ETOC: The EH domain is a highly conserved protein–protein interaction domain involved in endocytosis. The EH domains of yeast endocytic proteins, Pan1p, End3p, and Ede1p, have a redundant function and are required for efficient recruitment of several endocytic proteins to sites of endocytosis in order to facilitate clathrin coat assembly.
Abstract
Clathrin-mediated endocytosis involves a coordinated series of molecular events regulated by interactions among a variety of proteins and lipids through specific domains. One such domain is the Eps15 homology (EH) domain, a highly conserved protein–protein interaction domain present in a number of proteins distributed from yeast to mammals. Several lines of evidence suggest that the yeast EH domain–containing proteins Pan1p, End3p, and Ede1p play important roles during endocytosis. Although genetic and cell-biological studies of these proteins suggested a role for the EH domains in clathrin-mediated endocytosis, it was unclear how they regulate clathrin coat assembly. To explore the role of the EH domain in yeast endocytosis, we mutated those of Pan1p, End3p, or Ede1p, respectively, and examined the effects of single, double, or triple mutation on clathrin coat assembly. We found that mutations of the EH domain caused a defect of cargo internalization and a delay of clathrin coat assembly but had no effect on assembly of the actin patch. We also demonstrated functional redundancy among the EH domains of Pan1p, End3p, and Ede1p for endocytosis. Of interest, the dynamics of several endocytic proteins were differentially affected by various EH domain mutations, suggesting functional diversity of each EH domain.
INTRODUCTION
Clathrin-mediated endocytosis is a coordinated series of molecular events, including cargo loading, formation and invagination of coated pits, and vesicle formation (Geli and Riezman, 1998; Engqvist-Goldstein and Drubin, 2003; Sorkin, 2004; Toret and Drubin, 2006). Recent live-cell imaging studies revealed the detailed timing of protein recruitment to sites of clathrin-mediated endocytosis in budding yeast and mammalian cells (Merrifield et al., 2002; Kaksonen et al., 2003, 2005; Ehrlich et al., 2004; Newpher et al., 2005). In yeast, Syp1p, a SGIP1-α–related protein, and Ede1p, a Eps15-related protein, are the earliest proteins known to arrive at sites of endocytosis (Kaksonen et al., 2005; Toshima et al., 2006; Stimpson et al., 2009). Clathrin is also considered to be part of the early coat module, but the precise timing of its recruitment at sites of endocytosis has not been determined because a large proportion of clathrin is localized to highly dynamic intracellular compartments, including the trans-Golgi network, and its cortical localization is masked by a strong internal signal (Kaksonen et al., 2005; Newpher et al., 2005). Ent1/2p and Yap1801/2p, which are related to the mammalian clathrin adaptors epsin and AP180/CALM, respectively, have both clathrin- and phosphatidylinositol 4,5-bisphosphate–binding domains and may function to recruit clathrin to sites of endocytosis (Wendland and Emr, 1998; Wendland et al., 1999; Newpher et al., 2005). One to two minutes after Syp1p and Ede1p arrive at such sites, the late coat module, including the Pan1p complex, appears and couples coat formation to actin polymerization in cooperation with Las17p, a yeast WASP, and Myo5p (Kaksonen et al., 2003; Sun et al., 2006).
These coordinated endocytic events are regulated by interactions among a variety of proteins and lipids through specific domains (Engqvist-Goldstein and Drubin, 2003). One such domain is the Eps15 homology domain (EH domain), which is an evolutionarily conserved protein–protein interaction domain present in a number of proteins distributed from yeast to mammals (Di Fiore et al., 1997; Santolini et al., 1999). The EH domain was originally identified as a domain comprising repeats of ∼100 amino acids in the N-terminal region of Eps15, which is a binding partner of α-adaptin, a component of the AP-2 complex and part of the clathrin-coated pit (Benmerah et al., 1995, 1996; van Delft et al., 1997). Several experiments, including phage-display screens and screening of a human fibroblast expression library, demonstrated that the EH domain binds to peptides containing the Asn-Pro-Phe (NPF) sequence (Salcini et al., 1997; Paoluzi et al., 1998). Structural analyses demonstrated that NPF residues are almost completely embedded in a conserved hydrophobic pocket within the EH domain, allowing close contact between the asparagine residue of the tripeptide and a highly conserved tryptophan residue in the EH domain (de Beer et al., 1998, 2000). Mutation of this conserved tryptophan residue dramatically impairs binding of the EH domains to NPF motifs, and the mechanism of EH domain/NPF motif interaction is likely conserved among most EH domains (Salcini et al., 1997; de Beer et al., 1998; Grant and Caplan, 2008).
In yeast, three EH domain–containing proteins (EHDPs)—Pan1p, End3p and Ede1p—have been reported to play important roles during endocytosis. Pan1p is another yeast Eps15-related protein. It contains two EH domains and forms a stable complex with End3p, which also has two EH domains (Raths et al., 1993; Benedetti et al., 1994; Tang et al., 1997; Wendland and Emr, 1998; Toshima et al., 2007). Previous studies showed that Pan1p and Ede1p bind to NPF motifs of the clathrin adaptors Yap1801/2p and/or Ent1/2p and that the dynamics of Pan1p and Ede1p depends on these interactions (Wendland and Emr, 1998; Howard et al., 2002; Aguilar et al., 2003; Maldonado-Baez et al., 2008). These observations suggest that the EH domain/NPF interaction between Eps15-related proteins and clathrin adaptors is conserved from yeast to mammals. Although genetic and cell-biological studies of EHDPs suggest a role for the EH domain in clathrin-mediated endocytosis, functional differences among the EH domains of Pan1p, End3p, and Ede1p and how these EHDPs regulate clathrin coat assembly are not fully understood.
In the present study we find that the EH domains of Pan1p, End3p, and Ede1p function redundantly for efficient assembly of the clathrin coat. By live-cell imaging of cortical patch dynamics, we demonstrate that the patch lifetimes of Pan1p, End3p, and Ede1p are increased when their EH domains are mutated. We also demonstrate that mutations of the EH domains cause a defect of cargo internalization and delay assembly of the clathrin coat without affecting assembly of the actin patch. Furthermore, the EH domains of Pan1p, End3p, and Ede1p are required for proper localization of the endocytic coat proteins Sla1p and Yap1801p, suggesting that the EH domains target these proteins to endocytic sites in order to facilitate clathrin coat assembly.
