ETOC: During yeast endocytic site formation, Ede1p (yeast Eps15), but not clathrin light chain, is important for the recruitment of most other early-arriving proteins to endocytic sites. Cargo and clathrin light chain may play roles in regulating the transition of endocytic sites out of the “intermediate coat” stage of endocytosis.
Abstract
The earliest stages of endocytic site formation and the regulation of endocytic site maturation are not well understood. Here we analyzed the order in which the earliest proteins are detectable at endocytic sites in budding yeast and found that an uncharacterized protein, Pal1p/Ydr348cp, is also present at the initial stages of endocytosis. Because Ede1p (homologue of Eps15) and clathrin are the early-arriving proteins most important for cargo uptake, their roles during the early stages of endocytosis were examined more comprehensively. Ede1p is necessary for efficient recruitment of most early-arriving proteins, but not for the recruitment of the adaptor protein Yap1802p, to endocytic sites. The early-arriving proteins, as well as the later-arriving proteins Sla2p and Ent1/2p (homologues of Hip1R and epsins), were found to have longer lifetimes in CLC1-knockout yeast, which indicates that clathrin light chain facilitates the transition from the intermediate to late coat stages. Cargo also arrives during the early stages of endocytosis, and therefore its effect on endocytic machinery dynamics was investigated. Our results are consistent with a role for cargo in regulating the transition of endocytic sites from the early stages of formation to the late stages during which vesicle formation occurs.
INTRODUCTION
Endocytosis is the process by which cells internalize proteins and lipids from the plasma membrane and molecules from the surrounding environment. Live-cell fluorescence microscopy in Saccharomyces cerevisiae and mammalian cells has revealed dynamics of the machinery that drives endocytosis (Gaidarov et al., 1999; Merrifield et al., 2002, 2005; Kaksonen et al., 2003; Rappoport and Simon, 2003; Rappoport et al., 2005; Ehrlich et al., 2004; Keyel et al., 2004; Yarar et al., 2005; Henne et al., 2010; reviewed in Perrais and Merrifield, 2005). Protein recruitment to yeast endocytic sites occurs in a highly predictable order, and in mammalian and yeast cells recruitment signatures of groups of endocytic proteins have also been identified (Kaksonen et al., 2005; Taylor et al., 2011). Knowledge of endocytic protein spatiotemporal dynamics has provided valuable insight into the mechanisms underlying endocytic vesicle formation in yeast and mammalian cells (Merrifield et al., 2005; Newpher et al., 2005; Sun et al., 2006; Barker et al., 2007; Ferguson et al., 2009; Liu et al., 2009; Smaczynska-de et al., 2010). In budding yeast, >40 proteins have been shown to form “patches” at endocytic sites. These proteins have been classified into the early, coat, WASP/Myo, amphiphysin, or actin modules based on when the proteins arrive at endocytic sites, the length of time they are detectable at the cell surface (termed patch “lifetime”), and their spatial dynamics at endocytic sites (Kaksonen et al., 2005; Carroll et al., 2009; Stimpson et al., 2009; summarized in Tonikian et al., 2009).
There has recently been much interest in the early stages of endocytic site formation in both yeast and mammalian cells, specifically focusing on proteins that are likely to be involved in site establishment (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009; Henne et al., 2010). In mammalian cells, the proteins Fer/Cip4 homology domain–only proteins 1 and 2 (FCHo1/2), Eps15, and intersectin are present during the earliest known stages of clathrin coat assembly and are believed to be important in the nucleation of endocytic sites (Henne et al., 2010). In comparison, Ede1p (an Eps15 homologue), Syp1p (an FCHo1/2 homologue), clathrin, and the AP2 complex are among the first proteins to appear at nascent endocytic sites in budding yeast (Kaksonen et al., 2005; Newpher et al., 2005; Toshima et al., 2006; Boettner et al., 2009; Carroll et al., 2009; Reider et al., 2009; Stimpson et al., 2009). However, the order in which these proteins arrive at endocytic sites with respect to each other, and their roles in early site formation, have not been thoroughly studied. An exploration of the early endocytic stages in budding yeast will elucidate mechanisms that regulate endocytic site establishment.
Proteins that arrive early during endocytic site formation have variable patch lifetimes that range from 30 s to 4 min, whereas later-arriving proteins have highly regular patch lifetimes that generally last 10–40 s (Kaksonen et al., 2005; Newpher et al., 2005; Carroll et al., 2009; Stimpson et al., 2009). This observation suggests that a transition point exists past which endocytic vesicle formation cannot progress until some event triggers recruitment and/or activation of later-acting components of the endocytic machinery that drive the process to completion. The transition point in yeast endocytosis may be similar to a proposed endocytic checkpoint acting during clathrin-coated pit (CCP) maturation in mammalian cells (Loerke et al., 2009). Studies in mammalian cells have shown that various endocytic cargo molecules and their adaptors have differing effects on CCP maturation and therefore might be influencing progression past an endocytic checkpoint (Ehrlich et al., 2004; Loerke et al., 2009; Mettlen et al., 2009, 2010). Because in yeast endocytic cargoes are believed to arrive during the early phase of endocytosis (Toshima et al., 2006), they may also have a role in promoting early endocytic-site progression. Furthermore, the absence of clathrin in yeast cells causes some later-arriving proteins to have shorter patch lifetimes and others to have extended lifetimes, suggesting that clathrin has a role in endocytic site maturation (Kaksonen et al., 2005; Newpher and Lemmon, 2006; Newpher et al., 2006). This complex role for clathrin is also evident in animal cells in which knockdowns of clathrin light chain a and b isoforms have different effects on CCP dynamics (Mettlen et al., 2009). A more comprehensive study of the roles of the early-arriving proteins and endocytic cargo in regulating early endocytic stages promises to provide insight into the initiation stage of endocytosis and how endocytic sites progress through the transition point.
Here we find that the previously unstudied protein Pal1p/Ydr348cp and the adaptor protein Yap1802p arrive together with Ede1p, Syp1p, clathrin, and the AP2 complex during the earliest stage of endocytic site formation. Whereas Ede1p is important for recruitment of most early-arriving proteins to endocytic sites, clathrin light chain is important for transitioning endocytic sites out of the intermediate coat stage marked by Sla2p and Ent1/2p and into the late stages of endocytic site internalization. We also provide evidence that cargo may regulate yeast endocytic site maturation.
