Abstract
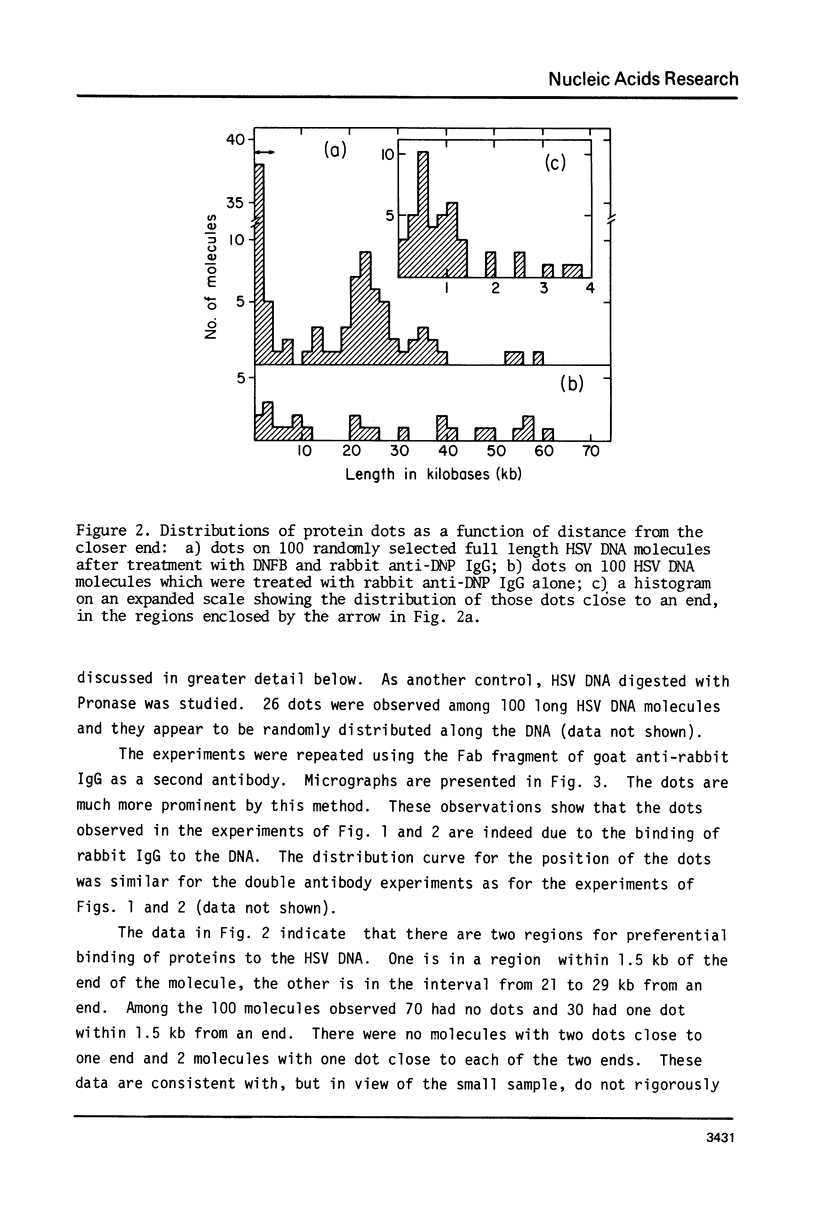
