ETOC: EPI64 is a TBC domain containing a microvillar protein that binds to Arf6 and induces actin-coated vacuole accumulation when overexpressed. EPI64 does this by stabilizing Arf6-GTP levels to induce clathrin-independent endocytosis and by preventing endosomes from maturing to the tubular endosome by lowering Rab8a GTP levels via association with JFC1.
Abstract
Cell function requires the integration of cytoskeletal organization and membrane trafficking. Small GTP-binding proteins are key regulators of these processes. We find that EPI64, an apical microvillar protein with a Tre-2/Bub2/Cdc16 (TBC) domain that stabilizes active Arf6 and has RabGAP activity, regulates Arf6-dependent membrane trafficking. Expression of EPI64 in HeLa cells induces the accumulation of actin-coated vacuoles, a distinctive phenotype seen in cells expressing constitutively active Arf6. Expression of EPI64 with defective RabGAP activity does not induce vacuole formation. Coexpression of Rab8a suppresses the vacuole phenotype induced by EPI64, and EPI64 expression lowers the level of Rab8-GTP in cells, strongly suggesting that EPI64 has GAP activity toward Rab8a. JFC1, an effector for Rab8a, colocalizes with and binds directly to a C-terminal region of EPI64. Together this region and the N-terminal TBC domain of EPI64 are required for the accumulation of vacuoles. Through analysis of mutants that uncouple JFC1 from either EPI64 or from Rab8-GTP, our data suggest a model in which EPI64 binds JFC1 to recruit Rab8a-GTP for deactivation by the RabGAP activity of EPI64. We propose that EPI64 regulates membrane trafficking both by stabilizing Arf6-GTP and by inhibiting the recycling of membrane through the tubular endosome by decreasing Rab8a-GTP levels.
INTRODUCTION
The polarized state of epithelial cells, in which the apical membrane has a protein and lipid composition distinct from that of the basolateral membrane, requires the delivery of membrane components to the appropriate domain. This polarity is achieved through both the selective sorting of biosynthetic cargo at the trans-Golgi network and endocytic pathways that internalize, sort, and deliver cargo to the appropriate surface. The intricate labyrinth of intracellular sorting pathways is guided in part by the recruitment and cross-talk of many small GTPase proteins in the Rab and Arf families and their respective effectors (Zerial and McBride, 2001). Signals are carried downstream by the recruitment of effector proteins that bind only to the active form of the GTPase, its GTP-bound state. GTPase activators, guanine nucleotide exchange factors (GEFs), and their negative regulators, GTPase-activating proteins (GAPs), play critical roles in membrane trafficking (Barr and Lambright, 2010; Donaldson and Jackson, 2011). By combining GTPases with their effectors, GEFs, and GAPs, the cell creates a cascade of information, enabling the complex trafficking and maturation of cargo and membranes through many different internal membrane compartments (Grosshans et al., 2006).
The initial step of membrane uptake from the plasma membrane involves either clathrin-dependent or clathrin-independent endocytosis (Maxfield and McGraw, 2004; Grant and Donaldson, 2009). Clathrin-independent endocytosis (CIE) can be further divided into dynamin-dependent and dynamin-independent pathways (Mayor and Pagano, 2007; Sandvig et al., 2011). Among the CIE pathways, the dynamin-independent, Arf6-dependent pathway has been featured in many recent reviews (Donaldson, 2003; Donaldson et al., 2009; D'souza-Schorey and Chavrier, 2006; Mayor and Pagano, 2007; Grant and Donaldson, 2009; Hansen and Nichols, 2009; Howes et al., 2010; Sandvig et al., 2011). Following endocytosis in this CIE pathway, Rab proteins regulate cargo transit through a number of endosomsal compartments, although how this is coordinated is largely unexplored. This pathway is of interest to us, as we previously described EPI64, a RabGAP protein with a Tre-2/Bub2/Cdc16 (TBC) domain that is an effector for Arf6 (Hanono et al., 2006). Here we provide evidence that EPI64 has an additional function in the Arf6-dependent pathway through regulating the levels of active Rab8a.
Arf6, the most divergent member of the Arf family of six small GTPases, regulates membrane trafficking and dynamics of the actin cytoskeleton at the plasma membrane (Donaldson, 2003; Donaldson and Jackson, 2011). Arf6 is N-terminally myristoylated and localized to the plasma and endosomal membranes. Arf6-GTP stimulates the formation of plasma membrane phosphatidylinositol 4,5-bisphosphate (PtIns(4,5)P2) by directly activating phosphatidylinositol 4-phosphate 5-kinase (PI(4)P5) and indirectly through phospholipase D to generate phosphatidic acid (Jovanovic et al., 2006), which together can dramatically increase CIE (Honda et al., 1999; Brown et al., 2001). Incoming endocytic vesicles can fuse with the Rab5a early endosome (Naslavsky et al., 2003) and then be recycled back to the plasma membrane through the Rab8a-dependent tubular endosome-recycling pathway (Hattula et al., 2002, 2006). Overexpressing a constitutively active Arf6 (Arf6-Q67L) causes membrane to be internalized without being recycled, resulting in an accumulation of PI(4,5)P2 and actin-coated vacuoles (Brown et al., 2001).
Rab8a has been linked in models to the Arf6-regulated endocytic pathway and has been shown to colocalize with Arf6-GDP on tubular endosomes, whereas expression of the constitutively active Arf6-Q67L blocks the formation of these tubules (Hattula et al., 2006). Rab8a is one of 63 Rab proteins present in human cells that collectively regulate multiple stages of intracellular transport, including vesicle formation, transportation, sorting, tethering, and fusion. Although Rab8a may function at more than one intracellular compartment, it is clearly involved in a late step of the Arf6-dependent CIE pathway (Wandinger-Ness and Deretic, 2008). In the current view of the Arf6-dependent CIE pathway, cargo is internalized into an actin-coated Arf6 early endosome at regions of high PI(4,5)P2 induced through Arf6 activation. The Arf6 early endosome then merges with the Rab5-marked early endosome, is transported to the Rab11-marked endocytic recycling compartment, moved in a Rab8a-dependent manner into the tubular endosome, and recycled back to the cell surface in a separate Arf6-dependent manner (Radhakrishna and Donaldson, 1997; Jovanovic et al., 2006). In addition, expression of Rab8a-Q67L in cultured cells leads to the formation of membrane protrusions through actin nucleation and polymerization, linking membrane recycling with cytoskeletal dynamics (Peranen et al., 1996; Hattula et al., 2002); these protrusions are suppressed by the coexpression of Arf6-Q67L (Peranen et al., 1996; Hattula et al., 2006). This suggests that Arf6 works upstream of Rab8a, and that these two small GTPases are interconnected in the recycling of membranes. Although MICAL-L1 has recently been implicated to regulate Rab8a function through direct binding to Arf6 (Rahajeng et al., 2012), no molecule has been described that coordinates the activities of Arf6 and Rab8a directly.
Rab8a functions in both endocytic recycling and exocytosis, and many of Rab8a's GAPs, GEFs, and effectors have been identified. Two GEFs (MSS4 and Rabin8; Burton et al., 1994; Hattula et al., 2002) and two GAPs (TBC1D30 and AS160; Ishikura et al., 2007; Yoshimura et al., 2007) are known. Among its effectors are FIP-2 (optineurin), which links Rab8a to the motor protein myosin-VI (Hattula and Peranen, 2000), and myosin-Vb, which binds Rab8a directly (Roland et al., 2007). Active Rab8a also binds several synaptotagmin-like proteins (JFC1, Noc2, rabphilin, Rim2; Fukuda, 2003; Hattula et al., 2006), as well as MICAL and MICAL-like proteins (MICAL-1, MICAL-L1, MICAL-L2; Fukuda et al., 2008; Yamamura et al., 2008). Rab8a activity and regulation are vital for endocytic and exocytic transport, cytoskeletal dynamics, and cell shape; a number of human diseases result from misregulation of its nucleotide state, including open-angle glaucoma, human microvillus inclusion disease, Bardet–Biedl syndrome, tumor cell invasion, and Niemann–Pick type C disease (Rezaie et al., 2002; De Marco et al., 2006; Bravo-Cordero et al., 2007; Linder et al., 2007; Sato et al., 2007). In the Rab8a conditional knockout mouse, basolateral markers of epithelial cells are properly localized, whereas apical peptidases and transporters accumulate in lysosomes, where they are degraded instead of being localized to the apical membrane. Mislocalization and degradation of these apical proteins leads to progressive wasting due to a reduction in nutrient uptake and eventual death (Sato et al., 2007). These data give clear evidence that Rab8a is an essential signaling molecule in epithelial cells, designed to route specific cargo to the apical membrane and to regulate apical cytoskeletal structure.
EPI64 (TBC1D10A) is a 508-residue protein that contains a TBC/RabGAP domain (Reczek and Bretscher, 2001) and is concentrated at the base of microvilli, where it associates with EBP50/ezrin/F-actin (Hanono et al., 2006). The TBC domain of EPI64 binds to Arf6, but EPI64 overexpression increases the cellular level of Arf6-GTP and therefore shows no GAP activity for Arf6 (Hanono et al., 2006). EPI64 has been shown to have GAP activity in vitro for Rab27a (Itoh and Fukuda, 2006) and Rab35 (Hsu et al., 2010). A mutation made in the predicted active site of the TBC domain of EPI64 (R160A) abolishes the known GAP activity for Rab27a (Itoh and Fukuda, 2006), but EPI64-R160A still binds to Arf6, and its overexpression still increases the cellular level of Arf6-GTP (Hanono et al., 2006).
In this study, we set out to explore in more detail the function of EPI64 and in particular the role of its RabGAP. This led us to find that EPI64 binds the Rab8a effector JFC1 and reduces the level of Rab8a-GTP to regulate Rab8a/Arf6-dependent membrane trafficking.