RESULTS
Arrival of proteins at the yeast early endocytic site
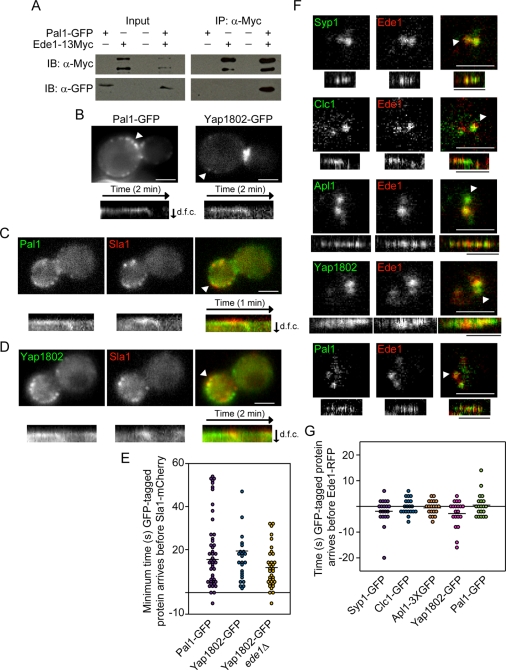
Ede1p, Syp1p, clathrin, and the AP2 complex are known to arrive at endocytic sites early in the pathway (Kaksonen et al., 2005; Newpher et al., 2005; Boettner et al., 2009; Carroll et al., 2009; Stimpson et al., 2009; Reider et al., 2009; Toshima et al., 2006). We predicted that the uncharacterized protein Ydr348cp might also be an endocytic protein because high-throughput studies have found that it has physical interactions with Ede1p, and Ydr348c-GFP localizes to the cell periphery and bud neck (Gavin et al., 2002, 2006; Huh et al., 2003). Furthermore, Ydr348cp is considered an orthologue of the Schizosaccharomyces pombe protein Pal1p, and both proteins share 37% identity over a conserved “Pal1” domain (Ge et al., 2005; Penkett et al., 2006; Finn et al., 2008). Pal1p is implicated in endocytosis since it is located at the cell periphery and has been shown to physically interact with the endocytic protein Sla2p (Ge et al., 2005). On the basis of this information, we here name S. cerevisiae Ydr348cp as “Pal1p.” We C-terminally tagged Pal1p with GFP and were able to confirm its interaction with a C-terminally 13Myc tagged-Ede1p by coimmunoprecipitation (Figure 1A). In live cells, Pal1–green fluorescent protein (GFP) forms dynamic patches at the cell surface (Figure 1B, left) that have an average lifetime of 65 ± 35 s (n = 50). When the cells are imaged in a medial focal plane, it is clear that these patches internalize from the cortex before disassembling (94%, n = 50), which is a characteristic of endocytic coat proteins (Kaksonen et al., 2005). A total of 94% of Sla1-mCherry patches contained Pal1-GFP (n = 50), indicating that Pal1-GFP is an endocytic protein present at essentially all endocytic sites (Figure 1C). Pal1-GFP also arrives at endocytic sites before Sla1-mCherry (94%, n = 47) (Figure 1E). On the basis of the length of time that Pal1p is present at endocytic sites, Pal1p can be considered as an early-arriving endocytic coat protein.
FIGURE 1:
Analysis of the yeast early endocytic site. (A) Protein lysates from yeast expressing Pal1-GFP and/or Ede1-13Myc were incubated with beads conjugated to an anti-Myc antibody. Pal1-GFP copurifies with Ede1-13Myc. IB, immunoblot; IP, immunoprecipitation. (B) Epifluorescence images of yeast expressing Pal1-GFP or Yap1802-GFP. Kymographs are of the patches indicated by the white arrowheads. (C) Epifluorescence images of a yeast cell expressing Sla1-mCherry and Pal1-GFP. Kymographs of the patch indicated by the white arrowhead are from two-color movies acquired at a rate of one frame per second. (D) Epifluorescence images of a yeast cell expressing Sla1-mCherry and Yap1802-GFP. Kymographs of the patch indicated by the white arrowhead are from two-color movies acquired at a rate of two frames per second. (E). Minimum amount of time the indicated GFP-tagged protein arrived at endocytic sites before Sla1-mCherry (Pal1-GFP, n = 47; Yap1802-GFP, n = 24; Yap1802-GFP ede1Δ, n = 35). In some instances, the GFP-tagged protein was present in the first frame of the movie, so the exact amount of time it was present before Sla1-mCherry arrived is unknown. Negative time represents instances when Sla1-mCherry arrived before the GFP-tagged protein. Horizontal bars represent average time for a group. (F) Two-color TIRF microscopy images of yeast expressing Ede1-RFP and Syp1-GFP, Clc1-GFP, Apl1-3XGFP, Yap1802-GFP, or Pal1-GFP. Kymographs of the patches indicated by the white arrowheads are from two-color movies. Black time bars, 30 s. (G) Amount of time the indicated GFP-tagged protein was detected at sites before Ede1-RFP was detected (n = 20 patches for each strain). Negative time represents instances when Ede1-RFP was detected before the GFP-tagged protein. GFP and RFP tagged proteins were first detected within 4 s of each other ≥80% of the time (n = 20 patches for each strain). Horizontal bars represent average time for a group. All white scale bars, 2 μm. d.f.c., distance from cortex.
Yap1801p and Yap1802p, homologues of mammalian AP180, are believed to recruit clathrin to the plasma membrane (Newpher et al., 2005) and therefore may act as early-arriving endocytic proteins. The dynamics of Yap1802p was analyzed to determine whether the yeast AP180s arrive early at endocytic sites. Yap1802-GFP patches have an average lifetime of 63 ± 26 s (n = 50) and, like Pal1p, move off the cortex before disassembly (98%, n = 50; Figure 1B, right). In a two-color analysis, Yap1802-GFP was present at 96% of Sla1-mCherry patches (n = 25), and Yap1802-GFP arrived at endocytic sites before Sla1-mCherry (100%, n = 24; Figure 1, D and E). Together these data place Yap1802p early in the coat module.
Dynamics and recruitment of many later-arriving endocytic proteins have been described (Kaksonen et al., 2003, 2005), which has been pivotal in our understanding of endocytic mechanisms. However, a detailed analysis of the arrival of early endocytic proteins, which could aid our understanding of the mechanism of site establishment and progression, is lacking. Two-color total internal reflection fluorescence (TIRF) microscopy was used to determine whether any regular order of recruitment of early proteins to endocytic sites could be detected relative to Ede1–red fluorescent protein (RFP) recruitment. Clathrin light chain (Clc1p), Apl1p, and Yap1802p were used as markers for clathrin, the AP2 complex, and the yeast AP180s, respectively. We found that Syp1-GFP, Clc1-GFP, Apl1-3XGFP, Yap1802-GFP, and Pal1-GFP appear at a similar time as Ede1-RFP in instances when both proteins were clearly visible (n = 20 patches for each strain; Figure 1F). There was no consistent pattern in whether the GFP-tagged proteins were detected before or after Ede1-RFP was detected; instead, our results suggest the early endocytic proteins arrive at the nascent endocytic site at approximately the same time (usually within 4 s; Figure 1, F and G). However, the fluorescence signal from these proteins is quite dim and the patches tend to fluctuate in intensity until their disassembly, precluding a definitive statement on whether there is any regular order of arrival. On the basis of these observations, we define a new early coat module comprised of clathrin, the AP2 complex, the yeast AP180s, and Pal1p (Figure 2). The early coat module proteins internalize with the vesicle before disassembly, which is distinct from the early module proteins (Ede1p and Syp1p) that disassemble before coat internalization (Stimpson et al., 2009).
