ETOC: Despite the use of antimitotic drugs, our understanding of the stress response, especially during mitotic arrest, is lacking. We report a molecular mechanism resulting in DNA damage during mitotic arrest that occurs via the apoptotic machinery but in the absence of cell death; this mechanism triggers p53 induction after cells slip from mitotic arrest.
Abstract
Mitotic arrest induced by antimitotic drugs can cause apoptosis or p53-dependent cell cycle arrest. It can also cause DNA damage, but the relationship between these events has been unclear. Live, single-cell imaging in human cancer cells responding to an antimitotic kinesin-5 inhibitor and additional antimitotic drugs revealed strong induction of p53 after cells slipped from prolonged mitotic arrest into G1. We investigated the cause of this induction. We detected DNA damage late in mitotic arrest and also after slippage. This damage was inhibited by treatment with caspase inhibitors and by stable expression of mutant, noncleavable inhibitor of caspase-activated DNase, which prevents activation of the apoptosis-associated nuclease caspase-activated DNase (CAD). These treatments also inhibited induction of p53 after slippage from prolonged arrest. DNA damage was not due to full apoptosis, since most cytochrome C was still sequestered in mitochondria when damage occurred. We conclude that prolonged mitotic arrest partially activates the apoptotic pathway. This partly activates CAD, causing limited DNA damage and p53 induction after slippage. Increased DNA damage via caspases and CAD may be an important aspect of antimitotic drug action. More speculatively, partial activation of CAD may explain the DNA-damaging effects of diverse cellular stresses that do not immediately trigger apoptosis.
INTRODUCTION
Prolonged mitotic arrest can lead to DNA damage and p53 induction (Lanni and Jacks, 1998; Dalton et al., 2007; Quignon et al., 2007), followed by cell cycle arrest or apoptosis. These processes may all contribute to the therapeutic activities of a drug like Taxol (paclitaxel), but the causal relationships between them and their consequences in the tumor environment remain elusive. For example, DNA damage can result in the expression of ligands on the cell surface that in turn recruit natural killer cells to remove the damaged cells (Gasser et al., 2005) and p53-driven senescence can up-regulate inflammatory cytokine production, resulting in tumor clearance via innate immune response (Xue et al., 2007). A better understanding of the causal relationships among mitotic arrest, DNA damage, and p53 could help improve therapeutic strategies that employ antimitotic drugs.
The tumor suppressor p53 is a central regulator of stress responses. When cells are stressed, for example by DNA damage, p53 becomes phosphorylated, resulting in its stabilization and induction of target genes that affect cell fate (Jin and Levine, 2001). These include key regulators of apoptosis (PUMA, NOXA) and cell cycle arrest (p21). p53 also induces MDM2, an E3 ligase that catalyzes p53 degradation, and together with other proteins, this circuit generates a series of uniform pulses of p53 in response to DNA damage induced by γ-irradiation and a graded pulse in response to UV irradiation (Batchelor et al., 2008, 2011). Different stresses therefore encode different p53 dynamics, and these dynamics may determine cell fate (Batchelor et al., 2011; Zhang et al., 2011).
Normal mitosis lasts ∼1 h, and the resulting daughter cells rapidly reenter the next cell cycle. Live imaging of cells undergoing normal mitosis revealed induction of a single pulse of p53 after mitotic exit in many cells, suggesting even normal mitosis can trigger the p53 circuit to some extent (Loewer et al., 2010). Cells can sense perturbations of mitosis and initiate a response. In noncancer cells, extension of prometaphase of mitosis from ∼30 min to 1.5–2 h with mitotic inhibitors, followed by washout and normal division, causes p53-dependent cell cycle arrest in the next G1 phase without obvious DNA damage (Uetake and Sluder, 2010). Prolonged mitotic arrest with the same drugs causes apoptosis during mitotic arrest in apoptosis-sensitive cell lines; in less sensitive lines, cells undergo slippage without division into tetraploid G1, which may be followed by p53-dependent arrest, apoptosis, or another round of mitosis (Rieder and Maiato, 2004; Gascoigne and Taylor, 2008; Orth et al., 2008). The cause of p53 induction and single-cell dynamics in these contexts remains unclear. In principle, the p53 circuit could detect mitotic defects by several proposed mechanisms: 1) a “mitotic clock” that senses the duration of mitosis, potentially involving p53 accumulation during mitosis (Blagosklonny, 2006, 2007; Uetake and Sluder, 2010); 2) a DNA ploidy or centrosome counter that acts after mitotic exit or slippage (Borel et al., 2002; Margolis et al., 2003), although the existence of an independent tetraploidy checkpoint has been challenged (Uetake and Sluder, 2004); and 3) detection of other errors or damage during or after mitotic exit or slippage (Barboule et al., 1997; Lanni and Jacks, 1998; Stewart et al., 2001).
In this paper, we investigate both the molecular mechanism resulting in DNA damage and p53 induction during and after mitotic arrest using primarily a potent and highly selective small molecule inhibitor of kinesin-5 (K5I, EMD534085; Orth et al., 2008; Schiemann et al., 2010), which prevents satisfaction of the spindle assembly checkpoint (SAC) by forestalling bipolar spindle assembly. An advantage of this type of arrest is that microtubule polymerization dynamics are not perturbed, minimizing effects of the perturbation outside prevention of SAC satisfaction. Additional mitotic drugs, including Taxol and nocodazole, were also used. We found that strong p53 induction occurs after slippage from prolonged arrest. This induction is significantly due to DNA damage during arrest caused by partial activation of apoptosis during mitotic arrest through the mitochondria, caspase-9 and -7, and inhibitor of caspase-activated DNase (ICAD).
RESULTS
p53 induction occurs after mitotic slippage and consists of two major types of dynamics
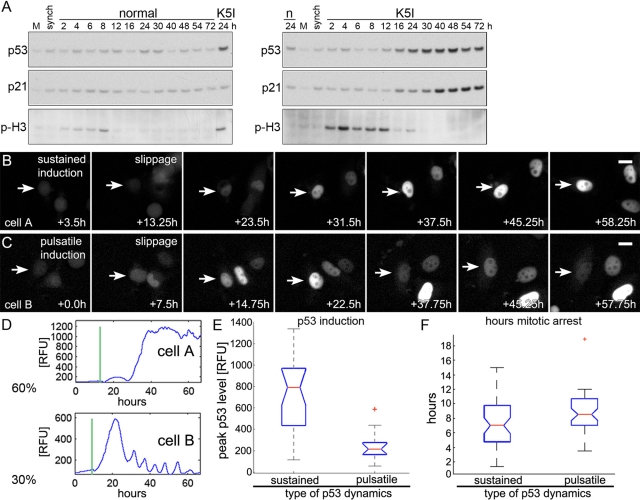
MCF7 cells, chosen because their p53 dynamics are well characterized (Batchelor et al., 2008, 2011; Loewer et al., 2010), were treated with saturating K5I (500 nM), resulting in mitotic arrest (Orth et al., 2008; Shi et al., 2008). MCF7 cells are apoptosis resistant, and leave prolonged mitotic arrest by slippage (Orth et al., 2008). To monitor p53 induction in a population of cells during or after mitotic arrest, we synchronized cells in late G2 using the highly selective Cdk1 inhibitor (Cdki), RO-3306 (Vassilev, 2006; Vassilev et al., 2006), and released them by washing into normal or K5I medium (Figure 1 and Supplemental Figure S1). Untreated cells initially passed through mitosis between 2 and 8 h postwashout and K5I-treated cells largely between 2 and 24 h, as determined by phospho-H3 levels (Figures 1A and S1). While p53 was not induced during arrest (when phospho-H3 levels are high) in K5I-treated cells, it showed strong and persistent induction a few hours after slippage, beginning at 16–24 h. The effector for cell cycle arrest, p21, was also induced, but with slightly delayed kinetics. We observed no induction of p53 or p21 in control cells. These data extend previous reports by showing that p53 and p21 induction occur only after mitotic slippage (Tao et al., 2007; Shi et al., 2008).