RESULTS
EPI64 is implicated in microvilli formation
We showed that EPI64 is present in apical microvilli of the epithelial JEG-3 cell line derived from human placenta (Reczek and Bretscher, 2001). We also reported that the N-terminal half of EPI64 that contains the TBC domain binds active Arf6 and that overexpression of EPI64 results in the accumulation of vacuoles in a small percentage of cells that are reminiscent of overexpression of active Arf6. However, how EPI64 manifests this phenotype remains unknown.
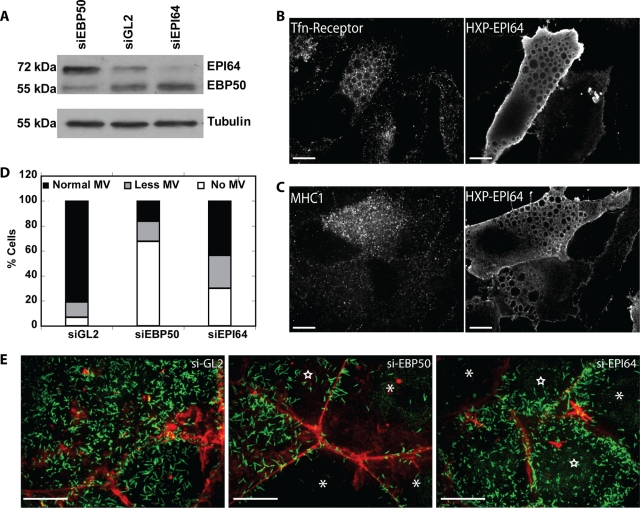
To explore the function of EPI64, we used small interfering RNA (siRNA) to knock down its expression in JEG-3 cells. At 48 h after transfection we observed a 70% reduction in EPI64 protein levels compared with control cells transfected with siRNA against luciferase (Figure 1A). Because Arf6 has been reported to regulate the trafficking of MHC1 but not transferrin (Radhakrishna and Donaldson, 1997), we examined MHC1 (Supplemental Figure S1A) and transferrin endocytosis in the EPI64-knockdown cells and found no significant differences in the rate of internalization or recycling. The lack of a defect in endocytosis could be due to the redundancy one of two other TBC1D10 isoforms potentially present in these cells. MHC1 endocytosis was also not affected by overexpression of EPI64 (Supplemental Figure S1A). We next looked at MHC1 and transferrin receptor localization in HeLa cells overexpressing HXP-EPI64. It was surprising that transferrin receptor was found associated with the EPI64 vacuoles (Figure 1B). MHC1 was also found associated with EPI64 vacuoles; however, unlike the transferrin receptor, MHC1 was also found as a haze inside the vacuoles in addition to being associated on the vacuoles (Figure 1C).
FIGURE 1:
EPI64 knockdown reduces microvilli (MV) on epithelial cells. (A) JEG-3 cells transfected with siRNA against EBP50, GL2, or EPI64 were lysed, resolved by SDS–PAGE, and a Western blot probed with antibodies to EPI64 and EBP50 or tubulin. (B, C) HeLa cells were transfected to express HXP-EPI64 and stained for either (B) transferrin receptor or (C) MHC1. Scale bars, 10 μm. (D) Quantification of the percentage of JEG-3 cells with normal microvilli (black bar), fewer microvilli (gray bar), and no microvilli (white bar) after transfection with siGL2, siEBP50, or siEPI64. (E) JEG-3 cells transfected to express siGL2 as a control (left), siEBP50 (middle), or siEPI64 (right) were stained for endogenous ezrin (green) and F-actin (red). Cells scored as having no MV are marked with asterisks, and cells with fewer MV are marked by stars. Scale bars, 10 μm.
The only overt phenotype we could discern in EPI64-depleted JEG-3 cells was a reduction in the number of cell surface microvilli, but the reduction was less than that seen after knockdown of the EPI64-interacting scaffolding protein EBP50 (Figure 1, D and E). Of interest, knockdown of EBP50 results in elevation of the level of EPI64, suggesting that EPI64 levels might be up-regulated to compensate for the loss of EBP50. Given the lack of a strong phenotype upon EPI64 knockdown, we chose to explore the basis of the robust phenotype of vacuole formation in cells overexpressing EPI64 to further probe the function of EPI64.
Overexpression of EPI64 induces the accumulation of actin-coated vacuoles
We proposed that EPI64 overexpression, due to the stabilization of Arf6-GTP that enhances PI(4)P 5-kinase activity, increases local PI(4,5)P2 synthesis to drive excessive Arf6-dependent membrane trafficking (Hanono et al., 2006). Because this phenotype is more often studied in HeLa cells than JEG-3 cells, we first examined what additional proteins associate with vacuoles in HeLa cells.
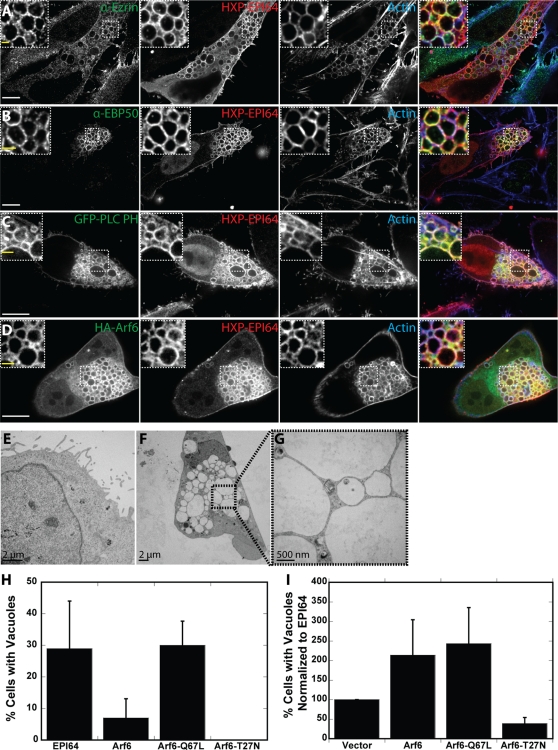
EPI64 was originally discovered as an EBP50-binding protein. EBP50 was identified as an ezrin-interacting protein (Reczek et al., 1997), and both EBP50 and ezrin are associated with the surface of the vacuoles induced by EPI64 overexpression (Figure 2, A and B). Actin, PI(4,5)P2 (as revealed by green fluorescent protein [GFP]-PLCδ-PH localization), and expressed HA-Arf6 were all localized to vacuoles in cells expressing HXP-EPI64 (Figure 2, C and D), similar to vacuoles previously studied in HeLa cells expressing Arf6-Q67L (Brown et al., 2001). Although the low abundance of EPI64 in HeLa cells makes endogenous localization in these cells challenging, endogenous EPI64 is localized in a punctate manner to vacuoles in JEG-3 cells expressing HA-Arf6-Q67L (Supplemental Figure S1B). Moreover, JEG-3 cells expressing Arf6-Q67L still contained vacuoles when EPI64 levels were knocked down by siRNA treatment, suggesting that EPI64 is acting upstream of Arf6 (Supplemental Figure S1, C and D). These vacuoles range in size from 1 to 10 μm in diameter and cluster together in one or more regions of cells. They ultimately lead to cell death, and so EPI64-overexpressing stable cell lines are not viable.
FIGURE 2:
EPI64 expression induces actin-coated vacuoles. (A–D) HeLa cells were transfected to express HXP-EPI64 and cotransfected to express (C) GFP-PLC PH or (D) HA-Arf6 for 24 h. Cells were fixed and stained for either (A) ezrin, (B) EBP50, or (D) the HA tag in green (first column), HXP tag in red (2nd column), and F-actin in blue (third column). Merge of all channels is shown in the fourth column. Insets show magnified regions labeled by dashed boxes. White scale bars, 10 μm; yellow scale bars in insets, 2 μm. (E–G) Thin section transmission electron microscopy (TEM) of HeLa cells transfected with HXP-EPI64 for 24 h. (E) TEM of cell with no vacuoles showing microvilli on cell periphery. (F) TEM of typical cell with vacuoles; the boxed region is enlarged in G. (H) Quantification of the percentage of cells transfected with HXP-EPI64, HA-Arf6, HA-Arf6-Q67L, or HA-Arf6-T27N that have vacuoles. Error bars are the SD from at least four separate experiments with >200 cells counted each time. (I) HeLa cells were cotransfected with HXP-EPI64 and vector control or HA-Arf6 constructs as indicated, and the number of cells with vacuoles was counted. Quantification of the percentage of cells with vacuoles was normalized to cells transfected to express EPI64. Error bars are the SD from at least four separate experiments with >200 cells counted each time.
To study the vacuoles at higher resolution, HeLa cells overexpressing HXP-EPI64 were examined by electron microscopy (EM) of thin sections (Figure 2, E–G). Cells without vacuoles have microvilli decorating their edges (Figure 2E), whereas cells containing vacuoles were devoid of microvilli (Figure 2F), an observation we also made at the light microscope level (Figure 2, A–D). Each vacuole is surrounded by a membrane, and we saw no evidence for fusion, as densely packed clusters of vacuoles seem to invaginate with their neighbors (Figure 2G). The vacuoles are devoid of densely staining material. Although we know from our light microscope images that these vacuoles are actin coated, no filaments could be resolved in our EM images.