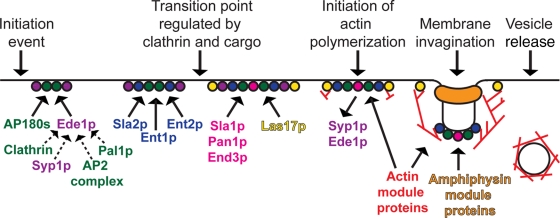
FIGURE 2:
Model for the temporal recruitment of endocytic proteins. The early module proteins (Ede1p and Syp1p; purple) and the early coat module proteins (clathrin, the AP2 complex, the yeast AP180s, and Pal1p; green) arrive earliest during endocytic site formation. Ede1p is important for the recruitment of Syp1p, clathrin, the AP2 complex, and Pal1p to endocytic sites. The dashed lines indicate that Ede1p may be involved directly or indirectly in endocytic protein recruitment. Next, the intermediate coat module proteins (Sla2p, Ent1p, and Ent2p; blue) arrive at endocytic sites. Endocytic sites seem to mature through a transition point regulated by clathrin and cargo before the late coat module proteins (Sla1p, Pan1p, and End3p; pink) and Las17p (a WASP/Myo module protein; yellow) are recruited to sites. Ede1p and Syp1p disassemble from endocytic sites at the start of actin polymerization. During membrane invagination, Sla2p, Ent1p, and Ent2p may be involved in connecting the actin network to the vesicle coat and/or plasma membrane.
Analysis of the roles of early-arriving proteins in endocytosis
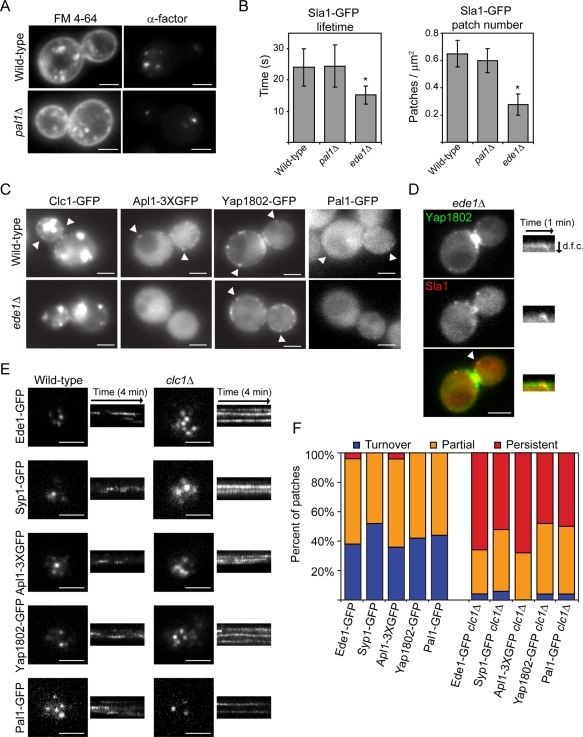
A more comprehensive analysis of the function of the early-arriving proteins is needed to better understand endocytic site formation and progression to later stages. Because the function of Pal1p in endocytosis has not been analyzed, the role of Pal1p in fluid-phase and receptor-mediated endocytosis was examined. Similar to what was observed in wild-type cells, pal1Δ yeast are able to internalize FM 4-64 and clear fluorescently labeled α-factor from the plasma membrane (Figure 3A). These results show that Pal1p is not required for bulk or receptor-mediated endocytosis. The dynamics of Sla1-GFP was examined in pal1Δ yeast to determine how Pal1p affects other components of the endocytic machinery. Unlike ede1Δ yeast, in which Sla1-GFP lifetimes and numbers of patches are known to decrease (Kaksonen et al., 2005; Stimpson et al., 2009), the average lifetimes (n = 50 patches) and numbers of Sla1-GFP patches (n = 20 cells) remain unchanged in pal1Δ yeast (Figure 3B).
FIGURE 3:
Analysis of the roles of Pal1p, Ede1p, and Clc1p in endocytosis. (A) FM 4-64 uptake (left) and fluorescent α-factor uptake (right) in the indicated strains after 10 min. (B) Lifetimes of Sla1-GFP patches ± SD (n = 50 patches; left) and Sla1-GFP patch number per cell surface area (μm2) ± SD (n = 20 cells; right) for the indicated yeast. Patch number was counted from maximum-intensity Z-projections of unbudded or large-budded cells. Movies used to generate lifetime data were acquired at a rate of one frame per second. Asterisk indicates a statistically significant decrease compared with wild-type (p < 0.0001). (C) Images of wild-type and ede1Δ yeast expressing Clc1-GFP, Apl1-3XGFP, Yap1802-GFP, or Pal1-GFP. White arrowheads indicate examples of cortical patches. (D) Images of an ede1Δ yeast cell expressing Yap1802-GFP and Sla1-mCherry. Kymographs of the patch indicated by the white arrowhead are from two-color movies. d.f.c., distance from cortex. (E) TIRF microscopy images of wild-type and clc1Δ yeast cell expressing Ede1-GFP, Syp1-GFP, Apl1-3XGFP, Yap1802-GFP, or Pal1-GFP. Kymographs are taken from 4-min movies. (F) Percentage of patches in the indicated strains that assemble and disassemble within a 4-min interval (turnover), are present throughout the TIRF microscopy movie (persistent), or are present in either the first or last frames of the movie (partial; n = 50 patches). All white scale bars, 2 μm.
Deleting SYP1, AP2 complex subunits, and YAP1801/2 does not cause readily detectable defects in endocytic dynamics, but these proteins are important for the internalization of certain cargoes (Huang et al., 1999; Kaksonen et al., 2005; Burston et al., 2009; Carroll et al., 2009; Reider et al., 2009; Stimpson et al., 2009). Ede1p and clathrin, however, are important for proper dynamics of later-arriving endocytic proteins and for efficient cargo uptake (Chu et al., 1996; Gagny et al., 2000; Kaksonen et al., 2005; Newpher and Lemmon, 2006; Stimpson et al., 2009). Therefore we decided to more thoroughly investigate the roles of Ede1p and clathrin in the initiation and maturation of early endocytic sites by assessing the localization and dynamics of early-arriving proteins in ede1Δ and clc1Δ yeast.
Clc1-GFP, Apl1-3XGFP, and Pal1-GFP all failed to form distinct, stable cortical patches in ede1Δ yeast (Figure 3C). This phenotype is similar to the behavior observed for Syp1-GFP in the absence of Ede1p (Reider et al., 2009; Stimpson et al., 2009). It is surprising that Yap1802-GFP forms patches in ede1Δ yeast (Figure 3C) and Yap1802-GFP still arrives before Sla1-mCherry in these cells (91%, n = 35; Figures 1E and 3D). Thus, Ede1p is important for the recruitment of most early-arriving proteins to endocytic sites, although Yap1802p can localize correctly in the absence of Ede1p. Of interest, Ede1-GFP patch lifetimes are unchanged in syp1Δ, apl1Δ, yap1802Δ, and pal1Δ yeast (n = 50 patches for each strain; results not shown).