FIGURE 1:
Prolonged mitotic arrest results in two types of postslippage p53 induction dynamics. (A) Mock, untreated (M), G2-synchronized cells (synch; see Materials and Methods), and synchronized cells released into normal (n) or K5I-medium (K5I) were immunoblotted for p53, p21, and the mitotic marker p-H3. p53 and p21 were not persistently induced to high levels in normal medium. Release into K5I resulted in p53 induction beginning at 12–16 h and p21 induction after 16 h. All blots in each panel are from the same gels and/or samples. (B–D) Live MCF7 cells expressing p53-Venus were imaged for >60 h. Times are from start of time lapse. First frame is start of mitotic arrest. No p53 increase was noted during mitotic arrest. Sixty percent of arrested cells showed sustained, high-level, p53-induction (cell A), while 30% showed a pulsatile response with a large first pulse (cell B). (B and C) Scale bars: 10 μm. See Movies S1 and S2 and note additional cells in the same microscopic field. Vertical lines in representative traces (D) mark mitotic slippage. RFU, relative fluorescence units. (E) Box-and-whisker plots of peak p53 levels in individual cells with sustained and pulsatile dynamics; p53 induction is significantly higher in cells with sustained induction. For cells in each group, the peak p53 intensity is determined, and the resulting values are assorted in ascending order. The boxes represent the second and third quartile of the population, the red horizontal line represents the median. Upper and lower “whiskers” represent data points from the first and fourth quartile that are within 1.5-fold of the interquartile range. Outliers are marked by red +. The notches about the median represent the 95% confidence interval. As the notches between the two compared populations do not overlap, the median values are significantly different (p < 0.0001 for sustained vs. pulsatile). (F) Box-and-whisker plots of hours arrested for the types of p53 dynamics. The medians are not significantly different. Forty cells were tracked: 26 showed a sustained induction, and 14 were pulsatile.
The precise kinetics and dynamics of p53 induction in individual cells depend on the nature of the upstream stress (e.g., γ-irradiation vs. ultraviolet [UV]) and may determine subsequent cell fates; p53 induction in individual cells after antimitotic drugs is unknown (Batchelor et al., 2009, 2011). To characterize p53 induction in cells after antimitotic drug treatment, MCF7 cells stably expressing p53-Venus (Batchelor et al., 2008) were treated with saturating K5I, and p53 was monitored using fluorescence time-lapse microscopy. As observed previously for parental MCF7 cells, nearly all (>95%) cells entered mitotic arrest and slipped into tetraploid G1 without cytokinesis and <5% died during mitotic arrest (Orth et al., 2008). After slippage, ∼90% remained in interphase for at least 96 h and often grew larger. During mitotic arrest, p53 levels in single cells appeared approximately constant, consistent with the results from immunoblotting (Figure 1, A–D). After slippage, ∼90% of cells showed strong p53 induction (Figure 1, B–D). Single-cell analysis revealed two separate types of p53 dynamics; ∼60% showed sustained, high-level induction of higher amplitude and longer duration than that induced by any kind of DNA damage previously characterized (Figure 1, B and D, cell A, and Supplemental Movie S1), and ∼30% showed uniform pulses similar to those induced by DNA double-strand breaks (Batchelor et al., 2008), but with a higher-amplitude first pulse (Figure 1, C and D, cell B, and Movie S2). Cells with sustained p53 induction showed significantly higher peak levels than cells with a pulsatile phenotype (Figure 1, D and E). The mitotic arrest duration for the two types of p53 induction was the same (Figure 1F). We obtained similar results from immunoblotting and fluorescence imaging of U2 OS and HCT116 cells treated with K5I, showing the generality of postslippage p53 induction and dynamics (Figure S2; unpublished data). Two structurally unrelated K5Is caused the same p53 induction (Figure S3A), and untreated control cells showed only brief, low-amplitude p53 pulses following normal cytokinesis, as previously reported (Figure S3A; Loewer et al., 2010). Interestingly, at the IC50 for the K5I we used (100 nM; Orth et al., 2008), some cells executed cytokinesis after a transient arrest (∼5 h), and some cells slipped after prolonged arrest as after treatment with 500 nM K5I. Those cells that divided showed low-amplitude induction similar to that of the control, and those that arrested and slipped showed induction similar to that of the 500 nM treatment (Figure S3A). p53 induction in live cells was also followed for other antimitotic small molecules, including Taxol (microtubule stabilizing; Movie S3) and nocodazole (microtubule destabilizing; Movie S4), and with inhibitors that target the mitotic Aurora kinases (VX-680; Movie S5) and polo-like kinase (BI-2536; Movie S6). These inhibitors have been shown to result in p53 induction by immunoblotting, but p53 induction has not been catalogued in live cells. As in mononucleated K5I postslip cells, p53 induction appeared to occur only after slippage, in this case in multinucleated postslippage cells. We previously used immunoblotting and showed p53 induction after Taxol (Shi et al., 2008), and here we compared K5I and nocodazole in two cell lines and found the timing and level of p53 induction to be similar (Figure S4).
Since K5I is thought to function only in mitotic spindle bipolarization, these data indicate the postslippage p53 induction is a consequence of prolonged mitotic arrest itself, stress due to slippage and/or tetraploidy or stress that occurred during arrest. To distinguish between these possibilities, we induced tetraploidy alone without mitotic arrest by blocking cytokinesis with cytochalasin-D or the compound Y-27632; these treatments did not show a p53 response (unpublished data). To alter the duration of arrest, we treated mitotis-arrested cells with RO-3306. Cdk1 activity is required to maintain mitotic arrest (Brito and Rieder, 2006), and addition of Cdki to mitosis-arrested cells triggers rapid exit to interphase without division, defined here as Cdki-triggered slippage (Hu et al., 2008). MCF7-Venus cells were arrested for 24 h with K5I, then slippage was triggered by Cdki. p53 was induced in these cells, but to dramatically lower levels than those observed in the same population after spontaneous slippage before Cdki or in slippage control cells (Figure S3B). The duration of mitotic arrest was not significantly different between the different conditions, but those in the triggered slippage population tended to be arrested for a shorter period (Figure S3C). Cluster plots were used to correlate arrest times with peak p53 induction (Figure S3, D and E). Regardless of arrest time, Cdki-triggered slippage cells have lower p53 induction. Contrary to triggered-slippage cells, those that slipped spontaneously before Cdki more often had higher p53 induction, regardless of arrest duration. The ability to induce p53 was not directly impaired by Cdki, as addition of the DNA-damaging drug neocarzinostatin (NCS) in conjunction with Cdki resulted in low-level induction (Figure S3B) similar to induction seen with NCS alone (Loewer et al., 2010). Further, Cdk1 inhibition and RO-3306 have been shown to not prevent DNA damage response or p53 induction (Morris et al., 2001; Chan et al., 2008). On the basis of the data, we hypothesize that p53 induction postslippage is not due to mitotic accumulation, mitotic slippage, or tetraploidy per se, but to stress during prolonged mitotic arrest. Because many Cdki-triggered slippage cells were arrested for long durations but did not induce p53 (Figure S3, D and E), we suspect the stress does not occur early or accumulate gradually during arrest, but instead occurs late in mitotic arrest, around the time of slippage.