Our model proposes that EPI64 acts upstream of Arf6, so we tested our hypothesis by exploring the effect of Arf6, constitutively active Arf6-Q67L, and dominant-negative Arf6-T27N on the accumulation of vacuoles with and without EPI64 coexpression. An overview of the results of these experiments, and all subsequent transfection studies, is given together with their statistical significance in Table 1. The vacuole phenotype is found in 29 ± 15% of HXP-EPI64–overexpressing HeLa cells 24 h after transfection, as seen by counting >5300 cells in 28 different preparations. Only 7 ± 5% of cells overexpressing HA-Arf6 contain vacuoles, whereas 30 ± 8% of cells overexpressing HA-Arf6-Q67L have vacuoles and HA-Arf6-T27N–overexpressing cells have none (Figure 2H). Because the percentage of cells that contain vacuoles in HXP-EPI64–overexpressing cells was somewhat variable between experiments, our results are henceforth normalized to the fraction of HXP-EPI64–expressing cells that contain vacuoles within the same experiment (see Materials and Methods). Coexpressing HA-Arf6 together with HXP-EPI64 resulted in an additive effect with 2.1 ± 0.90 times as many cells having vacuoles, as did coexpression of HXP-EPI64 and HA-Arf6-Q67L (2.4 ± 0.92 times as many cells) (Figure 2I; see Supplemental Figure S1E for expression levels). These similar data suggest that EPI64 is stabilizing the GTP-bound form of Arf6, since both constructs give similar results when expressed alone, and that EPI64 contributes to the vacuole phenotype in an Arf6-independent manner, as there is an additive effect when both proteins are coexpressed. When HXP-EPI64 and dominant-negative HA-Arf6-T27N are coexpressed, 39 ± 15% as many cells have vacuoles (Figure 2I). This further suggests that Arf6 lies downstream of EPI64.
TABLE 1:
Quantification of actin-coated vacuoles.
| Percentage of cells with vacuoles | SD | p value | |
|---|---|---|---|
| EPI64 | 29 | 15 | |
| Arf6 | 7 | 6 | 1.28E-02* |
| Arf6-Q67L | 30 | 8 | 3.99E-01* |
| EPI64 constructs | Normalized to EPI64 | ||
| EPI64-LA | 143 | 9 | 9.30E-02† |
| EPI64-R160A | 5 | 6 | 9.80E-09† |
| EPI64-Δ421-453 | 44 | 28 | 2.03E-03† |
| EPI64-Nterm | 0 | 0 | N.D. |
| EPI64-Cterm | 0 | 0 | N.D. |
| EPI64-Nterm + Cterm | 0 | 0 | N.D. |
| Coexpressed with EPI64 | Normalized to EPI64 | ||
| Rab4a | 93 | 20 | 4.38E-01† |
| Rab5a | 104 | 31 | 7.83E-01† |
| Rab7 | 116 | 42 | 4.00E-01† |
| Rab8a | 35 | 22 | 3.39E-12† |
| Rab10a | 105 | 23 | 7.50E-01† |
| Rab11a | 71 | 25 | 1.85E-01† |
| Rab22a | 90 | 13 | 3.02E-01† |
| Rab27a | 84 | 39 | 3.74E-01† |
| Rab35 | 204 | 43 | 5.13E-02† |
| Rab8a-Q67L | 23 | 15 | 3.49E-04† |
| Rab8a-T22N | 72 | 33 | 1.12E-01† |
| MICAL-L1 | 113 | 14 | 2.47E-01† |
| Rabin8 | 25 | 10 | 7.23E-04† |
| JFC1 | 171 | 77 | 4.42E-02† |
| JFC1-E41A | 94 | 16 | 5.54E-01† |
| Arf6 | 214 | 90 | 8.53E-02† |
| Arf6-Q67L | 243 | 93 | 5.39E-02† |
| Arf6-T27N | 39 | 15 | 3.95E-03† |
| Coexpressed with EPI64-Δ421-453 | Normalized to EPI64 | ||
| Myc-JFC1 | 106 | 16 | 1.44E-03‡ |
| Myc-JFC1-E41A | 33 | 14 | 6.28E-01‡ |
| mCh-Rab8a | 4 | 4 | 2.25E-02‡ |
| Coexpressed with Arf6-Q67L | Normalized to Arf6-Q67L | ||
| Rab8a | 15 | 10 | 4.71E-04§ |
| Rab8a-Q67L | 29 | 25 | 1.10E-02§ |
| Rab8a-T22N | 133 | 37 | 1.81E-01§ |
| Coexpressed with Arf6 | Normalized to Arf6-Q67L | ||
| Rab8a | 4 | 2 | 2.72E-02¶ |
| Rab8a-Q67L | 8 | 10 | 1.18E-01¶ |
| Rab8a-T22N | 69 | 24 | 1.18E-02¶ |
*p value, two-sample equal variance to EPI64.
†p value, two-sample unequal variance to normalized EPI64.
‡p value, two-sample equal variance to EPI64-Δ421-453.
§p value, two-sample unequal variance to normalized Arf6-Q67L.
¶p value, two-sample equal variance to Arf6.
EPI64 GAP activity is required for the accumulation of actin-coated vacuoles
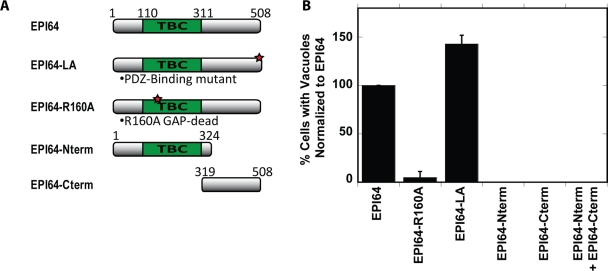
To determine what features of EPI64 are required to induce the accumulation of vacuoles, we explored the effects of expressing different constructs of EPI64 (Figure 3A; see Supplemental Figure S2, A–F, for images and expression levels). Cells expressing full-length HXP-EPI64 or HXP-EPI64-LA, a mutant in which the addition of a C-terminal alanine blocks EPI64's interactions with EBP50 (Hanono et al., 2006), robustly formed vacuoles (Figure 3B). Therefore the linkage of EPI64 to EBP50, and consequently to ezrin, is not necessary for inducing the vacuole phenotype and might even be hindering it slightly. Both ezrin and EBP50 were found localized to vacuoles formed when overexpressing HXP-EPI64-LA, indicating an EPI64-independent recruitment mechanism (data not shown). Expression of HXP-EPI64-R160A, which has a mutation in the arginine finger that destroys GAP activity (Itoh and Fukuda, 2006), induced vacuoles in very few cells (5 ± 6%; Figure 3B). The R160A mutation does not affect the interaction between Arf6 and EPI64 (Hanono et al., 2006), indicating that the EPI64 GAP activity is needed to induce the accumulation of vacuoles independent of binding to Arf6. To confirm this, an alternative mutation was made to inactivate the GAP activity of EPI64 (EPI64-D157A; Itoh and Fukuda, 2006). Expression of this mutant induces vacuoles in a reduced number of cells, similar to the phenotype observed with the R160A mutant, reinforcing the conclusion that the GAP activity is required to induce vacuoles (data not shown). To test whether GAP activity and Arf6-GTP binding are sufficient to induce vacuoles, HXP-EPI64-Nterm containing the TBC domain was overexpressed. This construct was unable to produce vacuoles in cells (Figure 3B), and the expressed protein was cytosolic (Supplemental Figure S2C); however, it was localized to the vacuoles formed in HA-Arf6-Q67L–expressing cells, most likely by association with active Arf6 (Supplemental Figure S2G). Expression of the complementary C-terminal region containing the last 189 residues of EPI64 (HXP-EPI64-Cterm) also failed to induce vacuoles and did not localize to HA-Arf6-Q67L vacuoles (Supplemental Figure S2H). These results implied that vacuole formation requires both the N- and C-terminal domains of EPI64. We tested this hypothesis; however, when these two domains were expressed together, vacuoles were still not formed, indicating that the N- and C-terminal halves of the protein need to be linked to induce vacuole formation. Collectively, these findings indicate that vacuole formation requires the GAP activity of the N-terminal domain linked to the C-terminal domain, but that the C-terminal PDZ binding motif is not necessary.
FIGURE 3:
The GAP activity of full-length EPI64 is required to induce vacuoles. (A) Schematic of EPI64 constructs used in this study. (B) HeLa cells were cotransfected with the HXP-EPI64 constructs listed, and the number of cells with vacuoles was counted. Quantification of the percentage of cells with vacuoles was normalized to cells transfected to express EPI64 alone. Error bars are the SD from at least three separate experiments with >200 cells counted each time.
Rab8a suppresses vacuole accumulation in cells overexpressing EPI64
Because vacuoles are not formed by overexpression of the HXP-EPI64-R160A mutant lacking RabGAP activity, vacuole formation by wild-type EPI64 likely involves inhibition of a target Rab protein. We reasoned that if the target Rab was coexpressed with wild-type EPI64, this might overcome EPI64's RabGAP activity and thereby provide a sufficient level of active Rab to compromise vacuole formation. We used HeLa cells expressing HXP-EPI64 in a screen to identify the target Rab protein by its ability to suppress the actin-coated vacuole phenotype.