The role of clathrin in early endocytic site establishment and maturation was studied using a clathrin light chain–deletion mutant. Ede1-GFP, Apl1-3XGFP, Yap1802-GFP, and Pal1-GFP are able to form cortical patches in clc1Δ yeast, indicating that clathrin light chain is dispensable for endocytic site establishment (Figure 3E). Although previous studies found that Syp1-GFP patch localization was reduced in clc1Δ yeast (Boettner et al., 2009), we observe that Syp1-GFP is able to form cortical patches in clc1Δ yeast (Figure 3E). However, the dynamics of Ede1-GFP, Syp1-GFP, Apl1-3XGFP, Yap1802-GFP, and Pal1-GFP patches are perturbed in clathrin light chain mutants: very few “turnover” patches (patches that assemble and disassemble within a 4-min period and are likely to represent productive endocytic events) are observed when compared with wild-type cells (Figure 3, E and F). There is also an increase in the number of “persistent” patches (patches that persist for >4 min) in clc1Δ yeast for each of the early-arriving endocytic proteins (n = 50 patches for each strain). Patches that only assemble (but do not disassemble) or that only disassemble (but do not assemble) during a 4-min movie were termed “partial' patches.
Clathrin light chain is important for progression through the intermediate coat stage
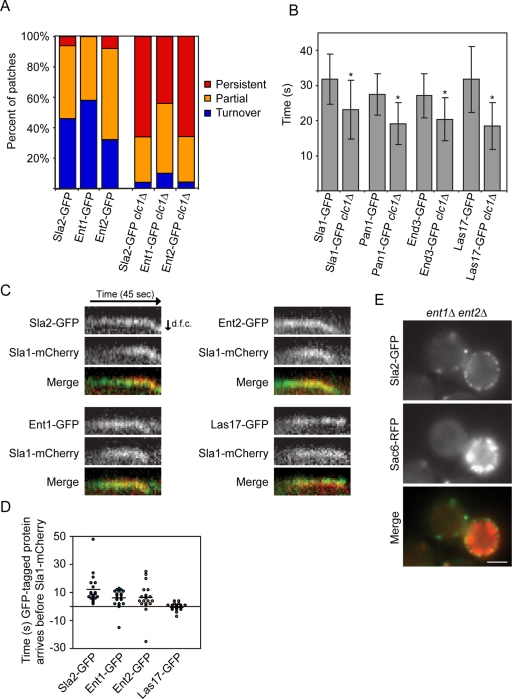
The increase in numbers of persistent patches formed by early-arriving endocytic proteins in clc1Δ yeast is similar to the phenotype previously observed for the later-arriving protein Sla2p, a Hip1R homologue, in clathrin knockouts (Newpher and Lemmon, 2006; Newpher et al., 2006). Therefore the recruitment and dynamics of later-arriving endocytic proteins were analyzed in clc1Δ yeast. Previous studies found that Sla2-GFP forms patches with extended lifetimes in clc1Δ yeast (Newpher and Lemmon, 2006; Newpher et al., 2006). We were able to repeat these findings (n = 50; Figure 4A). Of interest, we found that Ent1-GFP and Ent2-GFP also form more persistent patches in clc1Δ yeast (n = 50 patches for each strain; Figure 4A). In contrast to this result, Sla1-GFP and Las17-GFP were reported to have shorter lifetimes in clc1Δ yeast (Kaksonen et al., 2005; Newpher and Lemmon, 2006). We were able to reproduce this observation and furthermore found that Pan1-GFP and End3-GFP patches also have shorter lifetimes in clathrin light chain mutants (n = 50 patches for each strain; p < 0.0001 for each strain; Figure 4B). It should be noted, however, that Pan1-GFP and End3-GFP form a few persistent patches in clc1Δ yeast that are present throughout 90-s movies. Together these results show that clathrin light chain is important for the proper disassembly of Sla2p, Ent1p, and Ent2p.
FIGURE 4:
Sla2p, Ent1p, and Ent2p behave similarly in clc1Δ yeast, have similar localization dynamics, and display similar knockout phenotypes. (A) Percentage of patches in the indicated strains that assemble and disassemble within a 4-min interval (turnover), are present throughout the TIRF microscopy movie (persistent), or are present in either the first or last frames of the movie (partial; n = 50 patches). (B) Lifetime of Sla1-GFP, Pan1-GFP, End3-GFP, and Las17-GFP patches ± SD in wild-type or clc1Δ yeast (n = 50 patches). Movies used to generate lifetime data were acquired at a rate of one frame per second. Asterisk indicates a statistically significant decrease compared with wild type (p < 0.0001). (C) Kymographs from epifluorescence two-color movies of yeast expressing Sla1-mCherry and Sla2-GFP, Ent1-GFP, Ent2-GFP, or Las17-GFP. Movies were acquired at a rate of one frame per second. d.f.c., distance from cortex. (D) Amount of time the indicated GFP-tagged protein arrived before Sla1-mCherry (n = 20 patches for each strain). Negative time represents instances when Sla1-mCherry arrived before the GFP-tagged protein. Horizontal bars represent average time for a group. (E) Fluorescence images of an ent1Δ ent2Δ yeast cell expressing Sla2-GFP and Sac6-RFP. White scale bars, 2 μm.
Owing to their similar phenotypes in clc1Δ yeast, we examined Sla2p, Ent1p, and Ent2p more carefully. Previous studies found that Sla2-GFP arrives at endocytic sites before Sla1-mCherry (Newpher and Lemmon, 2006), and we were able to reproduce these results (100%, n = 20; Figure 4, C and D). Moreover, we now find that Ent1-GFP and Ent2-GFP also arrive at endocytic sites before Sla1-mCherry (90% for both, n = 20 patches for each strain; Figure 4, C and D). However, Sla1-mCherry and Las17-GFP arrive at endocytic sites at a similar time point (n = 20 patches for each strain; Figure 4, C and D). After finding that Sla2p, Ent1p, and Ent2p have a unique localization dynamics, we investigated whether these proteins also have similar functions in endocytosis. In our strain background ent1Δ ent2Δ yeast are viable, but we found that they form abnormal actin structures, marked by Sac6-RFP, that are similar to those previously reported in ent1Δ ent2Δ yeast expressing an ENTHY100R domain from Ent1p (Figure 4E) (Aguilar et al., 2006). The actin structures in ent1Δ ent2Δ yeast are also very similar to the elongated actin tails formed in sla2Δ yeast (Kaksonen et al., 2003). However, this phenotype is not caused by a lack of recruitment of Sla2p to endocytic sites in ent1Δ ent2Δ yeast (Figure 4E). These data suggest that Sla2p, Ent1p, and Ent2p act at a similar stage of endocytosis and should be considered components of a new, intermediate coat module (Figure 2).