DNA damage occurs late in mitotic arrest
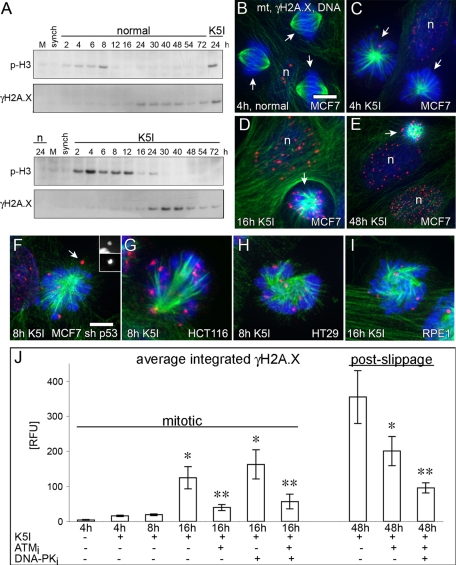
To gain insight into the nature of the p53-inducing stress, we checked p53 phosphorylation. We found evidence for Ser-15 and Ser-33 phosphorylation after K5I and nocodazole, consistent with a role for DNA damage-sensing kinases (Figure S5A; Shieh et al., 1997; Sanchez-Prieto et al., 2000). We therefore measured DNA damage in mitotic arrest and postslippage cells using three methods: γH2A.X immunoblotting, γH2A.X immunofluorescence, and the DNA comet assay (unpublished data). G2-synchronized cells were released into normal medium or K5I and immunoblotted for a mitotic marker (phospho-serine-10-histone-H3 [p-H3]) and γH2A.X. As the culture grew to high density in normal medium, we sometimes noted a low level of γH2A.X in the absence of p53 induction (Figure 2A, top panels), but it was not a consequence of the G2 synchronization (see synch lane in Figure 2A). Late in mitotic arrest or soon after slippage, beginning around 12–16 h after release into K5I, we observed a marked increase of γH2A.X (Figure 2A); U2 OS (unpublished data), HCT116 (Figure S2), and other cells also showed increased γH2A.X at late arrest/early slippage. Next quantitative γH2A.X immunofluorescence was used to measure DNA damage in single cells. Normal mitotic MCF7 cells exhibited very few γH2A.X foci (Figure 2, C and K). Mitosis-arrested cells after 4 and 8 h release also showed few foci. At 16 h K5I, numerous foci associated with condensed, mitotic chromosomes and with postslippage nuclei (Figure 2, D–F and K); Taxol treatment also resulted in foci late during mitotic arrest (Figure S6, A–D). Nocodazole-, VX-680-, and BI-2536-arrested cells also contained γH2A.X foci (Figure S7). The normal cellular and DNA morphology in these cells rules out that the DNA damage was caused by apoptosis. DNA damage late during mitotic arrest was p53-independent (Figure 2G) and is common in late mitotic arrest in both cancer and noncancer cell lines (Figure 2, H–J). The DNA damage persists, as most postslippage nuclei at 48 h K5I or 24 h Taxol contained high levels of γH2A.X (Figures 2, F and K, and S6). Interestingly, when γH2A.X is quantified in K5I-treated interphase cells, the DNA damage increases over time (Figure S8). For K5I cells, it is difficult to decipher whether a cell is postslippage based on phenotype, as pre- and postmitotic cells are both mononucleated. For Taxol, single-cell phenotype reports directly on pre- or postmitotic state, where only multinucleated cells are postslippage (Orth et al., 2011), where only multinucleated cells are postslippage. Quantification of multinucleated cells over time shows DNA damage tends to increase after slippage (Figure S6E)
FIGURE 2:
DNA damage after K5I initially increases late in mitotic arrest and involves DNA damage–response kinases, ATM and DNA-PK. (A) Mock, untreated (M), G2-synchronized, and cells released into normal (n) or K5I medium. Normal shows low-level γH2A.X, but K5I shows significantly higher levels beginning at mitotic slippage (decreased p-H3). All blots in each panel are from the same gels and/or samples. (B–E and J) Green, microtubules (mt); red, γH2A.X; blue, DNA. Only γH2A.X on the mitotic chromosomes or interphase DNA was quantified. n, nucleus; arrows, mitotic cells; RFU, relative fluorescence units. (C) Scale bar: 15 μm (applies also to B–E). Normal mitotic cells and mitosis-arrested cells at 4 and 8 h K5I show low γH2A.X (red), but 16 h K5I-treated cells shows high levels of γH2A.X. In 16 h arrested cells, γH2A.X is blocked by ATM inhibition (ATMi) but not by DNA-PK inhibition (DNA-PKi); combined ATMi and DNA-PKi at 16 h was similar to ATMi results. Postslippage γH2A.X at 48 h is high and was dependent on both ATM and DNA-PK. Values in (J) are average integrated fluorescence intensity per cell (± SE). At 4 h with no K5I, ATMi, or DNA-PKi, n = 15 cells, two experiments. All other conditions, n > 100 cells, ≥ two experiments. Mitotic conditions: *, p < 0.05 vs. 4 h control; **, p < 0.05 vs. 16 h K5I or 16 h K5I + DNA-PKi. Postmitotic conditions: *, p < 0.05 vs. 48 h K5I; **, p < 0.05 48 h K5I or 48 h K5I + ATMi. (F–I) γH2A.X occurs during mitotic arrest in different cell lines, is not p53-dependent (MCF7 sh p53), and occurs in cancer and noncancer cells (RPE1). Inset in (F) shows separate chromosome (top) and γH2A.X (bottom) channels for the γH2A.X foci (arrow). (F) Scale bar: 5 μm (applies also to G–I).
Two kinases, ataxia-telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK) are often involved in phosphorylating H2A.X during the DNA damage response to generate the γH2A.X epitope (Stiff et al., 2004). To define their role, we cotreated with selective inhibitors of each kinase (ATMi and DNA-PKi) and K5I, and quantified γH2A.X in mitotic arrest and postslippage cells (Figure 2K). ATMi blocked γH2A.X during mitotic arrest, but DNA-PKi did not, nor was there any additional effect during arrest when the two inhibitors were combined (Figures 2K and S5, C and D); ATMi also blocked γH2A.X in Taxol-arrested cells (Figure S6D). γH2A.X in 48h postslippage cells was decreased by ATMi and was decreased further upon coaddition of DNA-PKi (Figures 2K and S5, E and F). Phosphorylated, active ATM (P-ATM) and the DNA break site marker, 53BP1, localized in a foci pattern like γH2A.X in mitosis-arrested cells (Figure S5, G and I) and colocalized with postslippage γH2A.X foci (Figure S5, H and J). These data show that ATM and 53BP1 respond to DNA damage during mitotic arrest, and DNA-PK also participates after cells slip. We conclude that some event(s), typically during late mitotic arrest, and perhaps additionally after slippage, results in bona fide DNA damage.