RESULTS
Mutations in the EH domains of Pan1p, End3p, and Ede1p increase their lifetime at cortical patches
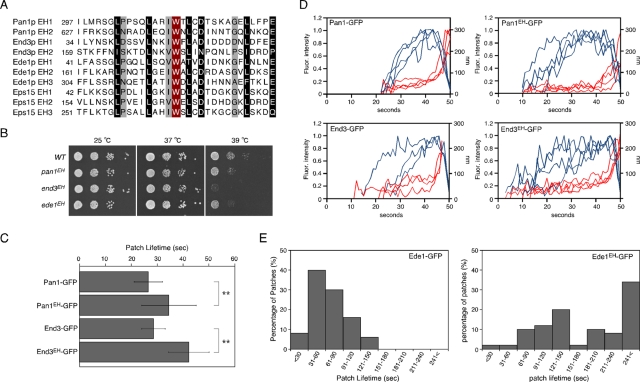
Several lines of evidence indicate that the yeast EHDPs Pan1p, End3p and Ede1p play important roles during endocytosis (Miliaras and Wendland, 2004). The EH domain is frequently present as tandem repeats within a single protein (Santolini et al., 1999; Grant and Caplan, 2008), and Pan1p, End3p, and Ede1p contain two or three EH domains, respectively, in their N-terminal regions (Figure 1A). Previous studies showed that mutation of a highly conserved tryptophan residue in the EH domain impairs its ability to bind to the NPF motif (Salcini et al., 1997; de Beer et al., 2000). To explore in detail the role of the EH domain in yeast endocytosis, we mutated all of the conserved tryptophan residues in the EH domains of Pan1p, End3p, or Ede1p to alanine and integrated them into their endogenous loci (termed pan1EH, end3EH, and ede1EH, respectively). Although pan1EH and ede1EH cells were viable at both 37 and 39°C, end3EH cells were temperature sensitive for growth at 39°C (Figure 1B). Because the second EH domain of End3p has only limited similarity and it is unclear whether it really functions as an EH domain (Santolini et al., 1999), we created additional mutants that were mutated in only the first or second EH domain of End3p. We found that both of the mutants were temperature sensitive for growth at 39°C (Supplemental Figure S1A), suggesting that the second EH domain might be important for the function of End3p. To examine the effects of EH domain mutation on EHDP dynamics, we tagged Pan1p, End3p, and Ede1p with green fluorescent protein (GFP) and examined the resulting effects. The GFP-tagged EH domain mutants were expressed at similar levels to the wild-type proteins (Supplemental Figure S1B). All of the GFP-tagged mutants were localized to cortical patches (Supplemental Figure S1, C–E), suggesting that the EH domain/NPF motif interactions are unnecessary for primary targeting of the EHDPs to cortical patches. Consistent with previous reports, Pan1-GFP and End3-GFP patches formed at the cell cortex with lifetimes of 26.5 ± 5.5 and 28.6 ± 4.5 s, respectively, culminating in inward movement (Figure 1C; Kaksonen et al., 2005). Pan1EH-GFP and End3EH-GFP patches formed and disappeared with the typical inward movement, but their lifetimes were increased to 34.6 ± 10.5 and 42.4 ± 7.8 s, respectively (Figure 1C). Tracking of individual patches showed that the distance and timing of the inward movement of Pan1EH-GFP or End3EH-GFP patches was almost the same as that of the wild-type proteins but that the time required to reach the maximum fluorescence intensity was increased (Figure 1D and Supplemental Figure S2), suggesting that mutation in EH domains might causes a delay in accumulation of EHDPs to endocytic sites. Ede1p is an early-arriving endocytic protein that appears at the cell cortex before Sla1p and has a lifetime ranging widely from ∼30 to 150 s (Figure 1E; Toshima et al., 2006; Stimpson et al., 2009). Similar to Pan1p and End3p patches, Ede1EH-GFP patches had a significantly increased lifetime, and ∼34% of them remained stationary at the plasma membrane for >240 s (Figure 1E). The EH domain therefore seems to be important for efficient recruitment of EHDPs to sites of endocytosis.
FIGURE 1:
Generation and characterization of EH mutants in Pan1p, Ede1p, and End3p. (A) Sequence alignments of the EH domains in Pan1p, End3p, Ede1p, and mouse Eps15. Residues chosen for mutagenesis are highlighted in red. Residues conserved highly and moderately throughout the family are indicated in black and gray, respectively. (B) The end3EH mutant was temperature sensitive for growth at 39°C. A dilution series of cells was plated on YPD medium and incubated at 25, 37, or 39°C. (C) Average lifetimes of Pan1-GFP and End3-GFP ± SD in wild-type and mutant cells. Data were taken from 2-min movies with a 1-s frame interval. n = 50 patches for each strain. **p < 0.001. (D) Quantification of fluorescence intensity (red) and distance from site of patch formation (blue) as a function of time for patches of indicated GFP-tagged proteins. Each curve represents data from one patch. Behavior of three independent patches was plotted for each strain. (E) Distribution of Ede1-GFP and Ede1EH-GFP patch lifetimes. Movies were taken with a 1-s frame interval. n = 50 patches for each strain.
Dynamics of the Sla1p patch and actin patch in EH domain mutants
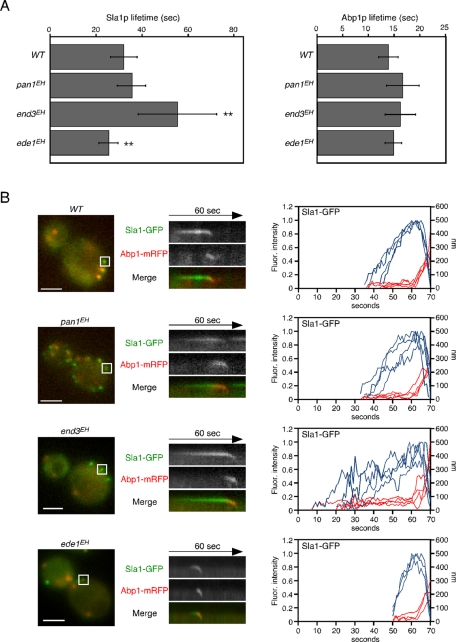
We next examined whether the dynamics of the late coat module and actin patch were affected in the EH domain mutants. We used Sla1-GFP and Abp1–monomeric red fluorescent protein (mRFP) as markers to follow the dynamics of the late coat module and actin patch, respectively (Kaksonen et al., 2003). First, we measured the mean lifetimes of both Sla1-GFP and Abp1-mRFP in each mutant. Of interest, the lifetimes of Sla1-GFP patches were differentially affected by different EH domain mutants. In pan1EH cells, Sla1-GFP patches had an average lifetime of 35.9 ± 6.0 s, similar to Sla1-GFP patches in wild-type cells (32.3 ± 5.8 s; Figure 2A). In contrast, the lifetime of Sla1-GFP patches was significantly increased in end3EH cells (55.7 ± 17.1 s) and slightly decreased in ede1EH cells (25.7 ± 4.1 s; Figure 2A). In contrast, all the mutants showed Abp1-mRFP lifetimes similar to that of wild-type cells (Figure 2A). Next we carried out simultaneous imaging of Sla1p and Abp1p to analyze the dynamic behavior of these proteins in live cells. In wild-type cells, Sla1-GFP was immediately followed by a burst of Abp1-mRFP recruitment, as shown in kymographs generated across a single pixel-wide line of an individual patch (see boxed area in Figure 2B). In all mutants, similar dynamics of Sla1-GFP and Abp1-mRFP patches were observed, except that the time required to reach the maximum fluorescence intensity was increased in end3EH mutants or decreased in ede1EH mutants (Figure 2B). Taken together, these data indicate that the EH domain is required for regulation of Sla1p coat assembly but not for actin-driven membrane invagination and vesicle internalization.
FIGURE 2:
Dynamics of the Sla1p patch and actin patch in EH domain mutants. (A) Average lifetimes of Sla1-GFP (left) and Abp1-mRFP (right) ± SD for indicated strains. Data were taken from 2-min movies with a 1-s frame interval. n = 50 patches for each strain. **p < 0.001. (B) Left, localizations of Sla1-GFP and Abp1-mRFP in live cells. Middle, kymograph representations of Sla1-GFP and Abp1-mRFP from the boxed area of strains as indicated. Right, quantification of fluorescence intensity (blue) and distance from the site of patch formation (red) as a function of time for patches of Sla1-GFP. Each curve represents data from one patch. Behavior of three independent patches was plotted for each strain. All movies were taken with a 1-s frame interval for both Sla1-GFP and Abp1-mRFP. The data in A were imaged as single-color movies using optimal GFP filter sets, whereas those in B were imaged as two-color simultaneous movies using narrow band-pass filter sets to completely separate GFP and mRFP fluorescence. Scale bars, 2 μm.