Cargo may play a role in progression through a regulatory transition point controlling endocytic site maturation
After establishing how early-arriving proteins affect endocytic site formation and maturation, we next sought to investigate the role of cargo in this process, since cargo also arrives early at endocytic sites (Toshima et al., 2006). The dynamics of the endocytic machinery was monitored in a secretion mutant, sec18-1ts, in which cell surface levels of endocytic cargo can be reduced. The endocytic cargo protein GFP-Snc1, which cycles continuously between the plasma membrane and endosomal compartments (Lewis et al., 2000), disappears from the plasma membrane of sec18-1ts yeast within 20 min of shifting to 37°C (Figure 5A). This indicates that within the first 20 min of shifting sec18-1ts yeast to 37°C, nearly all of the GFP-Snc1 is endocytosed from the plasma membrane and further secretion to the plasma membrane is blocked. After the yeast are incubated for a longer period of time at 37°C (30 min), the number of persistent Ede1-GFP, Clc1-GFP, and Sla2-GFP patches increases in sec18-1ts mutants when compared with wild-type cells (n = 50 patches for each strain; Figure 5B). The decrease in the number of dynamic Ede1-GFP patches in sec18-1ts mutants could be reversed by returning the yeast to 25°C for 20 min, thereby indicating that the cells remained viable (n = 50; Figure 5B). Furthermore, the number of Sla1-GFP patches in sec18-1ts mutants decreases when the yeast are incubated for 45 min at 37°C (n = 20 cells; p < 0.0001; Figure 5H). These results imply that endocytic sites present in sec18-1ts mutants do not properly mature when the yeast are shifted to 37°C for at least 30 min. This defect in endocytic patch dynamics in sec18-1ts mutants at the restrictive temperature is accompanied by a defect in FM 4-64 uptake (Figure 5C). The endocytic defects in the sec18-1ts mutant are likely not caused by a depletion of plasma membrane phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) because there is no detectable change in the distribution of GFP-2XPH(PLCδ), which is a marker for this lipid (Figure 5D; Stefan et al., 2002).
FIGURE 5:
Endocytic defects in sec18-1ts yeast. (A) Images of wild-type or sec18-1ts yeast expressing GFP-Snc1. Yeast were imaged at room temperature (RT) or incubated at 37°C for 20 min before imaging. (B) Percentage of patches in the indicated strains that assemble and disassemble within a 4-min interval (turnover), are present throughout the TIRF microscopy movie (persistent), or are present in either the first or last frames of the movie (partial; n = 50 patches). Wild-type and mutant strains were incubated at 37°C for 30 min before imaging, except for yeast incubated at 37°C for 30 min and then shifted to 25°C for 20 min before imaging (Ede1-GFP sec18-1ts shift 25°C). (C) FM 4-64 uptake in strains incubated at 37°C for 30 min and then labeled with FM 4-64 for 20 min at 37°C before imaging. (D) Images of wild-type and sec18-1ts yeast expressing GFP-2XPH(PLCδ), which is a marker for PtdIns(4,5)P2. Yeast were incubated at 37°C for 30 min before imaging. (E) Images of Gap1-RFP overexpressed in wild-type or sec18-1ts yeast. Cells were incubated at 37°C for 45 min (i, iv) or 59 min (ii, v), or cells were incubated at 37°C for 45 min and then treated with 0.1% glutamate (wt/vol) for 14 min at 37°C (iii, vi). Insets are line scans of fluorescence intensity (y-axis; arbitrary units) along the three white lines shown. (F) Decrease in the fluorescence intensity (arbitrary units) ± SE of the mean (n = 20 cells) of plasma membrane–localized Gap1-RFP in sec18-1ts yeast incubated at 37°C. Gap1-RFP levels were measured at a 45-min and a 65-min time point in cells that were incubated with (glutamate) or without (untreated) 0.1% glutamate (wt/vol) during this time period. (G) Percentage of Ede1-GFP patches in sec18-1ts yeast overexpressing Gap1-RFP that are turnover, persistent, or partial patches (4-min TIRF microscopy movie; n = 50 patches). Yeast were incubated at 37°C for 55 min (untreated) or incubated at 37°C for 45 min and treated with 0.1% glutamate for 10 min at 37°C (glutamate) before imaging. (H) Sla1-GFP patch number per cell surface area (μm2) ± SD (n = 20 cells) for wild-type and sec18-1ts yeast overexpressing Gap1-RFP. Yeast were incubated at 37°C for 55 min (untreated) or incubated at 37°C for 45 min and treated with 0.1% glutamate for 10 min at 37°C (glutamate) before imaging. Patch number was counted from maximum-intensity Z-projections of unbudded or large-budded cells. All white scale bars, 2 μm.
To determine whether other secretion mutants also have endocytic defects, we examined endocytosis in sec1-1ts mutants (Novick et al., 1980). The endocytic cargo GFP-Snc1 clears from the plasma membrane of sec1-1ts yeast within 45 min of shifting to 37°C (Supplemental Figure S1A). This result indicates that GFP-Snc1 is endocytosed from the plasma membrane and further secretion is blocked within the first 45 min of shifting sec1-1ts yeast to 37°C. There is a following increase in the number of persistent Ede1-GFP patches in sec1-1ts mutants when the yeast are incubated at 37°C for a longer period of time (90 min; n = 50; Supplemental Figure S1B). The sec1-1ts mutants also have defects in Lucifer yellow uptake after an extended incubation at the restrictive temperature (Supplemental Figure S1C). Together, these results show that sec18-1ts and sec1-1ts mutants have defects in endocytosis.
In sec18-1ts and sec1-1ts yeast at the restrictive temperature, constitutively recycled cargoes are endocytosed from the plasma membrane and are not replaced. The decrease in endocytic cargo concentration at the plasma membrane might influence endocytic site progression through a regulatory transition point. To test this hypothesis, the amino acid permease Gap1-RFP was first overexpressed in sec18-1ts yeast. Gap1p is expressed at the cell surface when wild-type or sec18-1ts yeast are grown on a poor nitrogen source (Figure 5E, i, ii, iv, and v). However, Gap1p is cleared from the plasma membrane and trafficked to the vacuole in the presence of a preferred nitrogen source, such as glutamate (Roberg et al., 1997; Soetens et al., 2001). Within 14 min of adding 0.1% glutamate, plasma membrane Gap1-RFP levels decrease in wild-type cells and in sec18-1ts cells incubated at the restrictive temperature (Figure 5E, iii and vi). Gap1-RFP does not accumulate in the vacuole when glutamate is added to sec18-1ts yeast, likely due to the mutant's trafficking defects (Hicke et al., 1997). Although there is a decrease in plasma membrane Gap1-RFP levels over time in sec18-1ts mutants, there is a greater decrease in plasma membrane Gap1-RFP levels in sec18-1ts cells treated with glutamate compared with untreated sec18-1ts cells (n = 20 cells; p < 0.0001; Figure 5, E, insets, and F). The increase in persistent Ede1-GFP patches (n = 50 patches) and the decrease in number of Sla1-GFP patches (n = 20 cells) at 37°C in sec18-1ts cells are partially rescued when Gap1-RFP is targeted for endocytosis by the addition of glutamate (Figure 5, G and H). The defect in Ede1-GFP dynamics at 37°C in sec1-1ts yeast is also rescued when Gap1-RFP internalization is induced by adding glutamate (n = 50; Supplemental Figure S1, D and E). The defects in Ede1p patch dynamics (n = 50) and number of Sla1p patches (n = 20 cells) at 37°C in sec18-1ts yeast are not rescued when glutamate is added to yeast that are also lacking Gap1p, indicating that the Gap1p cargo is mediating this effect (results not shown). The lifetime of Ede1-GFP patches and the number of Sla1-GFP patches in wild-type cells overexpressing Gap1-RFP does not change when glutamate is added (data not shown). Our results indicate that endocytic defects of sec18-1ts and sec1-1ts yeast can be partially rescued by inducing internalization of an endocytic cargo.