Partial activation of apoptosis is required for DNA damage during mitotic arrest
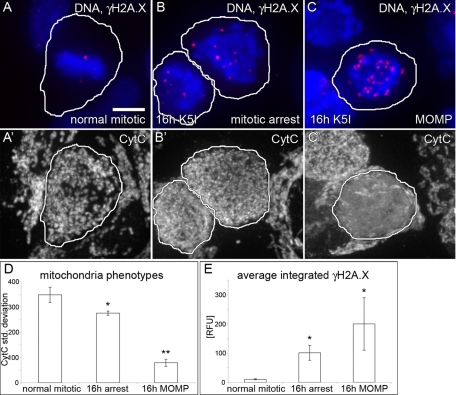
Although <10% of MCF7 cells undergo apoptosis during 72 h of K5I treatment (Orth et al., 2008), we considered the possibility that DNA damage during mitotic arrest could be due to activation of the apoptotic pathway. A defining event in intrinsic apoptosis is mitochondrial outer-membrane permeabilization (MOMP), resulting in release of cytochrome C (CytC) into the cytoplasm, and activation of caspase-9 followed by caspase-7; MCF7 cells lack caspase-3. To test for apoptosis and DNA damage in the same cell, we initially costained for DNA, γH2A.X, and CytC (Figure 3, A–C′). When CytC is released into the cytoplasm, its staining changes from punctate to homogeneous, which can be measured by the SD of the average fluorescence intensity per pixel (Goldstein et al., 2000). We quantified the CytC SD and γH2A.X in normal mitotic (Figure 3, A and A′) and 16-h mitosis-arrested cells (Figure 3, B and B′). The results reveal that there are three classes of cells and indicate that at 16 h K5I there are very few MOMP cells (Figure 3D). Normal mitotic cells showed a high SD and few γH2A.X foci (Figure 3, A, A′, D, and E). Mitosis-arrested cells at 16 h contained some punctate, mitochondria-associated CytC staining, but the contrast above background was lower, resulting in a small but significant decrease in CytC SD. This finding suggested some leakage of CytC into the cytoplasm, and it correlated with increased γH2A.X staining (Figure 3, B, B′, D, and E). Fewer than 1% of cells had undergone full MOMP at 16 h, and they showed essentially homogeneous CytC intensity with very low CytC SD and had, on average, greater γH2A.X than mitosis-arrested cells at the same time point, but there was significant cell-to-cell variability in this small population (Figure 3, C, C′, D, and E). Correlation plots of integrated CytC SD versus γH2A.X also show three heterogeneous but distinct populations of cells (Figure S9). Mitosis-arrested cells at 16 h did not stain for the apoptotic marker, cleaved PARP1, while post-MOMP cells often did (unpublished data). These data indicate that many cells at 16 h mitotic arrest show some CytC leakage and increased DNA damage, but clearly have not undergone full apoptosis based on normal DNA morphology and an absence of MOMP and cleaved PARP1.
FIGURE 3:
DNA damage during mitotic arrest occurs without full mitochondrial MOMP. (A–C′) DNA, γH2A.X and CytC staining in the same mitotic cells. (D) Quantification of the SD of the average CytC fluorescence pixel intensity in individual mitotic or MOMP cells (encircled) shows that 16 h mitosis-arrested cells (B′) are morphologically altered compared with normal (A′), and have a lower average CytC SD than normal mitotic cells, but a higher value than MOMP cells (C′). (E) Normal mitotic cells (A) have very low γH2A.X compared with 16 h arrest (B). MOMP cells (C) on average showed more DNA damage, but this was highly variable and not statistically significant in this small population (t test 16 h MOMP vs. 16 h arrest, 0.33). Average values are reported (± SE). (A) Scale bar: 5 μm (applies to all cell images). Control, n = 68; 16 h mitotic, n = 295; 16 h MOMP, n = 16; two experiments each condition. (D) *, p < 0.05 vs. control mitotic; **, p < 0.05 vs. 16 h mitotic and normal mitotic. (E) *, p < 0.05 vs. normal mitotic.
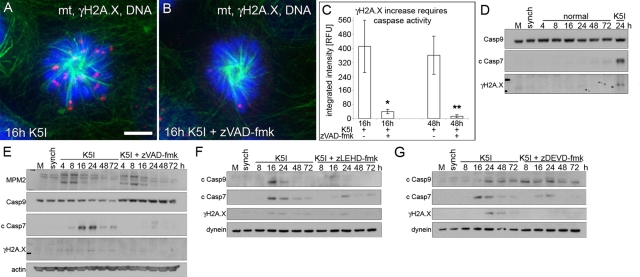
What is the molecular mechanism underlying DNA damage during prolonged mitotic arrest? As there was evidence of some CytC leakage, we explored the possibility that mitotic arrest might induce a partial activation of apoptosis. First, a role for caspases was tested using the pan-caspase inhibitor zVAD-fmk. Caspase inhibition significantly blocked γH2A.X foci in mitosis-arrested cells at 16 h and in postslippage cells at 48 h (Figure 4, A–C). DNA comet formation was also inhibited by zVAD-fmk (unpublished data). To determine the hierarchy and timing of caspase activation and to confirm the caspase-dependence of DNA damage, we used selective caspase inhibitors and immunoblotting. Release of synchronized cells into normal medium did not result in caspase activation or γH2A.X increase (Figure 4D). In contrast, a decrease of procaspase-9 and accumulation of cleaved caspase-7 first occurred at 8–16 h after K5I treatment, a late mitotic arrest/early slippage based on mitotic protein monoclonal 2, MPM2 blotting (Figure 4E). Moreover, the loss of procaspase-9 and γH2A.X increase in K5I was blocked by zVAD-fmk (Figure 4E). Cleaved caspase-7 and γH2A.X increased initially at late arrest/slippage and remained detectable (Figure 4, E–G). Caspase-9 inhibition (zLEHD-fmk) resulted in reduced caspase-7 cleavage, correlating with decreased γH2A.X (Figure 4F). When caspase-7 was inhibited (zDEVD-fmk), caspase-9 remained activated, but there was a marked reduction of fully cleaved caspase-7, also correlating with decreased γH2A.X (Figure 4G). Single-cell quantification of γH2A.X confirmed the immunoblotting and showed that caspase-9– specific and caspase-7–specific inhibition each blocked DNA damage; caspase inhibition also reduced the number of γH2A.X foci (Figure S10, A and B). The number of foci at 16 h was the same for Taxol as for K5I (Figure S6F vs. Figure S10B). We confirmed the role of caspases in noncancer RPE1 (Figure S10C) and after nocodazole (Figure S4) and caspase cleavage after Taxol (Figure S6G; Shi et al., 2008). Together, these data demonstrate that caspase-9 activates caspase-7 late in mitotic arrest and that they are required for DNA damage, but these events happen without full apoptosis.