To clarify whether the NPF-binding pocket is responsible for most of the functions of the EH domain or whether the EH domain has other functionally important interactions, such as lipid binding (Naslavsky et al., 2007), outside the NPF-binding pocket, we deleted the entire EH domains of Pan1p, End3p, and Ede1p (termed pan1ΔEH, end3ΔEH, and ede1ΔEH, respectively) and tested the phenotypic consequences in these mutants. All of the EH deletion mutants were localized to cortical patches, although parts of End3ΔEH-GFP formed a large aggregate in the cytosol, confirming that the EH domains are unnecessary for primary targeting of the EHDPs to cortical patches (Supplemental Figure S3A). The ede1ΔEH mutants exhibited almost the same phenotype as the ede1EH mutant in terms of growth, localization, and Sla1p and Abp1p dynamics (Supplemental Figure S3, B, D–F). Similar to the pan1EH mutant, the pan1ΔEH mutant also showed no marked phenotype, except that lifetimes of Pan1p and Sla1p were slightly decreased compared with wild-type cell (Supplemental Figure S3, B, D–F). The NPF-binding pocket therefore seems to be responsible for the major EH domain functions in Pan1p and Ede1p. In contrast, in comparison to the end3EH mutant, the end3ΔEH mutant exhibited much more severe phenotypic features, which resembled those of the end3-null mutant shown previously (Supplemental Figure S3, D–F; Kaksonen et al., 2005). Although Pan1p and Ede1p are relatively large proteins composed of 1480 and 1381 amino acids, respectively, and contain several domain structures, End3p is a small protein (349 amino acids) in which more than half of the structure is occupied by the EH domains. Thus the function of End3p besides the EH domain might be disrupted in the end3ΔEH mutant because of structural instability. In addition to the end3ΔEH mutant, we also examined the phenotype of another end3 mutant, end3 K47E/172E, in which two lysine residues that are potential sites for lipid binding (Naslavsky et al., 2007) are mutated. The end3 K47E/172E mutant exhibited almost the same phenotype as wild-type cells (Supplemental Figure S3, B and E–G), indicating that these sites are not critical for the function of End3.
Functional redundancy of EH domains for endocytic patch formation
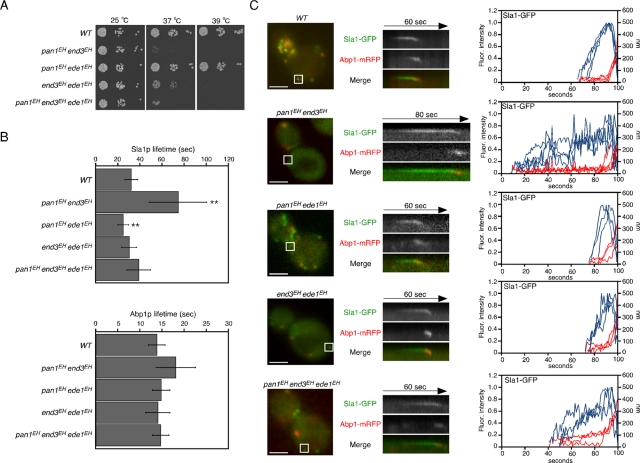
To investigate functional redundancy among EHDPs, we analyzed the effect of double or triple mutants on the dynamics of Sla1p or Abp1p. Although pan1EH end3EH cells were temperature sensitive for growth at 37°C, the growth of pan1EH ede1EH cells was almost the same as that of wild-type cells (Figure 3A). In addition, the lifetime of Sla1p patches was markedly increased in pan1EH end3EH cells, whereas the lifetime of Abp1p patches was little affected (Figure 3B). Because pan1EH cells have no apparent defects in growth or Sla1p patch lifetime, the function of the EH domains of Pan1p is probably substituted by those of End3p. It is intriguing that the lifetimes of Sla1p patches were decreased in double mutants, including the ede1EH mutation, even in end3EH cells, which showed a markedly increased Sla1p lifetime (Figures 2A and 3B), indicating that the EH domains of Ede1p have a dominant function in determining the lifetime of Sla1p patches. As shown in kymographs and particle-tracking analysis, inward movement of Sla1p appeared to be normal, but the times required to reach the maximum fluorescence intensity were changed in these double or triple mutants, similar to single mutants (Figure 3C).
FIGURE 3:
Redundant function of EH mutants in endocytic patch formation. (A) The pan1EH end3EH and triple mutants were temperature sensitive for growth at 37°C. A dilution series of cells was plated on YPD medium and incubated at the indicated temperature. (B) Patch lifetimes of Sla1-GFP (top) and Abp1-mRFP (bottom) ± SD in the indicated strains. Data were obtained from 2-min movies taken with a 1-s frame interval. n = 50 patches for each strain. **p < 0.001. (C) Left, localization of Sla1-GFP and Abp1-mRFP from the strains indicated. Middle, kymograph representations of Sla1-GFP and Abp1-mRFP from the boxed area for the indicated times. Right, quantification of fluorescence intensity (blue) and distance from site of patch formation (red) as a function of time for patches of Sla1-GFP. Each curve represents data from one patch. Behavior of three independent patches was plotted for each strain. All movies were taken with a 1-s frame interval for both Sla1-GFP and Abp1-mRFP. Scale bars, 2 μm.
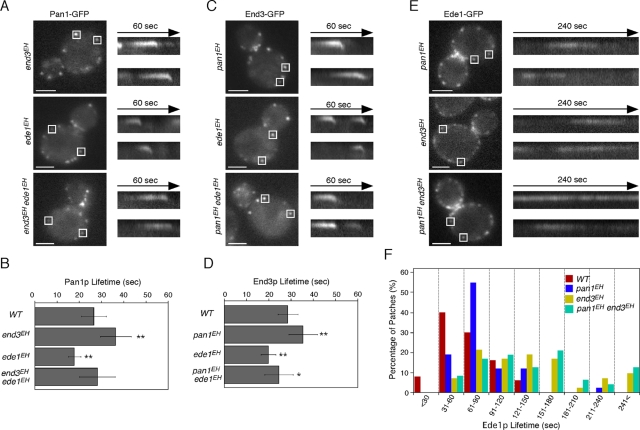
We next examined the dynamics of nonmutated Pan1p, End3p, and Ede1p in cells expressing mutant forms of the other EHDPs. Of interest, the dynamics of Pan1p and End3p were quite similar to those of Sla1p: the Pan1p and End3p patch lifetimes were increased in the end3EH or pan1EH mutants and decreased in the ede1EHmutant (Figure 4, A–D). This suggests that Pan1p and End3p are able to form a complex with Sla1p, components of the late endocytic coat module, and act together as reported previously (Tang et al., 2000), even though the functions of the EH domains are disrupted. Ede1p patch lifetimes were increased in all of the pan1EH, end3EH, and pan1EH end3EH mutants (Figure 4, E and F).
FIGURE 4:
Localization of EH domain proteins in EH domain mutants. (A, C, E) Localization of Pan1-GFP (A), End3-GFP (C), and Ede1-GFP (E) in cells expressing mutant forms of other EH domain proteins. Right, kymographs from the same movies. (B, D) Average lifetimes of Pan1-GFP and End3-GFP ± SD in mutant cells. Data were taken from 2-min movies with a 1-s frame interval. n = 50 patches for each strain. *p < 0.01, **p < 0.001. Scale bars, 2 μm. (F) Distribution of Ede1-GFP patch lifetimes in wild-type and indicated mutant cells. n = 50 patches for each strain. Movies were taken with a 1-s frame interval.
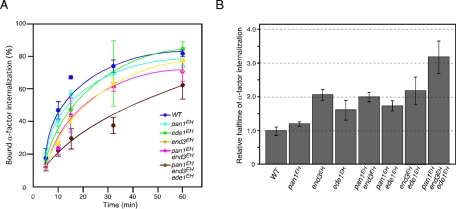
We further examined the effect on endocytic internalization by assessing the internalization of35S-labeled α-factor. end3EH cells exhibited a modest defect of α-factor internalization, whereas pan1EH or ede1EH cells had only a slight defect (Figure 5A), consistent with the finding that end3EH cells had a more severe phenotype in terms of growth and Sla1-GFP dynamics (Figures 1B and 2A). The relative α-factor internalization rates of the mutants are compared in the histogram shown in Figure 5B. Unexpectedly, combination of the end3EH and pan1EH mutations had no additive effect, although they exhibited a synthetic growth defect (Figures 3A and 5B). The end3EH mutant also had little additive effect on α-factor internalization when combined with ede1EH cells but a remarkable effect when combined with pan1EH ede1EH cells (triple mutant; Figure 5B). This result suggested that pan1EH and ede1EH cells exhibited a redundant function only when combined with end3EH cells. These results clearly demonstrate the presence of functional redundancy among these three EHDPs.