To further investigate this cargo-dependent effect, we added cycloheximide to yeast, which is another method reported to induce internalization of multispan plasma membrane proteins (Galan and Haguenauer-Tsapis, 1997; Lin et al., 2008). Cells incubated for 2 h in 50 μg/ml cycloheximide clear RFP-Snc1 and Fur4-GFP from the plasma membrane (data not shown). Cycloheximide-induced depletion of membrane cargo also results in an increase in the number of persistent Ede1-GFP patches (data not shown). Admittedly, cycloheximide effects on cell physiology are rather global, but this observation nonetheless is consistent with a role for cargo in modulating dynamics of the endocytic pathway.
DISCUSSION
In this study, we identified and analyzed the relative timing of recruitment of the earliest-arriving endocytic proteins and investigated the roles of these early proteins and cargo in endocytic dynamics.
Assembly of proteins at the early endocytic site
This work expanded the number of proteins known to arrive at the earliest stages of yeast endocytic site formation to include Yap1802p and the previously uncharacterized protein Pal1p. On the basis of their patch lifetimes and dynamics, Pal1p and Yap1802p have been classified as components of the early coat module.
Insight into the earliest stages of endocytosis was gained by defining which proteins are first detected at endocytic sites. We find that Ede1p, Syp1p, clathrin light chain, the AP2 complex, Yap1802p, and Pal1p appear at endocytic sites at about the same time. It is possible that a signal at the plasma membrane simultaneously recruits the early-arriving endocytic proteins and cooperative binding aids their stabilization at endocytic sites. It is also possible that some early-arriving endocytic proteins physically interact in the cytosol prior to associating with the plasma membrane, which has been observed for other endocytic proteins (Lundmark and Carlsson, 2004). Because the homologues of Ede1p and Syp1p (Eps15 and FCHo1/2, respectively) are also among the first proteins to arrive during CCP formation in mammalian cells (Henne et al., 2010), further investigation of the initial steps of endocytosis in both systems might reveal additional similarities.
Role of the early-arriving proteins in endocytosis
This work provided insight into how the early-arriving proteins contribute to endocytic site formation. PAL1 deletion mutants did not have detectable endocytic defects, which is consistent with the observation that many of the early-arriving proteins function in a cargo-specific manner (Burston et al., 2009; Carroll et al., 2009; Reider et al., 2009; Stimpson et al., 2009). Pal1p may also be a cargo-specific adaptor or it could act redundantly with other endocytic proteins.
Ede1p was hypothesized to be involved in endocytic site establishment because it affects the formation of productive endocytic sites (Kaksonen et al., 2005; Stimpson et al., 2009). However, previous studies only examined the dynamics of later-arriving proteins and therefore did not reveal the role of Ede1p in early stages of site formation. We reported here that clathrin light chain, the AP2 complex, and Pal1p do not stably associate with endocytic sites in ede1Δ yeast. In contrast, Yap1802p was recruited to sites in the absence of Ede1p, and Yap1802p alone was unable to recruit and stabilize the other early-arriving proteins at endocytic sites. Yap1802p might localize to endocytic sites in the absence of Ede1p because it has an AP180 N-terminal homology domain (Legendre-Guillemin et al., 2004; Maldonado-Baez et al., 2008), which may bind PtdIns(4,5)P2 on the plasma membrane. Conflicting reports exist over whether Syp1p is recruited to cortical patches in ede1Δ yeast (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). It is unclear why some early-arriving endocytic proteins are able to form patches in ede1Δ yeast, but our results confirm that Ede1p is an important factor in normal endocytic site initiation. Although knockdown of Eps15 in mammalian cells increased productive CCP lifetimes (Mettlen et al., 2009), which is a different phenotype than that observed in ede1Δ yeast, results from these experiments suggest that Eps15 and Ede1p are both important components of the endocytic machinery.
We find that all of the early-arriving endocytic proteins are able to form cortical patches in clc1Δ yeast. Clathrin light chain has been proposed to promote yeast endocytic site progression, possibly through its interaction with Sla2p's coiled-coil domain (Newpher and Lemmon, 2006; Newpher et al., 2006). This study confirms that clathrin light chain is important for site maturation and disassembly, and we find that it specifically regulates the transition from the intermediate to late coat stages (Figure 2). In mammalian cells, knockdown of the clathrin light chain b isoform increased the lifetimes of late abortive CCPs and decreased the lifetimes of productive CCPs (Mettlen et al., 2009). It is possible that yeast Clc1p may be more similar to the mammalian clathrin light chain b isoform because there is an increase in early endocytic protein lifetimes, as well as a decrease in late coat module protein lifetimes, in clc1Δ yeast. Although clathrin light chain is believed to be important for efficient heavy chain trimerization (Chu et al., 1996), it is important to note that small amounts of clathrin heavy chain may be present and functioning at endocytic sites in clc1Δ yeast. More research is needed to elucidate the roles of the different clathrin subunits in endocytic site regulation.
Comparing the phenotypes of clc1Δ and ede1Δ yeast reveals that the lack of clathrin at endocytic sites in ede1Δ yeast does not result in the same phenotypes observed in clc1Δ yeast. For example, we find that Sla2p and Ent2p form more persistent patches in clc1Δ yeast, but the lifetimes of these proteins are shortened in ede1Δ yeast (Stimpson et al., 2009). It is possible that small amounts of clathrin might localize to and promote maturation of endocytic sites in ede1Δ yeast. Alternatively, endocytic proteins that normally inhibit endocytic site maturation might not be recruited to endocytic sites, or they might be regulated differently, in ede1Δ yeast.
Formation of an intermediate coat module
Sla2p, Ent1p, and Ent2p arrive at endocytic sites before Sla1p, and ent1Δ ent2Δ yeast have similar phenotypes to sla2Δ yeast. These results have now led us to propose the existence of an intermediate coat module separate from the late coat module containing the Pan1 complex (Pan1p, Sla1p, and End3p; Figure 2; Kaksonen et al., 2003). The intermediate coat module may serve to link the early proteins involved in endocytic site formation to the later-arriving proteins that drive membrane invagination. Sla2p has been hypothesized to connect the force of actin polymerization to membrane invagination (Kaksonen et al., 2006), so Ent1p and Ent2p may also function in this step of endocytosis.