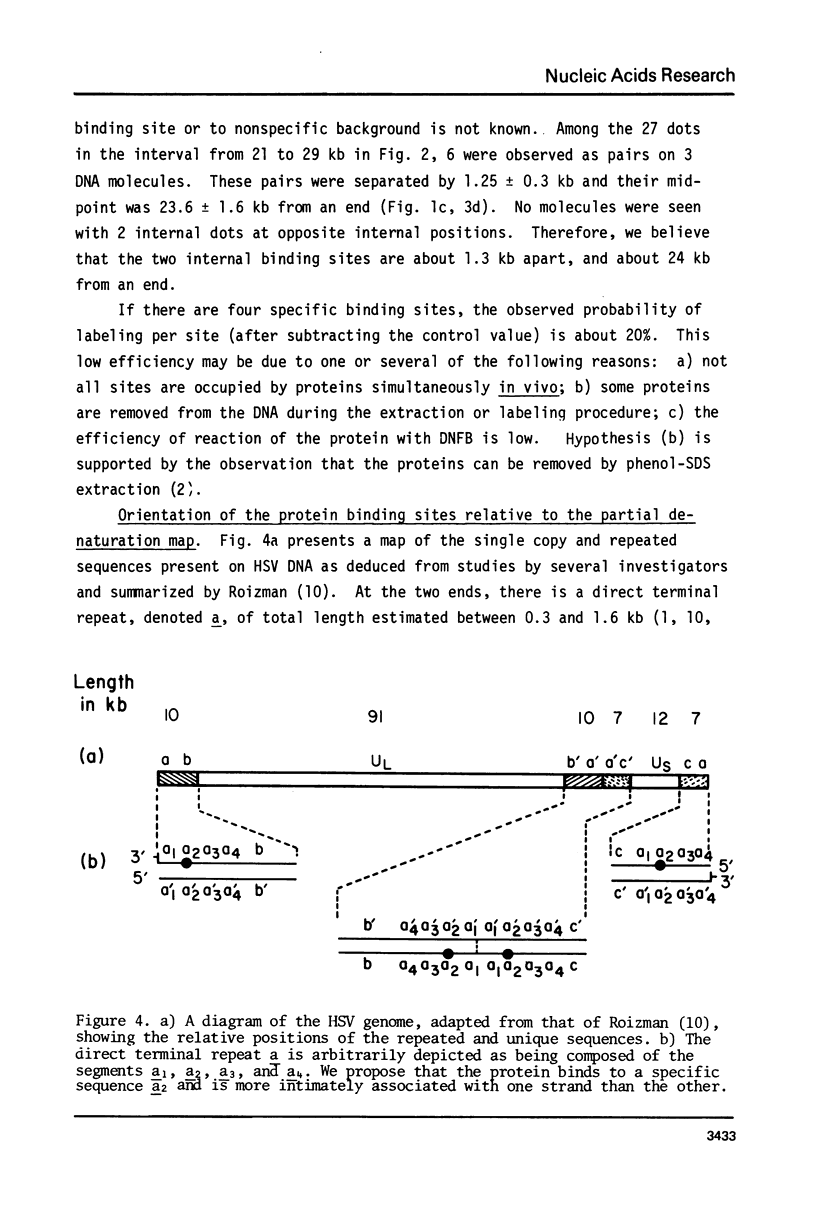
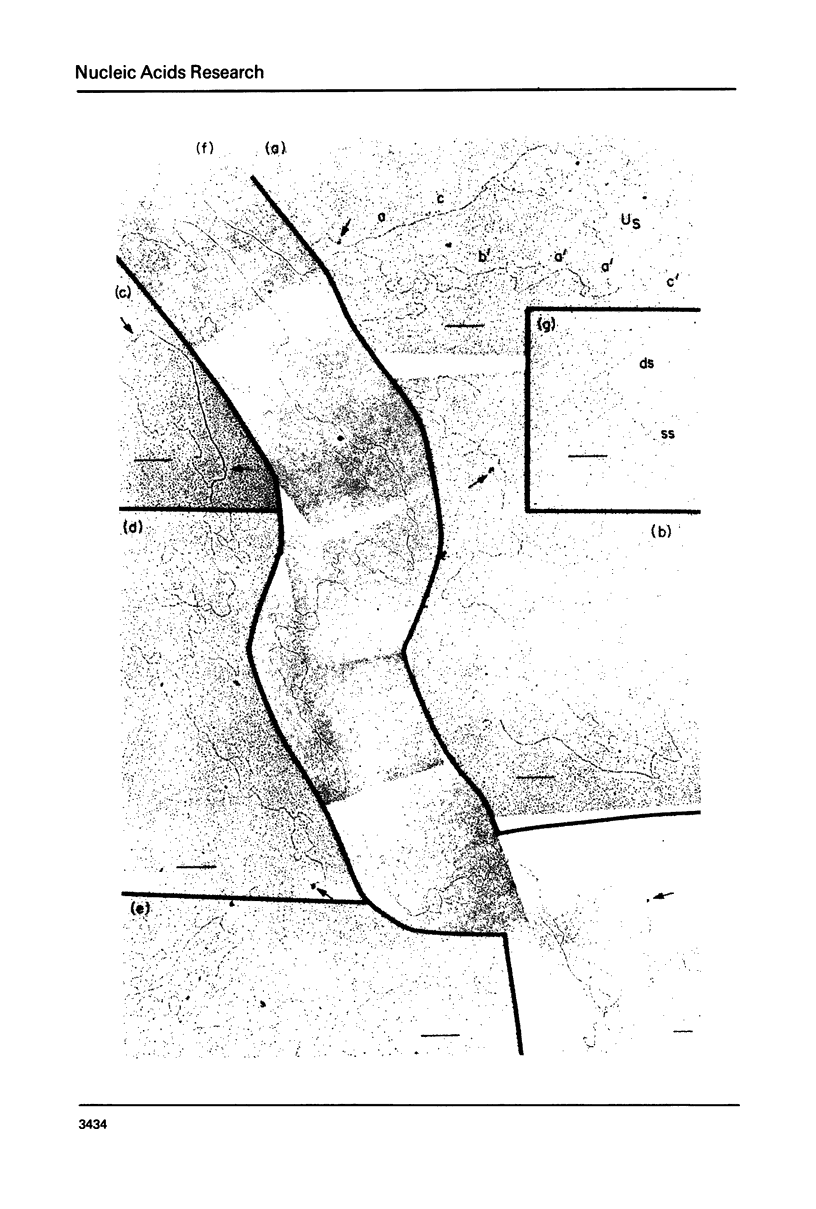