FIGURE 5:
Effects of EH mutations on endocytic internalization. (A) Radiolabeled α-factor internalization assays performed on cells expressing the indicated strains at 25°C. Each curve represents the average of three independent experiments, and error bars indicate the SD at each time point. (B) A histogram representing the relative half-time of the α-factor internalization rate calculated from an exponential curve fit on the assay shown in A. Error bars indicate the SD from at least three experiments.
Dynamics of the early endocytic component in EH domain mutants
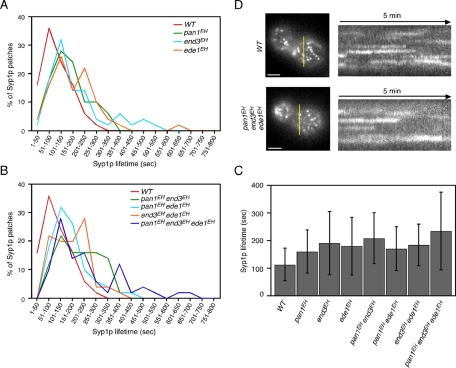
Similar to mammalian cells, clathrin in yeast is localized to cortical patches (Merrifield et al., 2002; Kaksonen et al., 2003, 2005; Newpher et al., 2005). Clathrin appears early at all sites of endocytosis, preceding Sla1p, and has a mean lifetime of ∼73.8 s (Kaksonen et al., 2005; Newpher et al., 2005). Therefore we next examined whether the overall process of clathrin coat assembly was affected by EH domain mutations. Because a large proportion of clathrin localizes to intracellular compartments, such as the trans-Golgi network, in addition to cortical patches and it is difficult to analyze the cortical localization precisely (Kaksonen et al., 2005; Newpher and Lemmon, 2006), we used another early-arriving endocytic protein, Syp1p, which has a long and variable lifetime similar to that of clathrin, as a marker of clathrin-coat assembly (Stimpson et al., 2009). To analyze extremely long lifetimes of Syp1p in the EH mutants (Figure 6), we tagged Syp1p with three tandem tagged copies of GFP (3GFP). The 3GFP-tagged Syp1p showed similar localization to Syp1-GFP at the cortical patches in addition to the bud neck, as reported previously (Supplemental Figure S4, A and B; Stimpson et al., 2009). The lifetime of Syp1p patches labeled by Syp1-3GFP in wild-type cells ranged from 50 to 350 s, with an average of 118.1 ± 51.0 s (Figure 6, A and C). The lifetime of Syp1-3GFP patches was a little longer compared with that of single GFP–tagged Syp1p (Stimpson et al., 2009) because of the enhanced fluorescence (Supplemental Figure S4C). All single mutants showed increased Syp1p patch lifetimes relative to wild-type cells, even in ede1EH cells (Figure 6, A and C, and Supplemental Figure S4D). These effects were enhanced modestly in the pan1EH end3EH double mutant and clearly in the triple mutant (Figure 6, B and C). For other double mutations, such as pan1EH ede1EH or end3EH ede1EH, no additive effect was observed (Figure 6C). It was noteworthy that these results were well consistent with those of α-factor internalization, shown in Figure 5B. Kymographs for these mutants showed that majority of Syp1p patches had extended lifetimes but regularly disappeared from the cell cortex in all mutants (Figure 6D).
FIGURE 6:
Dynamics of Syp1p patch in EH domain mutants. (A, B) Distribution of Syp1p patch lifetimes in the indicated single (A) or double and triple EH domain mutants (B). Data were taken from ∼13-min movies with a 4-s frame interval. (C) Average lifetimes of Syp1-GFP ± SD for indicated strains. (D) Left, localization of Syp1-3GFP from the indicated strains. Yellow bars mark where the kymograph was generated. Right, kymograph representations of Syp1-3GFP from 5-min movies. Scale bars, 2 μm.
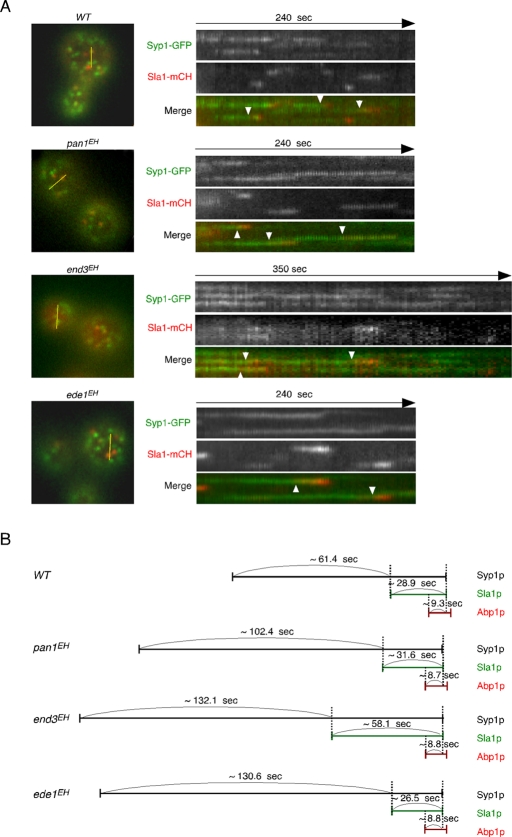
To determine the precise timing of Syp1p and Sla1p recruitment to the cortical patches, we imaged the cells expressing Syp1-3GFP and Sla1-mCherry. In wild-type cells, Syp1p appeared ∼61.4 s prior to Sla1p and stayed with Sla1p for ∼28.9 s at the cortical patch (Figure 7, A and B). Mutations in the EH domains of Pan1p caused the lifetime of Syp1p to increase but had almost no effect on the lifetimes of Sla1p and Abp1p (Figure 2A and 6B). In agreement with these results, in the pan1EH mutant, only the timing of Sla1p recruitment after the appearance of Syp1p at the cortical patch was delayed (Figure 7, A and B). Mutations in the EH domains of End3p caused an increment of the lifetimes of both Syp1p and Sla1p (Figure 2A and 6B) and, as a result, delayed the recruitment of Sla1p and extended the time of colocalization between Syp1p and Sla1p (Figure 7, A and B). In the ede1EH mutant, the lifetime of Syp1p patches was increased but that of Sla1p patches was decreased (Figure 2A and 6B), resulting in a significant delay of Sla1p recruitment and a decreased colocalization time (Figure 7, A and B). It follows from these results that assembly of the late coat module marked by Sla1p is delayed during endocytosis in all EH mutants, especially the end3EH and ede1EH mutants.
FIGURE 7:
Localization and temporal relationship of Syp1p and Sla1p patches in EH domain mutants. (A) Left, localization of Syp1-GFP and Sla1-mCherry (mCH) in live cells. Yellow bars mark where the kymograph was generated. Right, kymograph representations of Syp1-3GFP and Sla1-mCherry from 4- or 6-min movies. Arrowheads indicate examples of Syp1p patches that are joined by Sla1-mCherry. All movies were taken with a 3-s frame interval for both Syp1-GFP and Sla1-mCherry. Scale bars, 2 μm. (B) Temporal relationship of Syp1p, Sla1p, and Abp1p patches in EH domain mutants. Average time was obtained from at least 30 independent patches.
