M. Ethidium bromide as a probe of conformational heterogeneity of DNA in chromatin. The role of histone H1. Biochemistry. 1976 Jul 27;15(15):3301–3307. doi: 10.1021/bi00660a021. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Louis M. Ethidium bromide as a probe of chromatin structure. FEBS Lett. 1974 Mar 15;40(1):9–12. doi: 10.1016/0014-5793(74)80882-3. [DOI] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Lurquin P. F., Seligy V. L. Binding of ethidium bromide to avian erythrocyte chromatin. Biochem Biophys Res Commun. 1972 Feb 16;46(3):1399–1404. doi: 10.1016/s0006-291x(72)80131-1. [DOI] [PubMed] [Google Scholar]
- Lurquin P. F. The use of intercalating dye molecules in the study of chromatin structure. Chem Biol Interact. 1974 May;8(5):303–312. doi: 10.1016/0009-2797(74)90009-x. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Paoletti J., Magee B. B., Magee P. T. The structure of chromatin: interaction of ethidium bromide with native and denatured chromatin. Biochemistry. 1977 Feb 8;16(3):351–357. doi: 10.1021/bi00622a002. [DOI] [PubMed] [Google Scholar]
- Paoletti J. Relaxation of chromatin structure induced by ethidium binding: 1--Micrococcal nuclease digestion of the ethidium--chromatin complex. Biochem Biophys Res Commun. 1978 Mar 15;81(1):193–198. doi: 10.1016/0006-291x(78)91648-0. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Strätling W. H., Seidel I. Relaxation of chromatin structure by ethidium bromide binding: determined by viscometry and histone dissociation studies. Biochemistry. 1976 Nov 2;15(22):4803–4809. doi: 10.1021/bi00667a009. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., Wilhelm M. L., Erard M., Duane M. P. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978 Feb;5(2):505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L. Ultrastructure of chromatin subunits during unfolding, histone depletion, and reconstitution. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):43–55. doi: 10.1101/sqb.1978.042.01.007. [DOI] [PubMed] [Google Scholar]
- de Murcia G., Das G. C., Erard M., Daune M. Superstructure and CD spectrum as probes of chromatin integrity. Nucleic Acids Res. 1978 Feb;5(2):523–535. doi: 10.1093/nar/5.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]