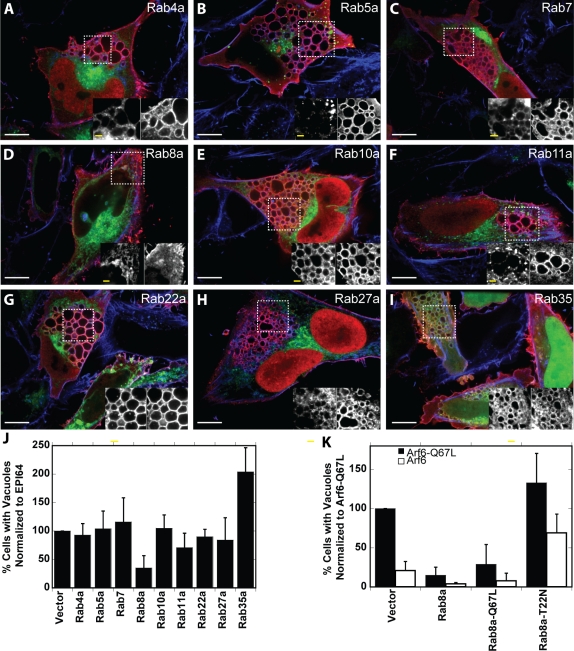
We chose to look at the effect of nine different Rab proteins (Rab4a, 5a, 7, 8a, 10a, 11a, 22a, 27a, 35a) that have been implicated in endocytic pathways (Zerial and McBride, 2001; Grant and Donaldson, 2009). The endosomal compartments with which each of these Rab proteins is associated is shown in the summary diagram later in this paper in Figure 9A. Cells coexpressing HXP-EPI64 and each of the nine GFP-Rab proteins were analyzed by immunofluorescence microscopy both to determine Rab localization (Figure 4, A–I) and to quantitate the percentage of cells that contain vacuoles (Figure 4J). Each of the GFP-tagged Rab proteins was expressed at a similar level and did not influence the expression level of HXP-EPI64 (Supplemental Figure S3A). Of the Rab proteins tested, Rab8a, Rab10a, Rab22a, and Rab35 colocalized with EPI64 on the actin-coated vacuoles (Figure 4, E, G, and I). Rab8a was the only GTPase tested to significantly suppress the vacuole phenotype, with 35 ± 22% of cells having vacuoles relative to HXP-EPI64 expression alone. Of the Rab proteins examined, Rab27a and Rab35 have been shown to be substrates of EPI64 in vitro (Itoh and Fukuda, 2006; Hsu et al., 2010). Expression of GFP-Rab27a had no effect on vacuole formation, whereas expression of GFP-Rab35 significantly enhanced vacuole formation, suggesting that it works antagonistically to Rab8a. These results suggest that Rab8a is a substrate for EPI64. It is interesting to note that a large screen recently tested the Rab substrate specificity of EPI64, with the single exception of Rab8a, due to technical difficulties with its purification (Hsu et al., 2010).
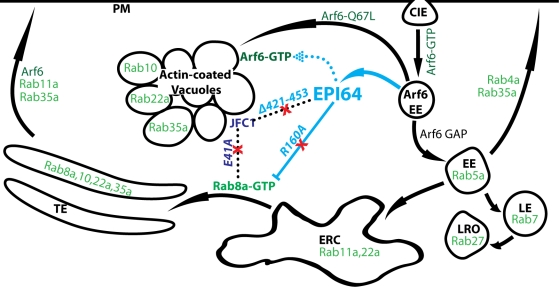
FIGURE 9:
EPI64 induces an accumulation of vacuoles by stabilizing Arf6-GTP and reducing Rab8a-GTP. Model showing the Rab proteins (green) that are involved in membrane-trafficking pathways and how EPI64 is believed to perturb the clathrin independent endocytosis (CIE) pathway (blue). Membrane-trafficking pathways are shown by arrows, and physical interactions are shown by dotted lines. Active Arf6-GTP at the plasma membrane (PM) induces CIE. An actin-coated Arf6 early endosome (EE) must hydrolyze Arf6-GTP to Arf6-GDP before it can merge/mature into the canonical Rab5a EE. The Rab5a EE can then develop into a late endosome (LE) or a lysosome-like organelle (LRO), be immediately recycled back to the PM, or go through the endosomal recycling center (ERC) to the tubular endosome (TE) before being recycled to the PM. Similar to expressing a constitutively active Arf6 (Arf6-Q67L), EPI64 (cyan) diverts this pathway to actin-coated vacuoles by binding to and stabilizing Arf6-GTP (dotted arrow) and by inhibiting Rab8a-GTP (dashed line) through binding to JFC1 (Navy blue). This creates an influx of membrane that is endocytosed through stabilization of Arf6-GTP and at the same time blocks the recycling of membrane to the TE by lowering Rab8a-GTP levels, resulting in an accumulation of actin-coated vacuoles that eventually cause cell death. Red X's mark mutations that suppress the actin-coated vacuoles from accumulating by either inhibiting EPI64 GAP activity (R160A) or disrupting the EPI64-JFC1-Rab8a complex (EPI64-Δ421-453 or JFC1-E41A).
FIGURE 4:
Rab protein colocalization with EPI64 and suppression of vacuoles. (A–I) HeLa cells were cotransfected to express HXP-EPI64 and GFP-Rab proteins (green) for 24 h. Cells were fixed and stained for HXP-tagged EPI64 (red) and F-actin (blue). Insets show magnified regions labeled by dashed boxes, with GFP-Rab protein on left and HXP-EPI64 on right. White scale bars, 10 μm; yellow scale bars in insets, 2 μm. (J) Quantification of the percentage of transfected cells with vacuoles was normalized to cells transfected with EPI64 alone. Error bars are the SD from at least three separate experiments with >200 cells counted each time. (K) HeLa cells were cotransfected to express either Arf6-Q67L (black bars) or Arf6 (white bars) together with vector DNA, Rab8a, Rab8a-Q67L, or Rab8a-T22N. Quantification of the percentage of cells with vacuoles was normalized to cells transfected to express Arf6-Q67L. Error bars are the SD from at least four separate experiments with >200 cells counted each time.
Our data suggest that EPI64 expression stabilizes elevated Arf6-GTP levels to enhance endocytic uptake into the cell. This results in the formation of vacuoles because recycling is inhibited by the RabGAP activity of EPI64 on Rab8a. To test whether a similar model holds for vacuole formation driven by Arf6 overexpression, we explored the effects of mCh-Rab8a expression on vacuole formation by Arf6 and dominant-active Arf6-Q67L (Figure 4K and Supplemental Figure S3B). Vacuole formation in Arf6-Q67L–expressing HeLa cells was greatly reduced by coexpression of mCh-Rab8a or constitutively active mCh-Rab8a-Q67L, whereas expression of GDP-bound mCh-Rab8a-T22N modestly enhanced vacuole formation. Expression of wild-type HA-Arf6 only induces a few vacuoles, but these are almost completely eliminated by mCh-Rab8a or mCh-Rab8a-Q67L expression and significantly enhanced by mCh-Rab8a-T22N expression (Figure 4K). It is important to note that cells overexpressing mCh-Rab8a-T22N by itself never contain vacuoles (data not shown). These data support our hypothesis that increasing Arf6 or diminishing Rab8a-GTP is not enough alone but that both need to be present to disrupt membrane trafficking to accumulate vacuoles.
Rab8a mutants and effector proteins affect vacuole formation by EPI64 overexpression
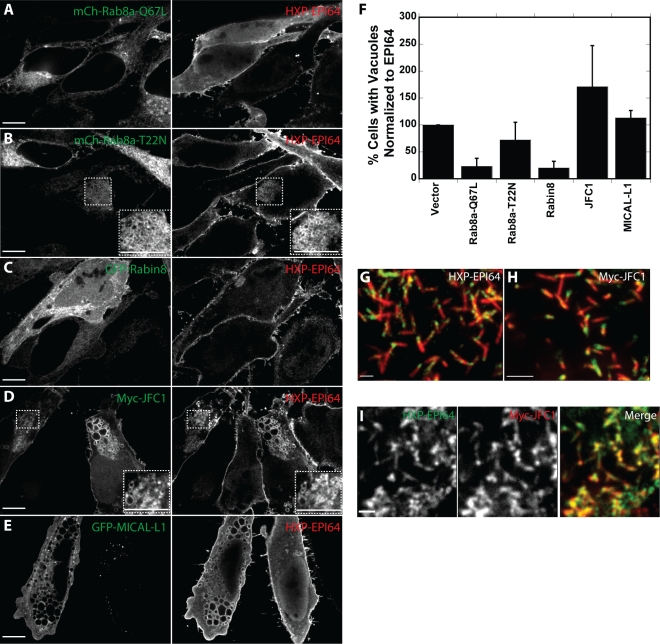
Because Rab8a coexpression with EPI64 reduced the percentage of cells with vacuoles, we explored the effects of modulating active Rab8 levels on this effect by coexpressing Rab8a mutants and Rab8a effectors with EPI64 (Figure 5, A–E, supplemental Figure S4A). Expression of constitutively active mCh-Rab8a-Q67L (Figure 5A) suppressed vacuole formation only slightly more efficiently than overexpression of mCh-Rab8a; expression of inactive mCh-Rab8a-T22N (Figure 5B) had no significant effect (Figure 5F).
FIGURE 5:
Rab8a mutants and associated proteins enhance or suppress the EPI64-induced vacuole phenotype. (A–E) HeLa cells were cotransfected to express mCherry-Rab8a-Q67L (A), mCherry-Rab8a-T22N (B), GFP-Rabin8 (C), Myc-JFC1 (D), or GFP-MICAL-L1 (E) (left) and HXP-EPI64 (right) for 24 h. Insets in B and D show magnified areas designated by dashed boxes. Scale bars, 10 μm. (F) Quantification of the percentage of cells with vacuoles was normalized to cells transfected to express EPI64 alone. Error bars are the SD from at least three separate experiments with >200 cells counted each time. (G, H) JEG-3 cells were transfected to express HXP-EPI64 (G) or Myc-JFC1 (H) and stained for HXP and Myc, respectively, in green, and endogenous ezrin in red. (I) JEG-3 cells transfected with both HXP-EPI64 in green (left) and Myc-JFC1 in red (middle). Scale bars, 2 μm.