The frequency of endocytic internalization is decreased in the triple mutant
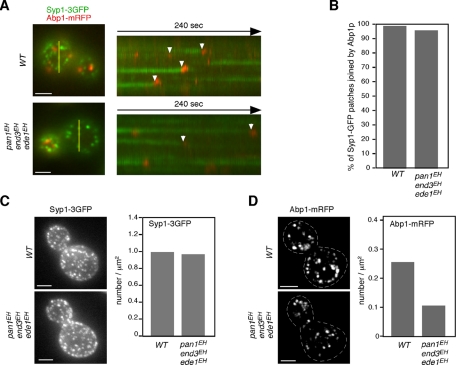
To examine whether Syp1p patches, which have extended lifetimes in the triple mutants, are eventually internalized or disappear without internalization, we coexpressed Syp1-3GFP and Abp1-mRFP and examined their localization on the cell surface. In wild-type cells, most of the Syp1-3GFP patches were joined by Abp1-mRFP and then disappeared (99.2%, n = 129; Figure 8, A and B). Similarly, it was observed that most Syp1-3GFP patches, despite having greatly extended lifetimes (>300 s) in the triple mutant, became colocalized with Abp1-mRFP for a short time and then disappeared (96.1%, n = 129; Figure 8, A and B). This observation indicates that most of the Syp1-3GFP–labeled endocytic sites in the triple mutants are capable of being internalized into the cytosol.
FIGURE 8:
All Syp1p patches are eventually internalized in triple mutants. (A) Epifluorescence images of a single cell coexpressing Syp1-3GFP and Abp1-mRFP. Yellow bars mark where the kymograph was generated. Right, kymograph representations of Syp1-3GFP and Abp1-mRFP from 3-min movies. Arrowheads indicate examples of Syp1p patches that are joined by Abp1-mRFP. Scale bars, 2 μm. (B) Percentage of the Syp1-3GFP patches that were eventually joined by Abp1-mRFP. (C, D) Maximum-intensity projections of Z stacks of wild-type and triple-mutant cells labeled with Syp1-3GFP (C) or Abp1-mRFP (D). The Z series was acquired through the entire cell at 0.2-μm intervals. Dotted lines represent outline of cells. Quantification of Syp1-3GFP or actin patches/μm2 ± SD in wild-type and triple mutant cells (n = 50 cells for each strain). Scale bars, 2 μm.
Having previously reported that ede1Δ mutants form fewer Sla1p-labeled endocytic sites than wild-type cells (Stimpson et al., 2009), we assessed the number of endocytic sites in the triple mutants. Maximum-intensity Z projections of live cells were analyzed to determine the average number of patches per surface area. Patch densities were quantified only in the mother cells, in which individual patches were distinguishable. As shown in Figure 8C, in the triple mutant, there was no decrease in the number of Syp1p patches per surface area relative to wild-type cells. We further examined whether EH domain mutations could affect the frequency of endocytotic internalization by comparing the number of Abp1-mRFP patches—a marker of actin-driven membrane invagination and endocytotic vesicle internalization. Maximum-intensity projections of Z stacks revealed that the number of endocytotic internalizations labeled with Abp1-mRFP decreased by ∼41% in the triple mutants (Figure 8D). This observation suggests that the frequency of endocytotic internalization is decreased in the triple mutants and that this might cause a reduction of α-factor internalization.
Effects of EH domain mutants on proteins containing the NPF motif
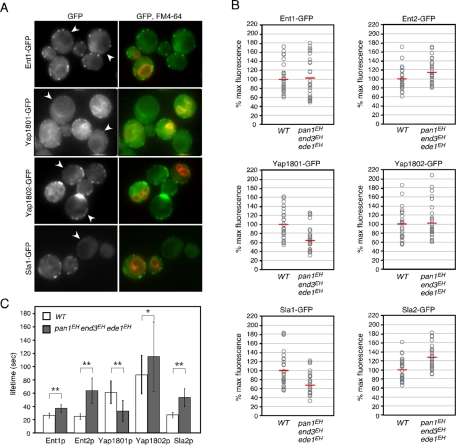
Having established that EH domains interact with NPF motifs (Salcini et al., 1997; de Beer et al., 1998; Paoluzi et al., 1998), we next examined whether EH domain mutations could affect the localization or dynamics of NPF motif–containing proteins. Ent1p and Ent2p, an essential pair of genes belonging to the epsin family (Chen et al., 1998; Wendland et al., 1999; Aguilar et al., 2003), contain two NPF motifs that interact with the EH domains of Pan1p and Ede1p (Aguilar et al., 2003). To evaluate precisely differences in the localization of NPF-containing proteins, we compared the triple mutants expressing Ent1-GFP or Ent2-GFP directly alongside wild-type cells (Figure 9A). To distinguish the wild-type and mutant cells, we treated the former with the vacuolar dye 3-triethylammoniumpropyl-4-p-diethylaminophenylhexatrienyl pyridinium dibromide (FM4-64). As shown in Figure 9, A and B, the fluorescence intensity of individual Ent1p or Ent2p patches was little affected, indicating that recruitment of these proteins was not dependent on the EH domains. In contrast, the lifetimes of Ent1p patches and Ent2p patches were affected differently by the EH domain mutants: Ent2p patches had a lifetime ∼2.5-fold longer in the triple mutants (64.0 ± 19.0 s) than in the wild-type cells (25.4 ± 4.4 s), whereas the Ent1p patch lifetime was extended only modestly (Figure 9C). We also examined the corresponding effect on two other NPF motif-containing proteins, Yap1801p and Yap1802p, which are homologues of the mammalian clathrin adaptor proteins CALM/AP180 (Wendland and Emr, 1998). Yap1802-GFP patches in the triple mutant also showed no marked change in fluorescence intensity but had a slightly extended lifetime relative to those in wild-type cells (Figure 9, A–C). In contrast to these proteins, however, the fluorescence intensity and lifetime of Yap1801-GFP were markedly decreased in the triple mutants (Figure 9, A–C). These results prompted us to examine other endocytic coat proteins, such as Sla1p and Sla2p. We found that Sla2-GFP patches exhibited a phenotype similar to that of Ent2-GFP patches, whereas the fluorescence intensities of Sla1-GFP patches were decreased in the triple-mutant cells relative to the wild-type cells (Figure 9, A–C). These results suggest that EHDPs seem important for the proper accumulation and dynamics of NPF-containing proteins at an endocytic patch. Because Sla1p contains a single NPF sequence at the C terminus (amino acids 2040–2042), we examined whether this NPF sequence is responsible for Sla1p localization. The GFP-tagged Sla1pNPF-AAA mutant in which the NPF sequence was converted to three alanine residues showed normal localization and lifetimes in wild-type cells (Supplemental Figure S5A). We also found that Sla1pNPF-AAA-GFP exhibited similar dynamics to wild-type Sla1p in the end3EH and ede1EH mutants (Supplemental Figure S5B), indicating that the NPF sequence is not required for Sla1p localization.
FIGURE 9:
Effects of EH domain mutations on endocytic proteins. (A) Localization of GFP-tagged endocytic proteins in wild-type and triple-mutant cells. Two strains were mixed and acquired in the same images. Only wild-type cells were labeled with FM4-64 in the right-hand images. Arrowheads indicate triple mutant cells. (B) The relative maximum fluorescence intensities of indicated GFP-fused proteins at cortical patches. Intensities of individual patches were compared in the same image by mixing wild-type and triple-mutant cells, which were distinguishable from FM4-64-treated wild-type cells. The values of intensities for wild-type and triple-mutant cells were divided by the mean for patch intensities in wild-type cells (n = 30), and the resulting values were multiplied by 100. Each open circle and horizontal red line represents intensity of a single patch and the mean of the intensities, respectively. (C) Patch lifetimes ± SD of GFP-tagged endocytic proteins in wild-type and triple mutant cells. Data were obtained from ∼3-min movies taken with a 1-s frame interval. n = 50 patches for each strain. *p < 0.01, **p < 0.001.