FIGURE 4:
DNA damage during mitotic arrest is dependent upon caspase-9 and -7. (A and B) Green, microtubules (mt); red, γH2A.X; blue, DNA. G2-synchronized cells released for 16 h into K5I or K5I with the caspase inhibitor zVAD-fmk show γH2A.X increase is caspase-dependent. Scale bar: 5 μm. (C) γH2A.X quantification of single cells shows caspase-dependent DNA damage in 16 h mitosis-arrested cells and 48 h postslippage cells. Average integrated fluorescence intensity (± SE). RFU, relative fluorescence units. n = 50+ for each condition; two experiments . *, p < 0.05 vs. 16 h K5I; **, p < 0.05 vs. 48 h K5I. (D) Mock, untreated (M) and G2-synchronized (synch) cells, and synchronized cells released into normal medium show no activation of caspase-9 or -7, as measured by loss of full-length caspase-9 (Casp9) or increase in cleaved caspase-7 (c Casp7). γH2A.X also did not increase compared with 24 h K5I. (E) Release into K5I shows loss of caspase-9, increased cleaved caspase-7, and increased γH2A.X in late arrest/early slippage (MPM2 blot) blocked by zVAD-fmk. MPM2 indicates several bands during mitosis. (F) Caspase-9 inhibition with zLEHD-fmk prevented caspase-9 and -7 activation (c Casp9 and c Casp7) and inhibited γH2A.X increase. (G) Caspase-7 inhibition with zDEVD-fmk did not prevent caspase-9 activation, but largely blocked caspase-7, resulting in decreased γH2A.X. Note: the slightly higher-migrating cleaved caspase-7 band in the presence of caspase inhibitors in (E, F, and G) represents incomplete processing (activation). All blots in each panel are from the same gels and/or samples. Dynein and actin are the loading controls.
Caspase-dependent cleavage of ICAD is required for DNA damage and p53 induction
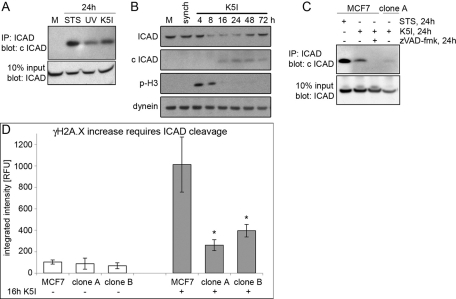
How could partial apoptosis and caspase activation induce DNA damage in arrested mitotic cells? An important activity of the executioner caspase-3 and -7 is to activate caspase-activated DNase (Lo Celso et al., 2009) via cleavage of ICAD, (Enari et al., 1998; Sakahira et al., 1998), which frees caspase-activated DNase (CAD) to cleave DNA. To test whether ICAD/CAD is involved in DNA damage during mitotic arrest, we first performed immunoprecipitation experiments to assay possible loss of full-length ICAD and gain of a cleaved ICAD product (c ICAD) after K5I treatment; we detected a partial cleavage of ICAD at 24 h K5I that was more than UV treatment but <24 h staurosporine (STS; Figure 5A). Cleavage of ICAD was observed with immunoblotting 8–16 h after release (Figure 5B); control cells showed no cleaved ICAD. Caspase inhibition with zVAD-fmk and expression of a noncleavable ICAD mutant (Enari et al., 1998; Sakahira et al., 1998) both blocked ICAD cleavage after 24 h K5I (Figure 5C, clone A). Importantly, ICAD was cleaved in late mitotic arrest, together with the first evidence of caspase-9 and -7 activation, just prior to increased DNA damage, mitotic slippage, and p53 induction (Figures 1, 2, and 4).
FIGURE 5:
Mitotic DNA damage occurs downstream of caspase-dependent, partial cleavage of ICAD. (A) Immunoprecipitation (IP) of ICAD followed by blotting for cleaved ICAD (c ICAD) at 24 h K5I showed less cleavage than 24 h 1 μM STS but more than UV treatment (see Materials and Methods). Ten percent input blot: ICAD shows total ICAD levels. (B) Mock, untreated (M) and G2-synchronized (synch) cells, and cells released into K5I showed decreased ICAD from 8 to 48 h and increased c ICAD from 16 h, correlating well with late mitotic arrest/slippage (p-H3). Dynein is the loading control. (C) Immunoprecipitation of ICAD followed by blotting for c ICAD showed partial cleavage of ICAD at 24 h K5I treatment is blocked by zVAD-fmk or expression of noncleavable ICAD (clone A). All blots in each panel are from the same gels and/or samples. (D) Noncleavable ICAD-expressing clone A and B showed the same background levels of γH2A.X as MCF7 CELLS. G2-synchronized clone A and B released into K5I for 16 h showed severely inhibited γH2A.X compared with MCF7 cells. Average integrated fluorescence intensity (± SE). RFU, relative fluorescence units. No K5I: MCF7, n = 31; clone A, n = 12; clone B, n = 16. 16 h K5I: MCF7, n = 210; clone A, n = 166; clone B, n = 142; two experiments each condition. *, p < 0.05 vs. MCF7 + 16 h K5I.
To test whether ICAD cleavage was required for DNA damage during mitotic arrest, we used two engineered cell lines (clones A and B) that stably express noncleavable ICAD. Both clone A and clone B showed a dramatic reduction of γH2A.X in arrested cells at 16 h K5I (Figure 5D). These data show that most DNA damage during late mitotic arrest is ICAD/CAD-dependent. Since inhibition of CAD by the ICAD mutant is probably not complete, it is difficult to determine whether this is the only damage pathway.
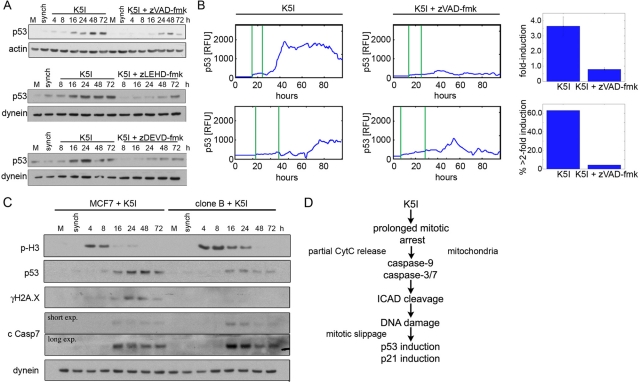
Returning to our initial observations on p53, we tested whether the caspase–CAD–DNA damage pathway is the cause of postslippage p53 induction. Caspase inhibition with zVAD-fmk reduced the induction of p53 in immunoblots, and zLEHD-fmk or zDEVD-fmk showed the same effect (Figures 6A, S4C, and S10C). Consistently, we observed that zVAD-fmk inhibited the induction of p53-Venus in single, postslippage cells (Figure 6B), as did zDEVD-fmk (unpublished data). The remaining p53 induction was similar to that seen following normal mitosis. Caspase inhibition also blocked p53 induction after Taxol (Movie S7). Noncleavable ICAD-expressing clones also significantly blocked DNA damage and p53 induction (Figures 6C and S11). Importantly, caspase-7 was cleaved in cells expressing mutant ICAD, indicating that ICAD cleavage occurs downstream of caspase-7 activation but upstream of p53 induction (Figure 6C). Taken together, these data indicate that p53 induction postslippage results from DNA damage during late mitotic arrest after partial activation of the intrinsic apoptosis pathway (Figure 6D).