Rabin8 is a GEF for Rab8a that elevates the level of active Rab8a by catalyzing the exchange of GTP for GDP (Hattula et al., 2002). Coexpression of GFP-Rabin8 with EPI64 (Figure 5C) greatly reduced the percentage of cells with vacuoles (Figure 5F), consistent with the model that active Rab8a is necessary to cycle membranes from the vacuoles back to the plasma membrane.
Two effectors of Rab8a-GTP were used in coexpression assays with EPI64 to assess their potential role in vacuole formation. JFC1 is a 567-residue protein that binds Rab8a-GTP through its N-terminal synaptotagmin-like protein (Slp) homology domain (SHD; Hattula et al., 2006). JFC1 is a member of the Slp family of proteins and was originally identified as a novel C2 domain protein that interacted with NADPH oxidase (McAdara Berkowitz et al., 2001) and as an effector that binds Rab27a (Strom et al., 2002). The second Rab8a effector we used is molecule interacting with CasL-like 1 (MICAL-L1; Fukuda et al., 2008; Yamamura et al., 2008). To explore whether these effectors influence vacuole formation induced by EPI64 overexpression, we coexpressed each with HXP-EPI64 in HeLa cells.
Overexpression of Myc-JFC1 enhances the percentage of cells with vacuoles compared with cells overexpressing HXP-EPI64 alone (Figure 5F). GFP–MICAL-L1 overexpression had little effect (Figure 5F). Myc-JFC1 was strongly associated with the formed vacuoles (Figure 5D), whereas MICAL-L1 showed a modest association (Figure 5E). The lack of an effect of MICAL-L1 on vacuole formation is not surprising, as it is normally associated with the tubular endosome, where it recruits Eps15 homology domain protein 1 (Sharma et al., 2009) and may be downstream of vacuole formation. By contrast, the localization of JFC1 has not been reported, but we found that it associates with EPI64 at the periphery of HeLa cells. To explore this in more detail, we examined the localization of JFC1 in JEG-3 cells, where it is highly enriched at the base of microvilli, just like EPI64 (Figure 5, G and H). Moreover, HXP-EPI64 and Myc-JFC1 colocalize very well in this region (Figure 5I). Our data indicate that active Rab8 and its effector JFC1 modulate the formation of vacuoles induced by EPI64 overexpression and that JFC1 and EPI64 colocalize.
EPI64 binds to the Rab8a effector JFC1
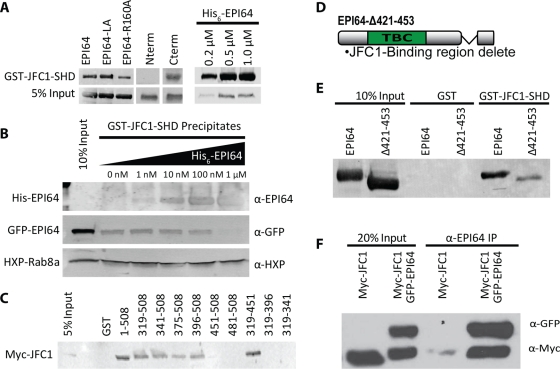
On the basis of the colocalization of JFC1 with EPI64, we explored whether the two proteins interact directly. Initial experiments showed that EPI64 expressed in HeLa cells could be recovered on a construct consisting of glutathione S-transferase (GST) fused to the N-terminal 120 residues of JFC1 encompassing the SHD (denoted GST-JFC1-SHD). GST-JFC1-SHD expressed in bacteria and immobilized on glutathione beads precipitates HXP-EPI64, HXP-EPI64-LA, or HXP-EPI64-R160A from HeLa cell lysates (Figure 6A). To explore which region of EPI64 binds JFC1, we examined the ability of GST-JFC1-SHD to recover HXP-EPI64-Nterm (residues 1–324) or HXP-EPI64-Cterm (residues 319–508). This experiment indicated that JFC1 associates with the C-terminal region of EPI64. Moreover, GST-JFC1-SHD precipitated His6-EPI64 purified from Sf9 cells in a concentration-dependent manner (Figure 6A). Thus the N-terminal domain of JFC1 binds directly to the C-terminal region of EPI64. This was surprising since the C-terminal half of EPI64 is largely uncharacterized, aside from the last four residues, which comprise a PDZ binding motif.
FIGURE 6:
The C-terminal region of EPI64 binds to JFC1. (A) GST-JFC1-SHD was used to assess the binding of HXP-EPI64 constructs expressed in HeLa cells lysate (left) or with recombinant His6-EPI64 purified from insect cells (right). Membranes were blotted with antibody against HXP tag for precipitates from HeLa cell lysates or against EPI64 for the recombinant protein. (B) GST-JFC1-SHD was preincubated with increasing concentrations of His6-EPI64 and then used to precipitate protein from lysate of HeLa cells expressing HXP-Rab8a and GFP-EPI64. Membranes were blotted with antibodies against EPI64, GFP, or HXP as shown. (C) GST or GST-EPI64 truncations immobilized on beads were used to precipitate Myc-JFC1 from HeLa cell lysate. Membranes were blotted with antibodies against Myc-tag. (D) Domain structure of EPI64-Δ421-453 construct designed to inhibit binding to JFC1. (E) GST or GST-JFC1-SHD was used to precipitate overexpressed HXP-EPI64 or HXP-EPI64-Δ421-453 from HeLa cell lysate. Membranes were blotted with antibodies against HXP tag. (F) HeLa cells were transfected to express either Myc-JFC1 or Myc-JFC1 and GFP-EPI64. An antibody against EPI64 was used to immunoprecipitate EPI64 from HeLa cell lysate. Twenty percent lysates or recovered proteins were resolved on SDS–PAGE, and membranes were blotted with antibodies against GFP and Myc.
Rab8-GTP binds to the same N-terminal SHD region of JFC1 that interacts with EPI64. To determine whether JFC1-SHD can interact with Rab8a and EPI64 simultaneously, we used GST-JFC1-SHD to pull down both HXP-Rab8a and GFP-EPI64 from a HeLa cell lysate while titrating in 0–1 μM recombinant His6-EPI64 (Figure 6B). The amount of Rab8a recovered is independent of the amount of His6-EPI64 added into the extract, whereas the amount of GFP-EPI64 recovered diminishes as additional His6-EPI64 is added. Thus recombinant EPI64 competitively inhibits binding of GFP-EPI64 but not HXP-Rab8a, demonstrating that the binding sites on JFC1 for EPI64 and Rab8a are distinct and that Rab8a and EPI64 can bind JFC1 simultaneously.
Our next goal was to identify the region within the C-terminal part of EPI64 (319–508) that is required for interaction with JFC1. We made GST-tagged constructs with truncations from both the N-terminal and C-terminal ends and used them to precipitate Myc-JFC1 from a HeLa cell lysate (Figure 6C). This analysis showed that EPI64 residues 319–396 and 451–508 were not necessary for the interaction (Figure 6C). In additional experiments, GST-JFC1-SHD was able to precipitate GFP-EPI64 residues 319–481 and GFP-EPI64 residues 424–508 from HeLa cells (Supplemental Figure S5A). The combined results suggest that the 28-residue region 424–451 in EPI64 is necessary for binding to JFC1. Consistent with this, HXP-EPI64 lacking residues 421–453 (HXP-EPI64-Δ421-453; Figure 6D) expressed in HeLa cells had a greatly diminished ability to be recovered by GST-JFC1-SHD (Figure 6E).
To determine whether EPI64 and JFC1 interact in vivo, we attempted to coimmunoprecipitate them from JEG-3 cells due to the higher level of endogenous EPI64 in these cells. This proved quite difficult since antibodies for both proteins are limited. Nonetheless, we were able to immunoprecipitate endogenous EPI64 from JEG-3 cells together with a small amount of endogenous JFC1. Moreover, expression of GFP-EPI64 at a level approximately equivalent to that of endogenous EPI64 resulted in about twice as much JFC1 recovered in the EPI64 immunoprecipitate (Supplemental Figure S5B). Because our signal was low using endogenous proteins, we expressed Myc-JFC1 with and without GFP-EPI64 in HeLa cells in an attempt to see a more robust interaction. Endogenous EPI64 immunoprecipitated a trace of Myc-JFC1, whereas in GFP-EPI64–expressing cells, Myc-JFC1 was recovered very efficiently (Figure 6F).
The EPI64-JFC1-Rab8a complex contributes to the vacuole phenotype
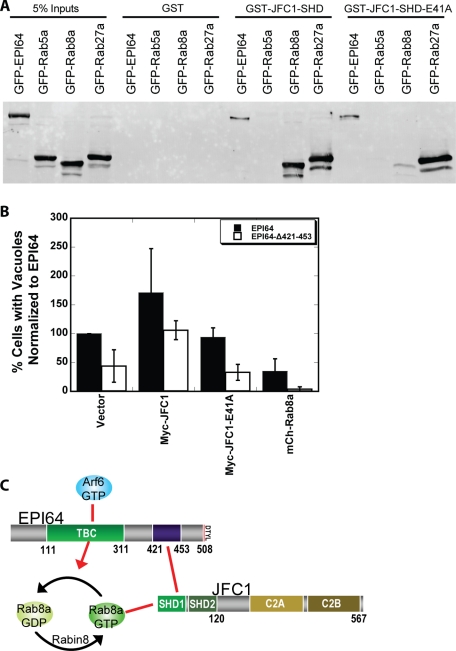
EPI64 binds JFC1 through its SHD, and JFC1 can simultaneously interact with active Rab8a. To test how important each of these interactions is to the formation of vacuoles, we wished to use mutants that uncouple EPI64 from JFC1, and JFC1 from Rab8a. As described, EPI64-Δ421-453 uncouples EPI64 from JFC1; we next sought to uncouple JFC1 from Rab8a. On the basis of the homologous complex of Slac2 bound to Rab27a (Kukimoto-Niino et al., 2008), we designed and made a point mutation in the SHD domain of JFC1 (E41A) that we predicted would disrupt the JFC1–Rab8a interaction. GST-JFC1-SHD or GST-JFC1-SHD-E41A purified on glutathione agarose beads was used to precipitate GFP-tagged EPI64, Rab5a, Rab27a, or Rab8a expressed in HeLa cells (Figure 7A). Both GST-JFC1-SHD constructs precipitated EPI64 and Rab27a but not Rab5a from HeLa cell extracts. By contrast, Rab8a was not precipitated by GST-JFC1-SHD-E41A but was by wild-type GST-JFC1-SHD.