DISCUSSION
EH domains of End3p, rather than those of Pan1p, play a critical role in endocytosis
In this study we mutated all of the EH domains of yeast endocytic EHDPs—Pan1p, End3p, or Ede1p—and evaluated their effects on several endocytosis-related processes, including recruitment of endocytic proteins to cortical patches, cargo internalization, and clathrin coat assembly. Previous studies revealed that native Pan1p and End3p form a stable complex with 1:1 stoichiometry (Tang et al., 1997; Toshima et al., 2007) and that therefore these proteins may function as a complex containing four EH domains. Among these EH domains, those of End3p seem to be relatively important since the end3EH mutant has more severely defective growth and endocytosis than the pan1EH mutant. Unexpectedly, the pan1EH mutant exhibited only minor endocytic defects, even though PAN1 is an essential gene and a variety of pan1 mutants have been reported to exhibit severe defects of the actin cytoskeleton and/or endocytosis (Tang and Cai, 1996; Tang et al., 2000; Wendland et al., 1996; Toshima et al., 2005). This result differs from a previous observation that the lifetime of Pan1p was significantly extended when its EH domain was mutated (Maldonado-Baez et al., 2008). Maldonado-Baez et al. (2008) showed that ∼60% of Pan1EH-GFP patches exhibited greatly extended lifetimes ranging from 75 to >120 s, whereas the remaining ∼40% had slightly extended lifetimes (45 ± 5 s). The different lifetimes of Pan1EH-GFP patches might be caused by differences in their manner of expression; in their experiment, Pan1EH-GFP was expressed via its endogenous promoter using a single-copy plasmid in pan1Δ cells, whereas we expressed Pan1EH-GFP from an endogenous locus. Any defects in the pan1EH mutant are probably masked by End3p because they work together as a complex and have redundant functions (Tang et al., 1997; Toshima et al., 2007).
Role of the EH domains of Ede1p in endocytosis
As opposed to the pan1EH or end3EH mutant, we found a decrease of ∼20.5% in the lifetime of the Sla1-GFP patch in the ede1EH mutant relative to wild-type cells. This effect is in agreement with earlier observations of Sla1-GFP in ede1Δ cells (Kaksonen et al., 2005; Stimpson et al., 2009). Of interest, the ede1EH mutant also decreased the lifetime of the Sla1-GFP patch even when combined with the pan1EH or end3EH mutant. Moreover, prolonged Sla1-GFP patch lifetime observed in pan1EH end3EH double mutant (Figure 3, B and C) was reduced when the double mutant was combined with the ede1EH mutant. These results suggest that the EH domains of Ede1p have a dominant role in determining the lifetime of the Sla1p patch. Although it has not been clarified how Ede1p regulates Sla1p localization, physical interaction might be important, as the third SH3 domain of Sla1p interacts with Ede1p (Tonikian et al., 2009). In contrast to the Sla1p patch, the lifetimes of the Ede1p and Syp1p patches were increased in ede1EH cells, suggesting that the total time required for completion of clathrin coat assembly is increased in the ede1EH mutant.
The presence of several phenotypes that are distinct between ede1EH and ede1Δ cells suggests a specific function of the Ede1p EH domain. For instance, in ede1Δ cells, Syp1p was strongly concentrated at the necks of buds, and cortical Syp1p was diffusely and unstably localized with polarization toward the daughter cell (Stimpson et al., 2009), whereas ede1EH mutants, as was the case in wild-type cells, exhibited cortical localization of Syp1p but had a lifetime that was increased approximately twofold. Another distinct phenotype was that deletion of the EDE1 gene reduced the number of endocytic sites (Stimpson et al., 2009), whereas the ede1EH mutant had a normal number of Syp1p patches, similar to the triple mutant (data not shown). Thus these observations suggest that the EH domains of Ede1p have a role in efficient assembly of the clathrin-coated pit but not in the regulation of Syp1p localization or endocytic site formation.
EH domains are required for early coat assembly
Although mutation of EH domains caused defects in coat proteins dynamics, no apparent effect was observed in Abp1p, a marker of actin-driven membrane invagination. This result is of interest because mutation of endocytic proteins can often affect the entire process of endocytosis. For instance, deletion of CHC1—the clathrin heavy chain—reduces the patch lifetimes of several coat proteins and, in contrast, extends the patch lifetime of Abp1p (Kaksonen et al., 2005; Newpher and Lemmon, 2006). end3Δ cells exhibited prolonged patch lifetimes for both coat proteins and Abp1p (Kaksonen et al., 2005). The pan1-15TA mutant, in which major potential Prk1p phosphorylation sites in Pan1p are mutated to alanine, exhibited a large abnormal actin structure, including several endocytic proteins (Toshima et al., 2005). Therefore our observation suggests a specific role of EH domain–mediated interactions in the process of early clathrin coat assembly.
How does mutation of EHDPs affect early coat assembly? Because Ede1 was previously characterized as a binding partner of Syp1p (Reider et al., 2009; Stimpson et al., 2009), it seems reasonable to suppose that Syp1p lifetime is increased in ede1EH cells because Ede1EH patch lifetime is increased. However, it is still unclear why the lengthened Syp1p lifetime is also observed in the pan1EH and end3EH mutants. By tracking individual Syp1p patches in the pan1EH and end3EH mutants, we found that the time required to reach the maximum fluorescence intensity was not increased in these mutants (Supplemental Figure S4D), although Syp1p patch intensities were quite irregular, as reported previously (Stimpson et al., 2009). This observation suggests that early coat assembly might be suspended until recruitment of late coat proteins such as Yap1801p and Sla1p begins. How late coat module determines lifetimes of early coat proteins is an important question for the future.
EH domains are required for Yap1801p recruitment
Among the yeast NPF proteins, only Yap1801p exhibited a decrease of both fluorescence intensity and lifetime at cortical patches in the triple EH domain mutant. Yap1801p is the yeast homologue of AP180/CALM family proteins, which are associated with clathrin assembly activity (Wendland and Emr, 1998). AP180 and CLAM have similar roles in facilitating assembly of the clathrin coat and controlling the size of clathrin-coated vesicles (Ahle and Ungewickell, 1986; Ye et al., 1995; Ye and Lafer, 1995; Morgan et al., 1999; Meyerholz et al., 2005). It is intriguing that Morgan et al. (2003) reported that squid Eps15 stimulated clathrin assembly by AP180 via NPF motif/EH domain interaction, although Eps15 itself is not capable of polymerizing clathrin. The role of yeast AP180 proteins was unclear, as deletion of both YAP180 genes resulted in no apparent defect of endocytosis or growth (Huang et al., 1999), although a recent study showed that NPF motifs in Yap1801/2p and Ent1/2p share a complementary role in both viability and endocytosis (Maldonado-Baez et al., 2008). It is unclear how a decreased level of Yap1801p at cortical patches affects clathrin coat assembly, whereas defective recruitment of some additional factors might cooperatively cause defects. It was reported that some EH domains recognize other peptide motifs, including Phe–Trp (FW), Trp–Trp (WW), and Ser–Trp–Gly (SWG) (Paoluzi et al., 1998). The EH domain of End3p was also shown to interact with peptides containing the His–(Thr/Ser)–Phe (HTF/HSF) motif (Paoluzi et al., 1998). Using the PatMatch (Yeast Genome Pattern Matching) program, we found that the FW sequence(s) exists in Chc1p, yeast clathrin heavy chain, and Apl1p, a large subunit of the clathrin-associated protein complex (AP-2). We also found that two WW sequences are present in Sla1p. These motifs would be attractive targets for further investigation of how EHDPs regulate clathrin-coated pit assembly.
Function of EH domains in endocytosis is conserved in both yeast and mammals
There are several lines of evidence to suggest that mammalian Eps15p has a similar function to yeast EHDPs. First, it was shown that clathrin-coated pit assembly is perturbed by overexpression of the Eps15 mutant lacking the second and third EH domains (Benmerah et al., 1999). A previous study demonstrated that EH domains are required but are not sufficient for targeting Eps15 to clathrin-coated pits (Benmerah et al., 2000). Knockdown of Eps15 increases the lifetime of clathrin, indicating that Eps15 is critical for efficient assembly of the clathrin coat (Mettlen et al., 2009). Thus the role of the EH domain in the endocytic pathway seems to be conserved between yeast and animal cells.