FIGURE 6:
p53 induction after mitotic DNA damage is caspase- and ICAD/CAD-dependent. (A) Immunoblotting of mock, untreated (M) and G2-synchronized (synch) cells, and cells released into K5I showed strong, persistent induction of p53 after release that was significantly blocked by caspase-9 or -7 inhibition. Actin and dynein are loading controls. (B) Live MCF7 p53-Venus cells. First vertical green line in traces is entry into mitotic arrest, second is mitotic slippage. In K5I, p53 is often induced to sustained, high levels. Cells cotreated K5I and zVAD-fmk showed only transient induction of p53 to low levels. RFU, relative fluorescence units. Caspase inhibition resulted in decreased fold induction (average of all cells ± SE) in individual cells and in fewer cells with >2 fold induction. Fold induction is the ratio of the peak intensity to the lowest intensity in each cell. K5I, n = 32; K5I + zVAD-fmk, n = 49. (C) Mock, untreated (M), MCF7, and noncleavable ICAD clone B were G2-synchronized (synch) and released into K5I. When ICAD cleavage is blocked, both p53 induction and DNA damage (γH2A.X) are decreased. Importantly, caspase-7 remains activated after K5I treatment. Dynein is loading control. All blots in each panel are from the same gels and/or samples. (D) Summary of pathway causing DNA damage during mitotic arrest that results in postslippage p53 induction.
DISCUSSION
p53 dynamics during and after mitotic arrest
When and how p53 is induced in response to antimitotic drugs has been studied for years, but the causal mechanisms are unclear. Currently, three alternative models prevail: the mitotic clock, the DNA ploidy or centrosome number checkpoint, and the stress-response model. Our data argue against the mitotic clock, since this model predicts slow accumulation of p53 during mitotic arrest itself, which we did not observe by immunoblotting or live-cell imaging (Figures 1 and 6 and Movies S1–S6). Instead, we only saw convincing p53 induction postslippage, regardless of the drug used. We cannot rule out modifications on p53 or slow accumulation of p53 mRNA. Our single-cell analysis also argues against a DNA tetraploidy or centrosome-counting checkpoint, since we did not observe p53 induction in tetraploid cells formed after mitotic slippage triggering or cytokinesis inhibition (Figure S3; unpublished data); other studies have also disfavored this model (Uetake and Sluder, 2004). Our data do favor the stress-response model, with DNA damage as the inducer of high p53 levels after slippage. The data do not rule out other stress or damage contributing in parallel. A study that used brief mitotic delays followed by cell division in a noncancer line showed that p53-dependent cell cycle arrest did not depend on DNA damage (Uetake and Sluder, 2010). Similarly, we see little DNA damage during early mitotic arrest but significant damage during late arrest (Figure 2). p53 induction, presumably mostly in response to DNA damage in late mitotic arrest, appears delayed until after slippage. It is most likely that p53, generally an unstable protein, cannot accumulate to high levels when transcription is inhibited during mitosis and when translation is reduced. p53 induction is therefore delayed until translation increases following slippage. Once p53 was induced after K5I treatment, its dynamics showed a large cell-to-cell variation, possibly reflecting different degrees of DNA damage after partial activation of caspases. The sustained induction is unique, and the oscillatory pattern is most similar to the pulses after double-stranded damage (Batchelor et al., 2008). Treatment with Taxol, nocodazole, VX-680, and BI-2536 each resulted in p53 induction to some extent in multinucleated postslippage cells and, as with K5I, there was significant cell-to-cell variability (Movies S3–S6). Interestingly, p53 induction sometimes appeared different in separate multinuclei of the same cell. It will be important and interesting to compare and contrast p53 induction and dynamics after different antimitotic drugs and to link those results to molecular pathways and cell fate.
DNA damage as a stress response
DNA damage is often considered as a cause of cell stress, for example when it is directly induced by radiation or drugs that bind DNA (e.g., NCS). However, our work and some other studies suggest that it can also be a consequence of stress (Betti et al., 2005; Lovric and Hawkins, 2010). This dual role suggests several important possibilities: stresses that are not intrinsically genotoxic may be mutagenic and carcinogenic, DNA damage may play a rather general role in stress signaling, and DNA damage could become a self-perpetuating or self-amplifying state by positive feedback (Batchelor et al., 2011; Zhang et al., 2011). Postslip cells still retained some DNA damage 48–72 h after treatment with K5I (Figures 2 and 4). It is not clear whether this was due to failure of repair, additional stress, or caspases and/or CAD remaining active at some low level. Continued low activity of CAD could possibly contribute to maintenance of the senescence-like state (Dayton et al., 1989). Interestingly, CAD-based DNA damage results in terminal differentiation of muscle cells, which is akin to senescence in that the cells no longer proliferate (Larsen et al., 2010). CAD activation is often viewed as an all-or-nothing response, sentencing a cell to death. This study shows that it can also act to stress cells after antimitotic drugs without directly killing them.
Implication for antimitotic drug action
The role of p53 in the response of cancer cells to antimitotic drugs has been controversial (Meek, 2000; Bacus et al., 2001). Our study helps clarify this issue. In apoptosis-sensitive cells that die during mitotic arrest, p53 is likely not induced. However, in apoptosis-resistant MCF7 cells and other cells, it is induced postslippage by DNA damage and plays a central role in preventing the cell from entering another round of mitosis, probably by inducing p21 and other cell cycle inhibitors. We suspect apoptosis during mitotic arrest and CAD-mediated DNA damage without apoptosis share the same upstream activators, though we have not proven this. Likely upstream proapoptotic signals include degradation of the antiapoptotic protein Mcl-1 through E3-ligases (Inuzuka et al., 2011; Wertz et al., 2011) and regulation of the antiapoptotic protein Bcl-xL, which plays a protective role in epithelial cancer cells (Shi et al., 2011; Tan et al., 2011). Caspase-3–dependent cleavage of chromosomal-associated condesin Cap H is also involved in death during mitotic arrest (Lai et al., 2011). CytC release from mitochondria is generally viewed as all-or-nothing. We previously confirmed that MOMP occurs in cells that die during mitotic arrest (Huang et al., 2009) and now find evidence for limited MOMP late in mitotic arrest, without cell death.
We suggest, as have others studying etoposide and tumor necrosis factor-related apoptosis-inducing ligand, that partial apoptosis through ICAD/CAD is another cause of DNA damage (Hars et al., 2006; Lovric and Hawkins, 2010). Partial apoptosis is likely very important in cells with a poor apoptotic response that do not commit to death but incur DNA damage, and could explain why antimitotic drugs, notably the taxanes paclitaxel and docetaxel, show increased apoptosis in vitro and in vivo when combined with direct DNA damage by radiation or platinum agents (DeNardo et al., 1997; Sherman and Fine, 2001; Borghaei et al., 2008). As for K5I, Taxol resulted in DNA damage during mitotic arrest in the absence of apoptosis (Figure S6), and in the postslippage induction of p53 (Movie S3). There was also some continued DNA damage in postslippage Taxol cells (Figure S6). Given the effectiveness of taxanes on some cancers, and that it is more toxic to some cells compared with K5I (Shi et al., 2008), it will be important to study the mechanisms of damage in postslippage cells, where the multinucleated phenotype could contribute additional cell stress that is largely absent in mononucleated K5I postslippage cells.