FIGURE 7:
EPI64-JFC1-Rab8a complex is necessary for the vacuole phenotype. (A) GST, GST-JFC1-SHD, or GST-JFC1-SHD-E41A was used to precipitate GFP-EPI64, GFP-Rab5a, GFP-Rab8a, or GFP-Rab27a from HeLa cell lysates. Membranes were blotted with antibodies against GFP-tag. (B) HeLa cells were cotransfected to express either HXP-EPI64 (black bars) or HXP-EPI64-Δ421-453 (white bars) together with vector DNA, Myc-JFC1, Myc-JFC1-E41A, or GFP-Rab8a. Quantification of the percentage of cells with vacuoles was normalized to cells transfected to express EPI64 alone. Error bars are the SD from at least four separate experiments with >200 cells counted each time. (C) Cartoon showing the EPI64-JFC1-Rab8a complex and associated factors.
HXP-EPI64-Δ421-453 and Myc-JFC1-E41A localized similarly to their wild-type counterparts when expressed in HeLa cells (Supplemental Figure S6, A–E). However, HXP-EPI64-Δ421-453 expression was less effective at inducing vacuoles, with only 44 ± 28% of cells containing vacuoles compared with cells expressing HXP-EPI64. When HXP-EPI64-Δ421-453 was coexpressed with GFP-Rab8a, almost none of the cells (4 ± 4%) had vacuoles (Figure 7B). From these results, we suggest a model in which the interaction of EPI64 with JFC1 contributes to vacuole formation in complex with Rab8a (Figure 7C). In support of this model, overexpression of Myc-JFC1 enhanced vacuole formation, yet expression of JFC1-E41A, which cannot bind Rab8a, did not (Figure 7B). It is intriguing that when EPI64-Δ421-453 was coexpressed with JFC1, the percentage of cells with vacuoles was restored to wild-type levels (106 ± 16%), but not when JFC1-E41A was coexpressed (33 ± 14%) (Figure 7B). Thus JFC1 expression might affect the levels of active Rab8a, possibly by sequestering Rab8a-GTP, independent of JFC1's interaction with EPI64. These results show that uncoupling JFC1 from EPI64 (EPI64-Δ421-453) or from Rab8a (JFC1-E41A) reduces the EPI64-induced vacuole phenotype.
EPI64 reduces the levels of Rab8a-GTP in vivo
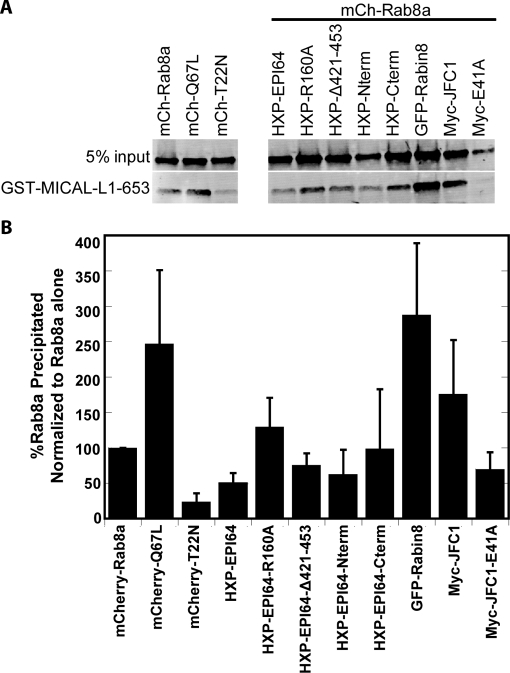
Our results suggest that EPI64 is a RabGAP for Rab8a. Ideally, we would have liked to test this directly with purified Rab protein, but Rab8a has been notoriously difficult to purify from bacteria cells (Bleimling et al., 2009) and is therefore often left out of Rab screens (Hsu et al., 2010). It is also often more useful to measure the effects of GAP activity in vivo, as many RabGAPs are quite promiscuous in vitro, with activity for many Rabs (Fukuda, 2011). Therefore we exploited the ability of the Rab8a effector MICAL-L1 to interact selectively with Rab8a-GTP. The C-terminal 653–863 residues of MICAL-L1 are conserved in the MICAL family of proteins and have been shown to bind Rab8a-GTP (Fukuda et al., 2008). We used bacterially expressed GST-MICAL-L1-653-863 (GST-MICAL-L1-653) immobilized on beads to recover Rab8a-GTP from HeLa cell extracts. We expressed mCh-Rab8a, mCh-Rab8a-Q67L, mCh-Rab8a-T22N, or mCh-Rab8a coexpressed with HXP-EPI64 constructs, GFP-Rabin8, or Myc-JFC1 and measured the amount of Rab8a-GTP recovered on GST-MICAL-L1-653 beads. The assay was validated by showing that GST-MICAL-L1-653 binds to expressed GTP-locked mCh-Rab8a-Q67L much better than to expressed nucleotide-free mCh-Rab8a-T22N (Figure 8A).
FIGURE 8:
EPI64 reduces the levels of Rab8a-GTP in HeLa cells. (A) GST-MICAL-L1-653 was used to precipitate mCherry-Rab8a, mCherry-Rab8a-Q67L, Rab8a-T22N, or mCherry-Rab8a coexpressed with the other proteins listed. MICAL-L1 binds specifically to GTP-bound Rab8a. Membrane was blotted with antibodies against Rab8a. (B) Quantification of the amount of Rab8a-GTP precipitated by GST-MICAL-L1-653 was normalized to the amount of Rab8a-GTP precipitated with only Rab8a overexpressed. Error bars are the SD from at least five experiments.
We used this system to determine whether EPI64 or JFC1 affected the level of active Rab8a in cells (Figure 8B). Coexpression of EPI64 with mCherry-Rab8a resulted in about half the control amount of mCherry-Rab8a-GTP being recovered, whereas coexpression of the GAP-dead EPI64-R160A had no effect. Accordingly, the RabGAP activity of EPI64 is able to reduce the level of Rab8a-GTP in vivo. Coexpression with EPI64-Δ421-453 or EPI64-Nterm resulted in a more modest reduction in mCherry-Rab8a-GTP recovered, and EPI64-Cterm had no significant effect. As expected for a GEF, Rabin8 increased the amount of Rab8a-GTP dramatically. Expression of the Rab8a effector JFC1, but not the JFC1-E41A mutant, also elevated the level of Rab8a-GTP, suggesting that JFC1 may be able to stabilize Rab8a-GTP. This indicates that both HXP-EPI64 and Myc-JFC1 expression have an effect on the levels of Rab8a-GTP but in different directions.
To test whether overexpression of Arf6 or Arf6 mutants had a direct or indirect effect on the regulation of Rab8a GTP levels by EPI64, we cotransfected cells to express mCh-Rab8a and HA-Arf6 with and without EPI64. Coexpression of Arf6, Arf6-Q67L, or Arf6-T27N did not influence the levels of Rab8a-GTP or affect the reduction of Rab8a-GTP levels seen when EPI64 was coexpressed (Supplemental Figure S7).
DISCUSSION
The actin-coated vacuole phenotype induced by the overexpression of Arf6-Q67L has been observed for a decade and has led to many insights into the CIE pathway (Brown et al., 2001). Our findings confirm that Arf6 function is tightly linked to Rab8a, as overexpression of Rab8a is able to suppress the accumulation of these vacuoles. This implies that the CIE pathway is regulated by a balance between these two small GTPases. We show that this balance can be regulated by EPI64, an Arf6 effector and probable Rab8a GAP (Figure 9). We further characterized EPI64 beyond its role as a microvillar maintenance protein to reveal it as a unique Arf6 effector protein found to lower Rab8a-GTP levels through its interactions with the Rab8a effector, JFC1. Similar to Rab8a's GEF, Rabin8, also a Rab11a effector (Knodler et al., 2010), is one of a growing number of examples of an effector/GEF/GAP cascade in membrane trafficking where one GTPase's effector is another GTPase's GEF or GAP. These cascades couple GTP-binding proteins to regulate flux and directionality to membrane trafficking pathways (e.g., Grosshans et al., 2006; for review, see Hutagalung and Novick, 2011).
Overexpression of EPI64 leads to the accumulation of PI(4,5)P2 and Arf6-containing vacuoles, which are coated with actin, EPI64, EBP50, and ezrin. These are all normal components of the plasma membrane, and, as suggested previously, these actin-covered vacuoles probably reflect excessive clathrin-independent endocytosis (Brown et al., 2001). It is clear from previous work in our laboratory that EPI64 is binding to Arf6 and stabilizing Arf6-GTP levels in a manner that is independent of its GAP activity. One possibility is that EPI64 binding locks Arf6-GTP in an active state; a second, more likely possibility is that EPI64 binds to Arf6 and sterically hinders Arf6 from associating with its GAP, delaying the hydrolysis of bound GTP to GDP. Without access to its GAPs, ACAP1 or ACAP2, Arf6 remains active as the actin-coated vacuoles bud from the plasma membrane into the cell. With Arf6 in its GTP-bound active state, PI(4,5)P2 concentrations remain high and the actin coat stays associated with the vacuoles. This prevents Rab5a recruitment to the vacuoles and further trafficking to the canonical Rab5a and EEA1 early endosomal pathways. Instead, these vacuoles cluster together and eventually overwhelm the normal functions of the cell, as it ceases to proliferate and eventually dies after ∼48 h.