MATERIALS AND METHODS
Yeast strains, growth conditions, and plasmids
The yeast strains used in this study are listed in Table 1. All strains were grown in standard rich medium (yeast extract/peptone/dextrose [YPD]) or synthetic medium supplemented with 2% glucose and appropriate amino acids. pan1EH, end3EH, or ede1EH mutant was integrated as follows: To create a pan1 integration plasmid, the XmnI–DraI fragment of the PAN1 gene was cloned into pBluescript II SK (pBS), and the SalI fragment of the LEU2 gene was inserted into the SalI site 154 base pairs upstream of the PAN1 open reading frame (ORF). The mutated MscI–NheI pan1 fragments were used to replace the PAN1 gene in the integration plasmid. To integrate pan1EH mutant at the endogenous locus, the integration plasmids were digested with SacI and XbaI and transformed into pan1Δ::HIS3/PAN1 diploid strains. Integrated pan1 mutants were selected on synthetic complete plates lacking leucine and sporulated to obtain pan1 mutants. To create end3 integration plasmids, the XbaI-ClaI fragment of the END3 gene was cloned into pBS, and the SmaI fragment of the URA3 gene was inserted into the EcoO109I site 150 base pairs upstream of the END3 ORF. The mutated XbaI–XhoI end3 fragments were used to replace the END3 gene in the integration plasmid. To integrate end3EH mutants at the endogenous locus, the integration plasmids were digested with XbaI and ClaI and transformed into end3Δ strains. To create ede1 integration plasmid, the SacI–XhoI fragment of the EDE1 gene was cloned into pBS, and the SmaI fragment of the LEU2 gene was inserted into the EcoRV site, which was made by amino acid substitutions, at 139 base pairs downstream of the EDE1 ORF. The mutated SacI–NdeI ede1 fragment was used to replace the EDE1 gene in the integration plasmid. To integrate ede1EH mutant at the endogenous locus, the integration plasmids were digested with SacI and XhoI and transformed into ede1Δ strains. Amino acid substitutions were made using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). pan1ΔEH mutant was constructed by inserting the PCR-amplified fragment (nucleotides [nt] 1074–1784) into the PAN1 fragment deleting EH domains and the region between the EH domains (nt 787–2063). Similarly, end3ΔEH, or ede1ΔEH mutant was constructed by inserting the PCR-amplified fragments (END3, nt 289–366; EDE1, nt 298–381 and 664–807) into the END3 or EDE1 fragment deleting EH domains and the region between the EH domains (END3, nt 16–663; EDE1, nt 19–1107). GFP and mRFP tags were integrated at the C terminus of each gene (Longtine et al., 1998). The triple-GFP tag was integrated at the C-terminus of the SYP1 gene as follows: The 3GFP fragment was subcloned into BamHI- and NotI-digested pBS (pBS-3GFP), and the NotI–SacII fragment, which contains the Saccharomyces cerevisiae ADH1 terminator and the HIS3MX6 module, was amplified by PCR using pFA6a-GFP (S65T)-HIS3MX6 (Longtine et al., 1998) as a template and inserted into NotI- and SacII-digested pBS-3GFP (pBS-3GFP-HIS3). To create an integration plasmid, a fragment of the SYP1 ORF (nt 2077–2610) was generated by PCR and cloned into the BamHI site of pBS-3GFP-HIS3. To integrate 3GFP at the C-terminus of the SYP1 gene, the integration plasmid was linearized by ClaI and transformed into yeast.
TABLE 1:
Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| JJTY0011 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::PAN1::LEU2 | Toshima lab |
| JJTY0059 | Matahis3-Δ200 leu2-3, 112 ura3-52 bar1Δ::LEU2 | Toshima lab |
| JJTY0147 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::PAN1::LEU2 PAN1-GFP::HIS3 | Toshima lab |
| JJTY0318 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 END3-GFP::HIS3 | Toshima lab |
| JJTY0369 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 | Toshima lab |
| JJTY0393 | Matahis3-Δ200 leu2-3, 112 ura3-52 SLA1-GFP::KanMX6 ABP1-RFP::HIS3 | Toshima lab |
| JJTY0943 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 | This study |
| JJTY0950 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 PAN1-GFP::HIS3 | This study |
| JJTY0954 | Matαhis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 SLA1-GFP::KanMX6ABP1-mRFP::HIS3 | This study |
| JJTY0958 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 bar1Δ::HIS3 | This study |
| JJTY0960 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::END3::URA3 | This study |
| JJTY0962 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 | This study |
| JJTY0972 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 SLA1-GFP::KanMX6ABP1-mRFP::HIS3 | This study |
| JJTY0976 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 bar1Δ::LEU2 | This study |
| JJTY0979 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3 | This study |
| JJTY0982 | Matαhis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3SLA1-GFP::KanMX6 ABP1-mRFP::HIS3 | This study |
| JJTY0983 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3 bar1Δ::LEU2 | This study |
| JJTY1074 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::EDE1::LEU2 | This study |
| JJTY1075 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 | This study |
| JJTY1077 | Matαhis3-Δ200 leu2-3, 112 ura3-52 ede1Δ::ede1EH::LEU2 SLA1-GFP::KanMX6ABP1-mRFP::HIS3 | This study |
| JJTY1079 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 bar1Δ::HIS3 | This study |
| JJTY1080 | Matαhis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 ede1Δ::ede1EH::LEU2SLA1-GFP::KanMX6 ABP1-mRFP::HIS3 | This study |
| JJTY1081 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 ede1Δ::ede1EH::LEU2SLA1-GFP::KanMX6 ABP1-mRFP::HIS3 | This study |
| JJTY1083 | Matahis3-Δ200 leu2-3, 112 ura3-52 end3Δ::end3EH::URA3 ede1Δ::ede1EH::LEU2 | This study |
| JJTY1085 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 | This study |
| JJTY1087 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 ede1Δ::ede1EH::LEU2 | This study |
| JJTY1088 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 ede1Δ::ede1EH::LEU2 bar1Δ::LEU2 | This study |
| JJTY1089 | Matahis3-Δ200 leu2-3, 112 ura3-52 end3Δ::end3EH::URA3 ede1Δ::ede1EH::LEU2 bar1Δ::HIS3 | This study |
| JJTY1090 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 SLA1-GFP::KanMX6 ABP1-mRFP::HIS3 | This study |
| JJTY1091 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 bar1Δ::HIS3 | This study |
| JJTY1227 | Matahis3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 YAP1802-GFP::KanMX6 | This study |
| JJTY1255 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 END3-GFP::URA3 | This study |
| JJTY1256 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 EDE1-GFP::HIS3 | This study |
| JJTY1257 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 EDE1-GFP::HIS3 | This study |
| JJTY1258 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 ENT1-GFP::HIS3 | This study |
| JJTY1259 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 ENT1-GFP::HIS3 | This study |
| JJTY1262 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 YAPLafer-GFP::HIS3 | This study |
| JJTY1263 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 YAP1801-GFP::HIS3 | This study |
| JJTY1928 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SYP1-3GFP::HIS3 ABP1-mRFP::KanMX6 | This study |
| JJTY1929 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 SYP1-3GFP::HIS3ABP1-mRFP::KanMX6 | This study |
| JJTY1930 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 SYP1-3GFP::HIS3ABP1-mRFP::KanMX6 | This study |
| JJTY1931 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 SYP1-3GFP::HIS3ABP1-mRFP::KanMX6 | This study |
| JJTY1932 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3SYP1-3GFP::HIS3 ABP1-mRFP::KanMX6 | This study |
| JJTY1933 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 ede1Δ::ede1EH::LEU2SYP1-3GFP::HIS3 ABP1-mRFP::KanMX6 | This study |
| JJTY1934 | Matahis3-Δ200 leu2-3, 112 ura3-52 end3Δ::end3EH::URA3 ede1Δ::ede1EH::LEU2SYP1-3GFP::HIS3 ABP1-mRFP::KanMX6 | This study |
| JJTY1935 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 SYP1-3GFP::HIS3 ABP1-mRFP::KanMX6 | This study |
| JJTY1936 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 ENT2-GFP::HIS3 | This study |
| JJTY1937 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 ENT2-GFP::HIS3 | This study |