Our data emphasize an emerging concept: antimitotic drugs not normally thought of as DNA damaging can lead to DNA strand breaks, as well as the expected single chromosome aneuploidy and tetraploidy that result from spindle damage. An unresolved question is how these DNA strand breaks contribute to treatment outcomes at the tumor level in vivo. p53 induction postslippage did not lead to strong apoptosis in MCF7 cells, though it might in other genetic contexts. Rather, it induced a senescence-like phenotype, in which cells were arrested for a prolonged period; another K5I also induced senescence (Nakai et al., 2009). This postslippage, senescence-like state might contribute to therapy in multiple ways, including essentially sterilizing the cell, inducing signals that recruit leukocytes to the tumor to contribute to clearance (Xue et al., 2007), and altering the tumor environment directly or indirectly. We need to better understand the role of partial apoptosis in the tumor context, and how it influences the therapeutic response.
MATERIALS AND METHODS
Cell lines
MCF7, U2 OS, HT29, and RPE1 were from ATCC (Manassas, VA); MCF7 sh-p53 was from R. Agami (The Netherlands Cancer Institute, Amsterdam); MCF7 p53-Venus was from G. Lahav (Harvard Medical School, Boston, MA); and p53 parental and null HCT116 were from B. Vogelstein (Johns Hopkins University, Baltimore, MD). MCF7 mutant mICADL-flag (noncleavable ICAD) was from this study. MCF7, U2 OS, HT29, RPE1, and HCT116 cells are wild type for p53 and are routinely used to study p53.
Small molecules and UV treatment
K5I EMD534085 (500 nM; Merck Serono, Darmstadt, Germany), K5I EMD596414 (500 nM; Merck Serono), Taxol (250 nM; Sigma-Aldrich, St. Louis, MO), nocodazole (500 nM; Tocris), VX-680 (250 nM; gift from N. Gray, Harvard Medical School, Boston, MA), BI-2536 (250 nM; gift from N. Gray), ATM inhibitor KU55933 (5 μM; Calbiochem, San Diego, CA), DNA-PK inhibitor NU7026 (10 μM; Sigma), RO-3306 (10 μM; Enzo Life Sciences, Farmingdale, NY), NCS (400 ng/ml, DNA double-strand break inducer; Sigma), zVAD-fmk (caspase inhibitor VI, 100 μM; Calbiochem), zLEHD-fmk (caspase-9 inhibitor, 50 μM; Calbiochem), zDEVD-fmk (caspase-3 inhibitor II, 50 μM; Calbiochem), STS (1 μM; Calbiochem), cytochalasin D (50 μM; Sigma), Y-27632 (10 μM; Sigma). UV treatment of cells was 30 J/m2 UV-C (254 nm).
Antibodies, immunoblotting, and immunoprecipitation
All antibodies were used per the manufacturer's recommendations: α-tubulin (DM1a; Sigma), β-actin (Sigma), p53 (DO1; Santa Cruz Biotechnology, Santa Cruz, CA), phospho-serine-15-p53 (Cell Signaling Technology, Danvers, MA), phospho-serine-33-p53 (Cell Signaling), p21 (Calbiochem), cytoplasmic dynein (Millipore, Billerica, MA), γH2A.X (Millipore and Cell Signaling), p-H3 (Millipore), MPM2 (Millipore), CytC (Promega, Madison, WI), caspase-9 (Cell Signaling), cleaved caspase-7 (Cell Signaling), phospho-serine-1981-ATM (Rockland Immunochemicals, Gilbertsville, PA), 53BP1 (Cell Signaling), cleaved PARP1 (Calbiochem), ICAD (Santa Cruz and Cell Signaling). For immunoblots, 10–20 μg total cell lysate was separated by SDS–PAGE in Nu-PAGE 4–12% gradient Bis-Tris gels (Invitrogen, Carlsbad, CA), transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA), incubated with antibody, washed, and detected using ECL Plus (Amersham, Piscataway, NJ). Immunoprecipitation of ICAD was performed using 2 μg anti-ICAD antibody (C-terminus; Santa Cruz) conjugated to 15 μl protein A beads (Affi-Prep; Bio-Rad). Cells were lysed in TNTE buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton-X100); 1000 μg total cell lysate was incubated with the beads, washed four times with TNTE buffer, eluted by boiling in sample buffer, separated by SDS–PAGE, and immunoblotted with anticleaved ICAD antibody (Cell Signaling).
Immunofluorescence staining, assays, and microscopy
Cells on glass coverslips were fixed for 10 min at room temperature in 3.7% formaldehyde/Dulbecco's phosphate-buffered saline (DPBS)/0.2% Triton-X100; washed 10 min in DPBS; blocked 30 min in 5% BSA/DPBS; incubated 45 min in primary antibody; washed 10 min; incubated 45 min with fluorescently conjugated Alexa Fluor 488, 568, or 647 secondary antibodies (Invitrogen); washed 10 min; stained 2 min in 1 μg/ml Hoechst (bis-benzamide; Sigma) or TO-PRO3 (Invitrogen); washed for 5 min; rinsed once in deionized water; and mounted in Prolong Gold antifade reagent (Invitrogen) on glass slides. Cells were imaged using a Nikon TE2000 with a spinning disk confocal at 100×, 1.40 numerical aperture oil immersion lens in Z-series at 0.5 μm optical sections using MetaMorph.
For γH2A.X quantification, mitotic chromosomes or interphase cell nuclei were segmented using the DNA stain, and γH2A.X-integrated fluorescence intensity on the chromosomes/DNA was measured using MetaMorph. For γH2A.X foci, all foci ≥ 2-fold above background fluorescence intensity were scored.
For quantification of mitochondrial/CytC phenotypes in mitotic cells, cells were identified using the DNA stain. MOMP cells were identified using the CytC and DNA channels. The region of interest corresponding to the area of each cell was defined, and the SD of the average pixel intensity was measured using MetaMorph (Huang et al., 2009). In healthy cells, CytC is sequestered in mitochondria and the SD is high. In cells that have undergone MOMP, the SD is dramatically decreased, because CytC is homogenously distributed throughout the cytoplasm. Statistics and plots were in Excel (Microsoft, Redmond, WA) and MatLab (Mathworks, Natick, MA).
For live imaging, MCF7, MCF7 p53-Venus, U2 OS p53-Venus, MCF7 sh-p53, HCT116, and HCT116 p53−/− were in glass bottom dishes (MatTek, Ashland, MA) and imaged at 20× in normal or drug-containing phenol red-free RPMI (Mediatech, Manassas, VA), at 10–15 min/frame in humidified air/5% CO2. Image analysis was performed as previously described (Batchelor et al., 2008; Loewer et al., 2010).
Supplementary Material
Acknowledgments
We thank S. Nagata (Kyoto University) for the mutant, non-caspase-cleavable ICAD plasmid (pEF-FLAG mICADL); the Nikon Imaging Core facility at Harvard Medical School; Soyeon Park, Aaron Groen, Edwin Tan, Yangzhong Tang, Eric Batchelor, Caroline Mock, and the Mitchison and Lahav laboratories; and Nathanael Gray for VX-680 and BI-2536. This research was supported by NIH grant CA139980 to T.J.M. and partially supported by NIH grant GM083303 to G.L. and the Charles A. King Trust fellowship to A.L.