Secretion and cargo may regulate progression of endocytic events
Previous studies found that several secretion mutants have defects in endocytosis (Riezman, 1985; Hicke et al., 1997), but how the two cellular processes are connected is not understood. We now find that endocytic sites are formed in sec18-1ts yeast, which contain the early-arriving and intermediate coat module proteins, but the sites do not progress to completion. We speculate that when cargoes are depleted from the yeast cell surface in a secretion mutant or by the addition of cycloheximide, the lack of cargo may activate a checkpoint, preventing the endocytic site from maturing past the intermediate coat stage (Figure 2). In further support of this model, we found that Ede1p lifetimes are shorter in yeast buds, which contain higher concentrations of cargo than mother cells (Layton et al., 2011). This model is also consistent with the observation that cargo normally arrives during the early stages of endocytosis (Toshima et al., 2006).
Depletion of endocytic cargo may not be the only reason why endocytosis is defective in sec18-1ts yeast. Adding glutamate to induce internalization of the cargo Gap1p in sec18-1ts yeast did not fully rescue the endocytic defects, suggesting that other factors may be involved in creating this phenotype. Although the distribution of PtdIns(4,5)P2 did not seem to change in sec18-1ts yeast, the lipid environment at the plasma membrane may be altered in secretion mutants (Yakir-Tamang and Gerst, 2009). Trafficking is perturbed in secretion mutants, and important machinery needed for endocytosis might not be recycled or delivered to the plasma membrane. It is also possible that in yeast exocytic events are linked to endocytic events directly. In fact, in mammalian cells compensatory endocytosis follows an exocytic event in some cells, and mechanisms linking exocytosis and endocytosis have also been proposed (Gundelfinger et al., 2003; Yao et al., 2009; Bai et al., 2010; Pechstein et al., 2010). Another interesting factor to consider is that signaling mechanisms could be contributing to the partial rescue of the endocytic defects observed in sec18-1ts yeast when Gap1p internalization is induced. For example, the addition of glutamate may cause the cargo Gap1p to activate a signaling pathway that alters the dynamics of endocytic proteins.
Studies in mammalian cells suggested that cargoes, along with other factors, regulate a checkpoint in clathrin-mediated endocytosis (Ehrlich et al., 2004; Loerke et al., 2009; Mettlen et al., 2009; Mettlen et al., 2010). For example, nascent CCPs may abort if cargoes have not been captured, and the capture of certain cargo/adaptor complexes can increase efficiency of CCP maturation (Ehrlich et al., 2004; Loerke et al., 2009; Mettlen et al., 2009). A slightly different mechanism may exist in yeast, in which endocytic sites may arrest at the intermediate coat stage if cargo and/or clathrin are not available to promote progression through an endocytic restriction point. In mammalian cells, different cargo proteins use different endocytic adaptors, and these factors may contribute to the heterogeneity in CCP dynamics and progression through the proposed endocytic checkpoint (Loerke et al., 2009; Mettlen et al., 2010). Furthermore, the clustering of transferrin receptors has been shown to increase CCP initiation (Liu et al., 2010). Perhaps future studies will establish whether other endocytic cargoes in yeast have regulatory capabilities and whether their different adaptor proteins are involved in endocytic site maturation. The results presented here increase our overall understanding of endocytic sites as we continue to discover similarities between yeast and mammalian endocytic machinery.
MATERIALS AND METHODS
Strains
Yeast strains used in this study are listed in Supplemental Table S1. Single C-terminal GFP and 13Myc tags were integrated into the chromosome as previously described (Longtine et al., 1998). C-terminal 3XGFP tags were created as previously described (Sun et al., 2007). Gene deletions were generated by replacing the gene open reading frame with Candida glabrata LEU2, URA3, or HIS3 cassettes. Yeast expressing GFP- and RFP-tagged Snc1p were transformed with the plasmids pRS416-GFP-SNC1 and pRS416-RFP-SNC1. Yeast were transformed with the plasmid pRS426-GFP-2XPH(PLCδ) to monitor PtdIns(4,5)P2 levels (Stefan et al., 2002).
Coimmunoprecipitation and immunoblotting
Coimmunoprecipitation was performed essentially as described previously (Peng and Weisman, 2008). Briefly, cell lysates were incubated with mouse anti-Myc antibody for 2 h and then incubated with protein G–Sepharose beads (GE Healthcare, Piscataway, NJ) for 2 h. The beads were washed with lysis buffer containing 10% glycerol, and then the proteins were eluted. Coimmunoprecipitation was assayed by immunoblot. To detect Pal1-GFP, the membrane was probed with 1:2000 rabbit anti-GFP antibody (Torrey Pines Biolabs, Secaucus, NJ). To detect Ede1-13Myc, the membrane was probed with 1:4000 mouse anti–Myc 9E10.
Microscopy
Yeast strains used for imaging were grown to log phase at 25°C in synthetic media lacking tryptophan (imaging media) and immobilized on concanavalin A–coated coverslips. The clc1Δ yeast and ent1Δ ent2Δ yeast strains were maintained as heterozygous diploids, which were sporulated and dissected before use. The spores were grown overnight in yeast extract/peptone/dextrose (YPD) and were then grown for 4 h in imaging media prior to imaging. Because sla2Δ yeast have growth defects, this strain was also grown overnight in YPD and then grown for 4 h in imaging media. The sec18-1ts and sec1-1ts yeast were grown overnight at 25°C and then incubated at 37°C for the time indicated and imaged in a 37°C temperature-controlled chamber. Yeast expressing Gap1-RFP were grown in synthetic media composed of yeast nitrogen base without amino acids and without ammonium sulfate (Difco, BD Biosciences, Franklin Lakes, NJ), 2% glucose, and 0.5% ammonium sulfate. The yeast were then incubated at 37°C for the time indicated before imaging. To induce Gap1-RFP uptake, the media was replaced with prewarmed imaging media supplemented with 0.1% glutamate (wt/vol) for the time indicated.
Olympus IX71 and IX81 microscopes with 100×/numerical aperture (NA) 1.4 objectives, Orca cameras (Hamamatsu, Hamamatsu, Japan), appropriate filter sets, and neutral density filters were used to image live yeast cells. Simultaneous two-color imaging was performed as described previously using a 488-nm argon-ion laser (CVI Melles Griot, Albuquerque, NM) to excite GFP and either a mercury lamp filtered through a 575/20-nm filter (Figures 1, C and D, 3D, and 4, C and E) or a 561-nm argon-ion laser (CVI Melles Griot; Figure 1F) to excite RFP or mCherry (Stimpson et al., 2009). TIRF microscopy was performed using an IX81 microscope with a 100×/1.65 NA objective and an adjustable angle laser beam, which was lowered to reduce background signal. Two-color TIRF microscopy in Figure 1F was performed by lowering the angle of both the 488- and 561-nm laser beams separately to reduce background signal. Movies were acquired at a rate of one frame every 2 s, unless indicated otherwise. Images were collected using MetaMorph software (Molecular Devices, Sunnyvale, CA) and processed using ImageJ (National Institutes of Health, Bethesda, MD). Patch lifetimes were calculated from patches that assembled and disassembled during the movie (turnover patches). Patch number was counted from maximum-intensity Z-projections of unbudded or large-budded cells. Z-Stacks were acquired through the entire cell at 0.15-μm intervals. Gap1-RFP fluorescence intensity was calculated by averaging the pixel intensity at four locations around the cell cortex.