We also observed that Rab8a, Rab10a, Rab22a, and Rab35a—all Rab proteins normally associated with the tubular endosome (Weigert et al., 2004; Kouranti et al., 2006; Roland et al., 2009)—are found associated with EPI64-induced, actin-coated vacuoles. This suggests that the actin-coated vacuoles are precursors of the tubular endosomes. Rab8a is a well-established marker of tubular endosomes (Roland et al., 2007; Sharma et al., 2009) and the only Rab protein that we observed to suppress vacuole accumulation and whose activity is diminished by EPI64 overexpression. Therefore it seems likely that the vacuoles are derived from the plasma membrane along the CIE pathway and have arrested as a precursor to the tubular endosome due to Rab8a inactivation. If this is the case, it implies that steps influenced by Rab10a, 22a, and 35a all occur prior to the Rab8a-dependent step. Moreover, the presence of these Rab proteins on the vacuoles is not enough to recycle these trapped membranes back the plasma membrane; only Rab8a was observed to suppress this phenotype. Combining our data with these earlier studies suggests that EPI64 induces vacuoles to accumulate due to enhanced endocytosis driven by elevated Arf6-GTP levels, together with the inhibition of the Rab8a pathway that tubulates membranes to facilitate recycling.
The TBC domain and PDZ-binding motif of EPI64 were identified from the sequence analyses undertaken when the protein was first purified from human placenta (Reczek and Bretscher, 2001). The C-terminal half of EPI64, responsible for linking the TBC domain with the last four PDZ-binding residues, was uncharacterized. Using the secondary structure threading algorithm PHYRE, we found no homology of this region of EPI64 to structures in the Protein Data Bank; moreover, this ∼200-residue stretch is predicted to be mainly unstructured. Although no apparent information is available about the secondary structure, the primary sequence within this region from residues 384–487 shows a large percentage of prolines (22%) and charged residues (25%). Polyproline motifs have often been identified as protein–protein interaction domains (Rath et al., 2005), but no proteins were previously identified to bind this region in EPI64. We found that EPI64 residues 421–453 were required to bind the SHD domain of JFC1, a domain known to be a Rab8a and Rab27 effector (Hattula et al., 2006). Furthermore, a construct of EPI64 with this region deleted had a significant loss of binding to JFC1. Our data show that binding of EPI64 to JFC1 enhances the induction of actin-coated vacuoles. This suggests that the binding of JFC1 to the C-terminal half of EPI64 is necessary to bring the TBC domain of EPI64 in close proximity to Rab8a to hydrolyze its bound GTP. As far as we are aware, this is the first example of a Rab effector binding to the relevant GAP protein to potentially enhance deactivation of the Rab.
JFC1 has localization very similar to that of EPI64, except that JFC1 is found at the tubular endosome and EPI64 is not. Here we identified JFC1 as a Rab8a-binding protein that recruits an Arf6 effector and possible Rab8a GAP, EPI64. This could lead to a negative feedback loop that increases the rate of Rab8a cycling. Active Rab8a could recruit JFC1, which can simultaneously associate with EPI64 to inactive Rab8a. This complex could reduce the amount of time that Rab8a is active at the apical membrane, where EPI64 is normally found localized to the base of microvilli. Because we do not see EPI64 present at the tubular endosome, Rab8a cycling rates may differ here greatly.
EPI64 is localized to the apical membrane and associates with the base of microvilli. Disrupting the interactions between the complex of microvillar components, EPI64-EBP50-ezrin, either by reducing endogenous levels of EPI64 through siRNA (this study) or expressing EPI64 uncoupled from the microvillar-localized EBP50-ezrin (Hanono et al., 2006), results in a reduction of microvilli; similarly, disrupting the interactions of the EPI64-JFC1-Rab8a complex leads to a loss of actin-coated vacuoles induced by EPI64 overexpression. It is through these multiprotein complexes that cells are able to precisely tune membrane structures and trafficking. This also suggests that altering the balance or location of Arf6/Rab8a activities compromises the presence of microvilli on cells. In addition, when EPI64 is overexpressed to induce actin-coated vacuoles, there is also a large reduction in microvilli, as seen both by immunofluorescence microscopy and thin-section electron microscopy. One of the main functions of microvilli is to increase the surface area of the cell for nutrient uptake. When there is a large influx of membrane into the cell that is not being recycled back to the plasma membrane, the cell must compensate somehow. We do not see a dramatic decrease in the overall size of the cell; this suggests that the cell is compensating for the influx of membranes by losing its microvilli. These data imply that two aspects of EPI64 function contribute to the presence of microvilli: first, its intrinsic ability to regulate the endocytic pathway, and second, the spatial localization where it confers this regulation. In the context of a polarized cell, this model would propose that EPI64 catalyzes uptake from the apical membrane through stabilizing Arf6-GTP and inhibits recycling in this region by deactivating Rab8a-GTP. Very little is known about how cells regulate the presence of microvilli on their surface, but our studies reveal that endocytic pathways are likely to be a crucial aspect of this regulation.
MATERIALS AND METHODS
siRNA, DNA constructs, and cloning
Many DNA constructs were obtained from generous scientists across the globe. Steve Caplan (University of Nebraska Medical Center, Omaha, NE) supplied the GFP-MICAL-L1 construct, Jim Goldenring (Vanderbilt University, Nashville, TN) supplied the mCherry-Rab8a and mCherry-Rab10 constructs, Genaro Patino-Lopez (National Institutes of Health, Bethesda, MD) supplied the GFP-Rab35 construct, and Johan Peranen (University of Helsinki, Helsinki, Finland) supplied the Myc-JFC1, GST-JFC1-SHD, and GFP-Rabin8 constructs. Ira Mellman (Yale University, New Haven, CT) supplied the Rab4a, Rab5a, and Rab11a constructs. Craig Roy (Yale University) supplied the Rab7 construct. John Hammer (National Institutes of Health) supplied the Rab27a construct. Rab22a was purchased as a clone from Open Biosystems (Thermo Biosystems, Huntsville, AL). The siRNAs targeting human EBP50 (5′-CGGCGAAAACGTGGAGAAG-3′), and luciferase GL2 (5′-CGUACGCGGAAUACUUCGA-3′) were obtained from ThermoFisher Scientific (Waltham, MA) and Applied Biosystems (Foster City, CA). SiRNA targeting human EPI64 (5′-CCACCGACGAACUCAGCUC-3′) was obtained from Dharmacon RNA technologies (Lafayette, CO). All other constructs were either described earlier (Hanono et al., 2006) or generated by PCR, QuikChange II mutagenesis by Stratagene (Santa Clara, CA), or subcloning techniques.
Antibodies and Western blotting
Protein samples were resolved by SDS–PAGE and transferred to Immobilon-FL (Millipore, Billerica, MA). All Western blots were imaged using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). Affinity-purified antibodies against human ezrin and EBP50 were described previously (Bretscher, 1989; Reczek and Bretscher, 1998). His6-tagged EPI64 was expressed and purified from Sf9 insect cells and used to elicit antibody production in rabbits from Pocono Rabbit Farm and Laboratory (Canadensis, PA). The antisera were affinity purified over GST-EPI64 expressed and purified from Escherichia coli bacteria and recognized a specific band at 64 kDa from cell extracts. Antibodies were purchased as follows: against the Xpress (HXP) epitope from Invitrogen or Santa Cruz Biotechnology (Santa Cruz, CA), against the HA epitope from Covance (Berkeley, CA), against the Myc epitope from Roche (Indianapolis, IN), against human Rab8a from BD Transduction Laboratories (Lexington, KY), against JFC1 and GFP from Santa Cruz Biotechnology, and against transferrin receptor from Molecular Probes (Invitrogen, Carlsbad, CA). Antibodies to MHC1 were purchased as an Alexa Fluor 488–conjugated HLA-A,B,C clone W6/32 from BioLegend (San Diego, CA). Alexa Fluor–conjugated secondary antibodies, Alexa Fluor 660–conjugated phalloidin, and Alexa Fluor 488–conjugated transferrin were purchased from Invitrogen. Horseradish peroxidase–conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA), and IRDye 680- and 800-conjugated secondary antibodies were from LI-COR Biosciences.
Scanning electron microscopy
Cells were grown and processed on 35-mm dishes. Fixation was accomplished using 2% glutaraldehyde in 0.1 M sodium cacodylate buffer for 1.5 h. Cells were then washed three times in 0.1 M sodium cacodylate buffer. After the final rinse, cells were postfixed on ice in 1% osmium tetroxide for 1 h. The cells were then rinsed again as described and dehydrated in a graded ethanol series. The cells were removed from the dishes using propylene oxide and pelleted in an Eppendorf tube. The cell pellet was rinsed two times in propylene oxide. The propylene oxide was exchanged for 1:1 propylene oxide/Spurr's resin for 30 min on a Ferris wheel. The mixture was exchanged for 100% Spurr's for overnight rotation on a Ferris wheel. The next day the resin was exchanged several times. After the final change, the resin was polymerized at 60°C. Thin sections were cut on a Leica UC6 ultramicrotome (Leica Microsystems, Buffalo Grove, IL) and stained with lead citrate and uranyl acetate. Grids were examined on an FEI (Hillsboro, OR) Morgagni transmission electron microscope. All images were taken with a digital camera by Hamamatsu (Hamamatsu, Japan), using software by Advanced Microscopy Techniques (Woburn, MA).