Abbreviations used:
- ATM
ataxia-telangiectasia mutated
- CAD
caspase-activated DNase
- CytC
cytochrome C
- DNA-PK
DNA-dependent protein kinase
- DPBS
Dulbecco's phosphate-buffered saline
- ICAD
inhibitor of caspase-activated DNase
- K5I
kinesin-5
- MOMP
mitochondrial outer-membrane permeabilization
- NCS
neocarzinostatin
- P-ATM
phosphorylated, active ATM
- p-H3
phospho-serine-10-histone-H3
- SAC
spindle assembly checkpoint
- STS
staurosporine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0781) on December 14, 2011.
REFERENCES
- Bacus SS, Gudkov AV, Lowe M, Lyass L, Yung Y, Komarov AP, Keyomarsi K, Yarden Y, Seger R. Taxol-induced apoptosis depends on MAP kinase pathways (ERK and p38) and is independent of p53. Oncogene. 2001;20:147–155. doi: 10.1038/sj.onc.1204062. [DOI] [PubMed] [Google Scholar]
- Barboule N, Chadebech P, Baldin V, Vidal S, Valette A. Involvement of p21 in mitotic exit after paclitaxel treatment in MCF-7 breast adenocarcinoma cell line. Oncogene. 1997;15:2867–2875. doi: 10.1038/sj.onc.1201469. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol Syst Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti CJ, Villalobos MJ, Jiang Q, Cline E, Diaz MO, Loredo G, Vaughan AT. Cleavage of the MLL gene by activators of apoptosis is independent of topoisomerase II activity. Leukemia. 2005;19:2289–2295. doi: 10.1038/sj.leu.2403966. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Prolonged mitosis versus tetraploid checkpoint: how p53 measures the duration of mitosis. Cell Cycle. 2006;5:971–975. doi: 10.4161/cc.5.9.2711. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6:70–74. doi: 10.4161/cc.6.1.3682. [DOI] [PubMed] [Google Scholar]
- Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci USA. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghaei H, et al. Phase II study of paclitaxel, carboplatin, and cetuximab as first line treatment, for patients with advanced non-small cell lung cancer (NSCLC): results of OPN-017. J Thorac Oncol. 2008;3:1286–1292. doi: 10.1097/JTO.0b013e318189f50e. [DOI] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, On KF, Chan WM, Wong W, Siu HO, Hau PM, Poon RY. The kinetics of p53 activation versus cyclin E accumulation underlies the relationship between the spindle-assembly checkpoint and the postmitotic checkpoint. J Biol Chem. 2008;283:15716–15723. doi: 10.1074/jbc.M800629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton WB, Nandan MO, Moore RT, Yang VW. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67:11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton MA, Nahreini P, Srivastava A. Augmented nuclease activity during cellular senescence in vitro. J Cell Biochem. 1989;39:75–85. doi: 10.1002/jcb.240390109. [DOI] [PubMed] [Google Scholar]
- DeNardo SJ, Kukis DL, Kroger LA, O'Donnell RT, Lamborn KR, Miers LA, DeNardo DG, Meares CF, DeNardo GL. Synergy of Taxol and radioimmunotherapy with yttrium-90-labeled chimeric L6 antibody: efficacy and toxicity in breast cancer xenografts. Proc Natl Acad Sci USA. 1997;94:4000–4004. doi: 10.1073/pnas.94.8.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- Hars ES, Lyu YL, Lin CP, Liu LF. Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Res. 2006;66:8975–8979. doi: 10.1158/0008-5472.CAN-06-1724. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Shi J, Orth JD, Mitchison TJ. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16:347–358. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- Lai SK, Wong CH, Lee YP, Li HY. Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death. Cell Death Differ. 2011;18:996–1004. doi: 10.1038/cdd.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric MM, Hawkins CJ. TRAIL treatment provokes mutations in surviving cells. Oncogene. 2010;29:5048–5060. doi: 10.1038/onc.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Meek DW. The role of p53 in the response to mitotic spindle damage. Pathol Biol (Paris) 2000;48:246–254. [PubMed] [Google Scholar]
- Morris EJ, Keramaris E, Rideout HJ, Slack RS, Dyson NJ, Stefanis L, Park DS. Cyclin-dependent kinases and P53 pathways are activated independently and mediate Bax activation in neurons after DNA damage. J Neurosci. 2001;21:5017–5026. doi: 10.1523/JNEUROSCI.21-14-05017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R, et al. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 2009;69:3901–3909. doi: 10.1158/0008-5472.CAN-08-4373. [DOI] [PubMed] [Google Scholar]
- Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–4616. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Tang Y, Shi J, Loy CT, Amendt C, Wilm C, Zenke FT, Mitchison TJ. Quantitative live imaging of cancer and normal cells treated with kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol Cancer Ther. 2008;7:3480–3489. doi: 10.1158/1535-7163.MCT-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F, Rozier L, Lachages AM, Bieth A, Simili M, Debatisse M. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 2007;26:165–172. doi: 10.1038/sj.onc.1209787. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Rojas JM, Taya Y, Gutkind JS. A role for the p38 mitogen-activated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res. 2000;60:2464–2472. [PubMed] [Google Scholar]
- Schiemann K, Finsinger D, Zenke F, Amendt C, Knochel T, Bruge D, Buchstaller HP, Emde U, Stahle W, Anzali S. The discovery and optimization of hexahydro-2H-pyrano[3,2-c]quinolines (HHPQs) as potent and selective inhibitors of the mitotic kinesin-5. Bioorg Med Chem Lett. 2010;20:1491–1495. doi: 10.1016/j.bmcl.2010.01.110. [DOI] [PubMed] [Google Scholar]
- Sherman WH, Fine RL. Combination gemcitabine and docetaxel therapy in advanced adenocarcinoma of the pancreas. Oncology. 2001;60:316–321. doi: 10.1159/000058527. [DOI] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269–3276. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhou Y, Huang HC, Mitchison TJ. Navitoclax (ABT-263) accelerates apoptosis during drug-induced mitotic arrest by antagonizing Bcl-xL. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Stewart ZA, Tang LJ, Pietenpol JA. Increased p53 phosphorylation after microtubule disruption is mediated in a microtubule inhibitor- and cell-specific manner. Oncogene. 2001;20:113–124. doi: 10.1038/sj.onc.1204060. [DOI] [PubMed] [Google Scholar]
- Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- Tan N, Malek M, Zha J, Yue P, Kassees R, Berry L, Fairbrother WJ, Sampath D, Belmont LD. Navitoclax enhances the efficacy of taxanes in non-small cell lung cancer models. Clin Cancer Res. 2011;17:1394–1404. doi: 10.1158/1078-0432.CCR-10-2353. [DOI] [PubMed] [Google Scholar]
- Tao W, South VJ, Diehl RE, Davide JP, Sepp-Lorenzino L, Fraley ME, Arrington KL, Lobell RB. An inhibitor of the kinesin spindle protein activates the intrinsic apoptotic pathway independently of p53 and de novo protein synthesis. Mol Cell Biol. 2007;27:689–698. doi: 10.1128/MCB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint.”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 2006;5:2555–2556. doi: 10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Liu F, Wang W. Two-phase dynamics of p53 in the DNA damage response. Proc Natl Acad Sci USA. 2011;108:8990–8995. doi: 10.1073/pnas.1100600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.