FM 4-64, fluorescent α-factor, and Lucifer yellow uptake assays
For FM 4-64 staining, cells were incubated with 8 μM FM 4-64 in imaging media for the time indicated and then imaged. HiLyte Fluor 488 α-factor (AnaSpec, Fremont, CA) uptake experiments were performed essentially as described previously (Toshima et al., 2006). Cells were grown to log phase in imaging media. The cells were pelleted and then resuspended to ∼40 OD/ml in imaging media and incubated with 200 ng of fluorescent α-factor on ice for 1 h. The cells were washed three times in ice-cold imaging media, warmed to room temperature, and imaged after the time indicated. Lucifer yellow uptake assays were performed as described previously (Baggett et al., 2003), but with some modifications. Cells were first grown to early log phase in YPD. Approximately 1 × 107 cells were pelleted and then resuspended in 90 μl of YPD. After incubation of the cells for 90 min at 37°C, 10 μl of 40 mg/ml Lucifer yellow CH dilithium salt (Invitrogen, Carlsbad, CA) was added. Cells were incubated for 90 min at 37°C and then washed four times in ice-cold 50 mM potassium phosphate buffer, pH 7.4, containing 10 mM NaN3 and 10 mM NaF. Cells were imaged in the wash buffer at room temperature.
Acknowledgments
We thank Randy Schekman, Aaron Cheng, Voytek Okreglak, Sandra Lemmon, Beverly Wendland, and lab members for providing strains, engaging in helpful discussions, and/or offering critical readings of the manuscript. D.G.D. acknowledges funding from the National Institutes of Health (GM50349).
Abbreviations used:
- CCP
clathrin-coated pit
- FCHo1/2
Fer/Cip4 homology domain–only proteins 1 and 2
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0108) on December 21, 2011.
REFERENCES
- Aguilar RC, et al. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc Natl Acad Sci USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggett JJ, Shaw JD, Sciambi CJ, Watson HA, Wendland B. Fluorescent labeling of yeast. Curr Protoc Cell Biol. 2003;Chapter 14 doi: 10.1002/0471143030.cb0413s20. Unit 4.13. [DOI] [PubMed] [Google Scholar]
- Bai J, Hu Z, Dittman JS, Pym EC, Kaplan JM. Endophilin functions as a membrane-bending molecule and is delivered to endocytic zones by exocytosis. Cell. 2010;143:430–441. doi: 10.1016/j.cell.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker SL, Lee L, Pierce BD, Maldonado-Baez L, Drubin DG, Wendland B. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol Biol Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, D'Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol. 2009;185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SY, Stirling PC, Stimpson HE, Giesselmann E, Schmitt MJ, Drubin DG. A yeast killer toxin screen provides insights into a/b toxin entry, trafficking, and killing mechanisms. Dev Cell. 2009;17:552–560. doi: 10.1016/j.devcel.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DS, Pishvaee B, Payne GS. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J Biol Chem. 1996;271:33123–33130. doi: 10.1074/jbc.271.51.33123. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ge W, Chew TG, Wachtler V, Naqvi SN, Balasubramanian MK. The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis. Mol Biol Cell. 2005;16:4124–4138. doi: 10.1091/mbc.E04-11-0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger ED, Kessels MM, Qualmann B. Temporal and spatial coordination of exocytosis and endocytosis. Nat Rev Mol Cell Biol. 2003;4:127–139. doi: 10.1038/nrm1016. [DOI] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Pypaert M, Rohrer J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KM, D'Hondt K, Riezman H, Lemmon SK. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–87. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Keyel PA, Watkins SC, Traub LM. Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:13190–13204. doi: 10.1074/jbc.M312717200. [DOI] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–25. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Liu AP, Aguet F, Danuser G, Schmid SL. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J Cell Biol. 2010;191:1381–1393. doi: 10.1083/jcb.201008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J Biol Chem. 2004;279:42694–42702. doi: 10.1074/jbc.M407430200. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Dores MR, Perkins EM, Drivas TG, Hicke L, Wendland B. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Mettlen M, Loerke D, Yarar D, Danuser G, Schmid SL. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J Cell Biol. 2010;188:919–933. doi: 10.1083/jcb.200908078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Stoeber M, Loerke D, Antonescu CN, Danuser G, Schmid SL. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol Biol Cell. 2009;20:3251–3260. doi: 10.1091/mbc.E09-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17:4343–4352. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Lemmon SK. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Pechstein A, Shupliakov O, Haucke V. Intersectin 1: a versatile actor in the synaptic vesicle cycle. Biochem Soc Trans. 2010;38:181–186. doi: 10.1042/BST0380181. [DOI] [PubMed] [Google Scholar]
- Peng Y, Weisman LS. The cyclin-dependent kinase Cdk1 directly regulates vacuole inheritance. Dev Cell. 2008;15:478–485. doi: 10.1016/j.devcel.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkett CJ, Morris JA, Wood V, Bahler J. YOGY: a Web-based, integrated database to retrieve protein orthologs and associated Gene Ontology terms. Nucleic Acids Res. 2006;34:W330–W334. doi: 10.1093/nar/gkl311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Rappoport JZ, Benmerah A, Simon SM. Analysis of the AP-2 adaptor complex and cargo during clathrin-mediated endocytosis. Traffic. 2005;6:539–547. doi: 10.1111/j.1600-0854.2005.00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport JZ, Simon SM. Real-time analysis of clathrin-mediated endocytosis during cell migration. J Cell Sci. 2003;116:847–855. doi: 10.1242/jcs.00289. [DOI] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Roberg KJ, Rowley N, Kaiser CA. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczynska-de R II, Allwood EG, Aghamohammadzadeh S, Hettema EH, Goldberg MW, Ayscough KR. A role for the dynamin-like protein Vps1 during endocytosis in yeast. J Cell Sci. 2010;123:3496–3506. doi: 10.1242/jcs.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetens O, De Craene JO, Andre B. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J Biol Chem. 2001;276:43949–43957. doi: 10.1074/jbc.M102945200. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir-Tamang L, Gerst JE. A phosphatidylinositol-transfer protein and phosphatidylinositol-4-phosphate 5-kinase control Cdc42 to regulate the actin cytoskeleton and secretory pathway in yeast. Mol Biol Cell. 2009;20:3583–3597. doi: 10.1091/mbc.E08-10-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CK, Lin YQ, Ly CV, Ohyama T, Haueter CM, Moiseenkova-Bell VY, Wensel TG, Bellen HJ. A synaptic vesicle-associated Ca2+ channel promotes endocytosis and couples exocytosis to endocytosis. Cell. 2009;138:947–960. doi: 10.1016/j.cell.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]