| JJTY1938 | Matα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 SLA2-GFP::HIS3 | This study |
| JJTY1939 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 SLA2-GFP::HIS3 | This study |
| JJTY1940 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3ede1Δ::ede1EH::LEU2 YAP1802-GFP::HIS3 | This study |
| JJTY2421 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SYP1-3GFP::HIS3 Sla1-mCherry::KanMX | This study |
| JJTY2422 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 pan1Δ::pan1EH::LEU2 Syp1-3GFP::HIS3Sla1-mCherry::KanMX | This study |
| JJTY2423 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 Syp1-3GFP::HIS3Sla1-mCherry::KanMX | This study |
| JJTY2424 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 Syp1-3GFP::HIS3Sla1-mCherry:: KanMX | This study |
| JJTY2426 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 END3-GFP::HIS3 | This study |
| JJTY2427 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU EDE1-GFP::HIS3 | This study |
| JJTY2428 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ede1Δ::ede1EH::LEU2 PAN1- GFP::HIS3 | This study |
| JJTY2429 | Matahis3-Δ200 leu2-3, 112 ura3-52 ede1Δ::ede1EH::LEU2 END3-GFP::HIS3 | This study |
| JJTY2430 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 PAN1-GFP::HIS3 | This study |
| JJTY2431 | Matahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 end3Δ::end3EH::URA3 EDE1-GFP::HIS3 | This study |
| JJTY2432 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 end3Δ::end3EH::URA3EDE1-GFP::HIS3 | This study |
| JJTY2433 | Matahis3-Δ200 leu2-3, 112 ura3-52 pan1Δ::pan1EH::LEU2 ede1Δ::ede1EH::LEU2END3-GFP::HIS3 | This study |
| JJTY2434 | Matahis3-Δ200 leu2-3, 112 ura3-52 end3Δ::end3EH::URA3 ede1Δ::ede1EH::LEU2 PAN1-GFP | This study |
Fluorescence microscopy
Fluorescence microscopy was performed using an Olympus IX81 microscope equipped with a 100×/numerical aperture 1.40 (Olympus, Center Valley, PA) objective and an Orca-AG cooled charge-coupled device camera (Hamamatsu, Hamamatsu, Japan), using MetaMorph software (Universal Imaging, West Chester, PA). Simultaneous imaging of red and green fluorescence was performed using an Olympus IX81 microscope, as described, and an image splitter (Dual-View; Optical Insights, Santa Fe, NM) that divided the red and green components of the images with a 565-nm dichroic mirror and passed the red component through a 630/50-nm filter and the green component through a 530/30-nm filter. Rhodamine–phalloidin staining of filamentous actin and FM4-64 staining were performed as described previously (Toshima et al., 2005).
35S-labeled α-factor internalization assay
Preparation and internalization of 35S-labeled α-factor was performed as described previously (Toshima et al., 2005). Briefly, cells were grown to an OD600 of 0.3 in 50 ml of YPD, briefly centrifuged, and resuspended in 4 ml of YPD containing 1% (wt/vol) bovine serum albumen, 50 mM KH2PO4, pH 6.0, and 20 μg/ml uracil, adenine, and histidine. After addition of 35S-labeled α-factor, cell aliquots were withdrawn at various time points and subjected to a wash in pH 1 buffer to remove surface-bound α-factor so internal α-factor could be measured or in pH 6 buffer to determine the total (internal and bound) α-factor. The amount of cell-associated radioactivity after each wash was determined by scintillation counting. Each experiment was performed at least three times.
Supplementary Material
Acknowledgments
We thank R. Kashikuma, E. Furuya, and M. Saito for construction of plasmids and strains. We also thank the members of the Toshima lab for sharing materials and for helpful discussions. This work was supported by Japan Society for the Promotion of Science (to J.Y.T.) and the Novartis Foundation (Japan), the Mitsubishi Foundation, the Naito Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Futaba Electronics Memorial Foundation, the Astellas Foundation for Research on Metabolic Disorders, the Hamaguchi Foundation for the Advancement of Biochemistry, the Takeda Science Foundation, and the Kurata Memorial Hitachi Science and Technology Foundation (to J.T.).
Abbreviations used:
- EH domain
Eps15 homology domain
- mRFP
monomeric red fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0380) on December 21, 2011.
REFERENCES
- Aguilar RC, Watson HA, Wendland B. The yeast epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- Ahle S, Ungewickell E. Purification and properties of a new clathrin assembly protein. EMBO J. 1986;5:3143–3149. doi: 10.1002/j.1460-2075.1986.tb04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci. 1999;112:1303–1311. doi: 10.1242/jcs.112.9.1303. [DOI] [PubMed] [Google Scholar]
- Benmerah A, Begue B, Dautry-Varsat A, Cerf-Bensussan N. The ear of alpha-adaptin interacts with the COOH-terminal domain of the Eps 15 protein. J Biol Chem. 1996;271:12111–12116. doi: 10.1074/jbc.271.20.12111. [DOI] [PubMed] [Google Scholar]
- Benmerah A, Gagnon J, Begue B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N. The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J Cell Biol. 1995;131:1831–1838. doi: 10.1083/jcb.131.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Poupon V, Cerf-Bensussan N, Dautry-Varsat A. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J Biol Chem. 2000;275:3288–3295. doi: 10.1074/jbc.275.5.3288. [DOI] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- de Beer T, Carter RE, Lobel-Rice KE, Sorkin A, Overduin M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science. 1998;281:1357–1360. doi: 10.1126/science.281.5381.1357. [DOI] [PubMed] [Google Scholar]
- de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pelicci PG, Sorkin A. EH: a novel protein-protein interaction domain potentially involved in intracellular sorting. Trends Biochem Sci. 1997;22:411–413. doi: 10.1016/s0968-0004(97)01127-4. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- Grant BD, Caplan S. Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic. 2008;9:2043–2052. doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KM, D'Hondt K, Riezman H, Lemmon SK. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Dores MR, Perkins EM, Drivas TG, Hicke L, Wendland B. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Mettlen M, Stoeber M, Loerke D, Antonescu CN, Danuser G, Schmid SL. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol Biol Cell. 2009;20:3251–3260. doi: 10.1091/mbc.E09-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Jin S, Augustine GJ, Lafer EM. Eps15 homology domain-NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J Biol Chem. 2003;278:33583–33592. doi: 10.1074/jbc.M304346200. [DOI] [PubMed] [Google Scholar]
- Morgan JR, Zhao X, Womack M, Prasad K, Augustine GJ, Lafer EM. A role for the clathrin assembly domain of AP180 in synaptic vesicle endocytosis. J Neurosci. 1999;19:10201–10212. doi: 10.1523/JNEUROSCI.19-23-10201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S. EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem. 2007;282:16612–16622. doi: 10.1074/jbc.M609493200. [DOI] [PubMed] [Google Scholar]
- Newpher TM, Lemmon SK. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17:6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E, Salcini AE, Kay BK, Yamabhai M, Di Fiore PP. The EH network. Exp Cell Res. 1999;253:186–209. doi: 10.1006/excr.1999.4694. [DOI] [PubMed] [Google Scholar]
- Sorkin A. Cargo recognition during clathrin-mediated endocytosis: a team effort. Curr Opin Cell Biol. 2004;16:392–399. doi: 10.1016/j.ceb.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Tang HY, Cai M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–4587. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates JR 3rd, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delft S, Schumacher C, Hage W, Verkleij AJ, van Bergen en Henegouwen PM. Association and colocalization of Eps15 with adaptor protein-2 and clathrin. J Cell Biol. 1997;136:811–821. doi: 10.1083/jcb.136.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:1564–1568. [PubMed] [Google Scholar]
- Ye W, Lafer EM. Bacterially expressed F1-20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission. J Neurosci Res. 1995;41:15–26. doi: 10.1002/jnr.490410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.