Transfections and immunofluorescence
HeLa and JEG-3 cells (American Type Culture Collection, Manassas, VA) were maintained as described (Hanono et al., 2006). HeLa cells were transfected for 24 h with DNA using Lipofectamine 2000 (Invitrogen) or for 48 h with siRNA using Lipofectamine RNAiMax (Invitrogen). JEG-3 cells were transfected for 48 h with siRNA using Lipofectamine RNAiMax and/or for 24 h with DNA using polyethylenimine. For immunofluorescence, cells grown on glass coverslips were fixed in 3.7% formaldehyde/phosphate-buffered saline (PBS) for 10 min. Cells were permeabilized in 0.2% Triton X-100 PBS for 5 min at room temperature and incubated in first primary, then secondary antibodies and additional markers (phalloidin) in 3% fetal bovine serum (FBS) in PBS. Coverslips were mounted onto glass slides using Vectashield (H-1000; Vector Laboratories, Burlingame, CA), and images were then acquired on a CSU-X spinning disk microscope (Intelligent Imaging Innovations, Denver, CO) with spherical aberration correction device, 63×, 1.4 numerical aperture objective on an inverted microscope (model DMI6000B; Leica) and an HQ2 CCD camera (Photometrics, Tucson, AZ), using SlideBook 5.0 (Intelligent Imaging Innovations). Images were also processed using SlideBook 5.0.
Endocytosis and recycling assays
For measuring initial rates of endocytosis by flow cytometry, cells were first washed twice with PBS containing 10 mM EDTA and incubated for 30 min in serum-free suspension minimal essential medium (SMEM) to force cells into suspension. Cells were collected and washed in cold, serum-free SMEM to halt endocytosis before nutating for 30 min at 4°C in cold, serum-free SMEM with 0.4 μg/ml MHC1 antibody conjugated to Alexa Fluor 488. Cells were washed twice in cold PBS containing 10 mM EDTA and then incubated with 37°C SMEM containing 10% FBS for the indicated time points. Cells were then washed in cold acid buffer to remove surface-bound antibody for 30 s before being washed in cold PBS and fixed in 3.7% formaldehyde for 10 min. Cells were then analyzed for the amount of fluorescent MHC1 internalized using a Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Brea, CA). For qualitative immunofluorescence cells all steps used PBS with no EDTA and MEM to allow cells to remain adherent to coverslips.
Scoring the vacuole phenotype of transfected HeLa cells
Transfected HeLa cells were viewed by immunofluorescence, and each transfected cell was scored for the presence of vacuoles or not. At least 200 cells were counted on each slide, and a control cell transfected with EPI64 was always included in the set of coverslips to be scored (or Arf6-Q67L for Figure 4K). The percentage of cells with vacuoles for each coverslip in a set was then normalized to the EPI64 coverslip in the same set by dividing the percentage of cells with vacuoles by the percentage of EPI64-transfected cells with vacuoles and multiplying by 100. This resulted in EPI64-transfected cells always being normalized to 100%. Because there was no variance for normalized EPI64, Student's t tests were done with a two-tailed distribution, assuming a two-sample unequal variance. p values are given in Table 1.
Scoring microvilli in Jeg-3 cells
Transfected JEG-3 cells were scored as described previously (Hanono et al., 2006). In brief, cells were transfected with siRNA for 48 h and then fixed and stained for ezrin. At least 200 cells were counted for each siRNA from four separate experiments. Cells were scored as having normal microvilli, fewer microvilli, or no microvilli.
Protein purifications
Expression of GST proteins was induced in Rosetta2 DE3 PlysS bacteria grown in Terrific Broth and cells disrupted by sonication in 500 mM NaCl, 20 mM Tris, pH 7.4, 0.2 mM EDTA, 1 mM dithiothreitol, and complete protease inhibitor, then cleared by centrifugation and affinity purified by binding to hydrated glutathione agarose (Sigma-Aldrich, St. Louis, MO). His-EPI64 was expressed in Sf 9 insect cells, which were then lysed in 25 mM Tris pH 7.4, 25 mM imidazole, 300 mM NaCl, 1 mM β-mercaptoethanol (β-ME), 1% Igepal, and a protease complete tablet by five strokes in a Dounce homogenizer and sonication. Protein was first purified over a HisTrapFF column on the fast protein liquid chromatography (GE Healthcare, Piscataway, NJ), followed by further purification over a HiTrapFFQ column (GE Healthcare).
In vitro binding assay
Purified GST-tagged proteins attached to glutathione agarose beads were used to precipitate expressed proteins out of HeLa cell lysate. HeLa cells grown on tissue culture plates were placed on ice and lysed in 100 mM NaCl, 20 mM Tris, pH 7.4, 1% Triton X-100, 5% glycerol, 5 mM β-ME, and a complete protease inhibitor by scraping. Cell lysates were cleared by centrifugation and nutated with 20 μl of glutathione agarose beads with attached GST-tagged protein. Beads were washed three times and resuspended in SDS 2X sample buffer and then boiled to elute the recovered protein.
In vivo Rab8a-GTPase assay
The in vitro binding assay was repeated using GST-MICAL-L1-653 attached to glutathione beads with the following changes: 5 mM MgCl2 was added to all buffers to preserve the GTP-bound state of Rab8a. Glutathione beads with attached GST-MICAL-L1-653 were washed well to remove any residual EDTA. The entire pull-down experiment was completed in 45 min to minimize the amount of Rab8a-GTP that was hydrolyzed to Rab8a-GDP after lysis.
Supplementary Material
Acknowledgments
We are very grateful to Steve Caplan, Jim Goldenring, Johan Peranen, Ira Mellman, and Craig Roy for providing plasmids, Janet Ingraffea (Cornell University, Ithaca, NY) for providing insect cells infected to express EPI64, and Bret Judson (Cornell University) for doing the electron microscopy. This work was supported by National Institutes of Health Ruth L. Kirschstein National Research Service Award GM-087959 (to D.H.) and National Institutes of Health Grant GM-36652-25 (to A.B.).
Abbreviations used:
- ClE
clathrin-independent endocytosis
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- mCh
mCherry
- MHC1
major histocompatibility complex 1
- MV
microvilli
- SHD
synaptotagmin-like protein (Slp) homology domain
- TBC
Tre-2/Bub2/Cdc16
- Tfn
transferrin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0521) on January 4, 2012.
REFERENCES
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleimling N, Alexandrov K, Goody R, Itzen A. Chaperone-assisted production of active human Rab8A GTPase in Escherichia coli. Protein Expr Purif. 2009;65:190–195. doi: 10.1016/j.pep.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Burns ME, Gatti E, Augustine GJ, De Camilli P. Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J. 1994;13:5547–5558. doi: 10.1002/j.1460-2075.1994.tb06892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem. 2006;281:16147–16156. doi: 10.1074/jbc.M601467200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Porat-Shliom N, Cohen LA. Clathrin-independent endocytosis: a unique platform for cell signaling and PM remodeling. Cell Signal. 2009;21:1–6. doi: 10.1016/j.cellsig.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem. 2003;278:15373–15380. doi: 10.1074/jbc.M212341200. [DOI] [PubMed] [Google Scholar]
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Nichols BJ. Molecular mechanisms of clathrin-independent endocytosis. J Cell Sci. 2009;122:1713–1721. doi: 10.1242/jcs.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Arffman A, Peranen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13:3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Honda A, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Howes MT, Mayor S, Parton RG. Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Curr Opin Cell Biol. 2010;22:519–527. doi: 10.1016/j.ceb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Hsu C, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Bilan PJ, Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem Biophys Res Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- Itoh T, Fukuda M. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J Biol Chem. 2006;281:31823–31831. doi: 10.1074/jbc.M603808200. [DOI] [PubMed] [Google Scholar]
- Jovanovic OA, Brown FD, Donaldson JG. An effector domain mutant of Arf6 implicates phospholipase D in endosomal membrane recycling. Mol Biol Cell. 2006;17:327–335. doi: 10.1091/mbc.E05-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008;16:1478–1490. doi: 10.1016/j.str.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Linder MD, Uronen RL, Holtta-Vuori M, van der Sluijs P, Peranen J, Ikonen E. Rab8-dependent recycling promotes endosomal cholesterol removal in normal and sphingolipidosis cells. Mol Biol Cell. 2007;18:47–56. doi: 10.1091/mbc.E06-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdara Berkowitz JK, Catz SD, Johnson JL, Ruedi JM, Thon V, Babior BM. JFC1, a novel tandem C2 domain-containing protein associated with the leukocyte NADPH oxidase. J Biol Chem. 2001;276:18855–18862. doi: 10.1074/jbc.M011167200. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Weigert R, Donaldson JG. Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol Biol Cell. 2003;14:417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen J, Auvinen P, Virta H, Wepf R, Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahajeng J, Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13:82–93. doi: 10.1111/j.1600-0854.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath A, Davidson AR, Deber CM. The structure of “unstructured” regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80:179–185. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher A. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J Cell Biol. 2001;153:191–206. doi: 10.1083/jcb.153.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland JT, Lapierre LA, Goldenring JR. Alternative splicing in class V myosins determines association with Rab10. J Biol Chem. 2009;284:1213–1223. doi: 10.1074/jbc.M805957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Pust S, Skotland T, van Deurs B. Clathrin-independent endocytosis: mechanisms and function. Curr Opin Cell Biol. 2011;23:413–420. doi: 10.1016/j.ceb.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Sato T, et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448:366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- Wandinger-Ness A, Deretic D. Rab8a. 2008. UCSD Nature Molecule Pages 24 April, doi:10.1038/mp.a001997.01.
- Weigert R, Yeung AC, Li J, Donaldson JG. Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol Biol Cell. 2004;15:3758–3770. doi: 10.1091/mbc.E04-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19:971–983. doi: 10.1091/mbc.E07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.