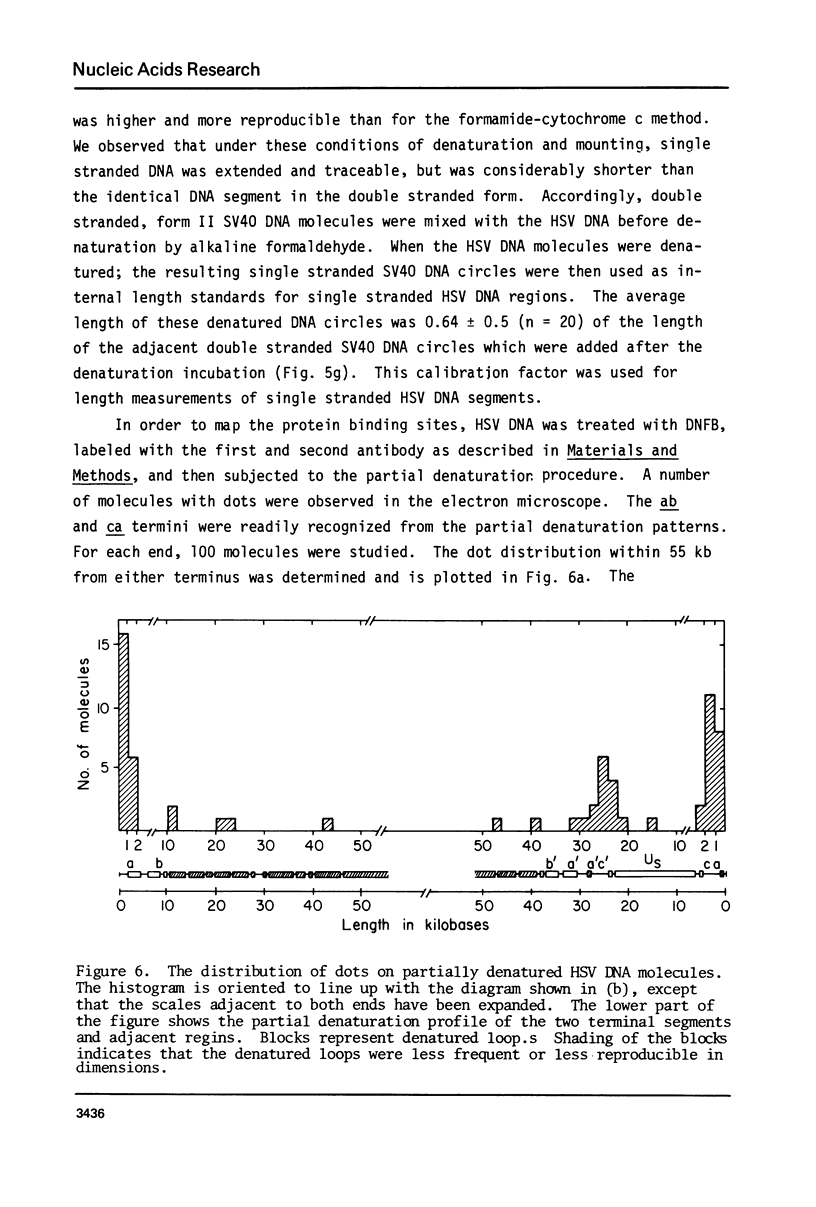
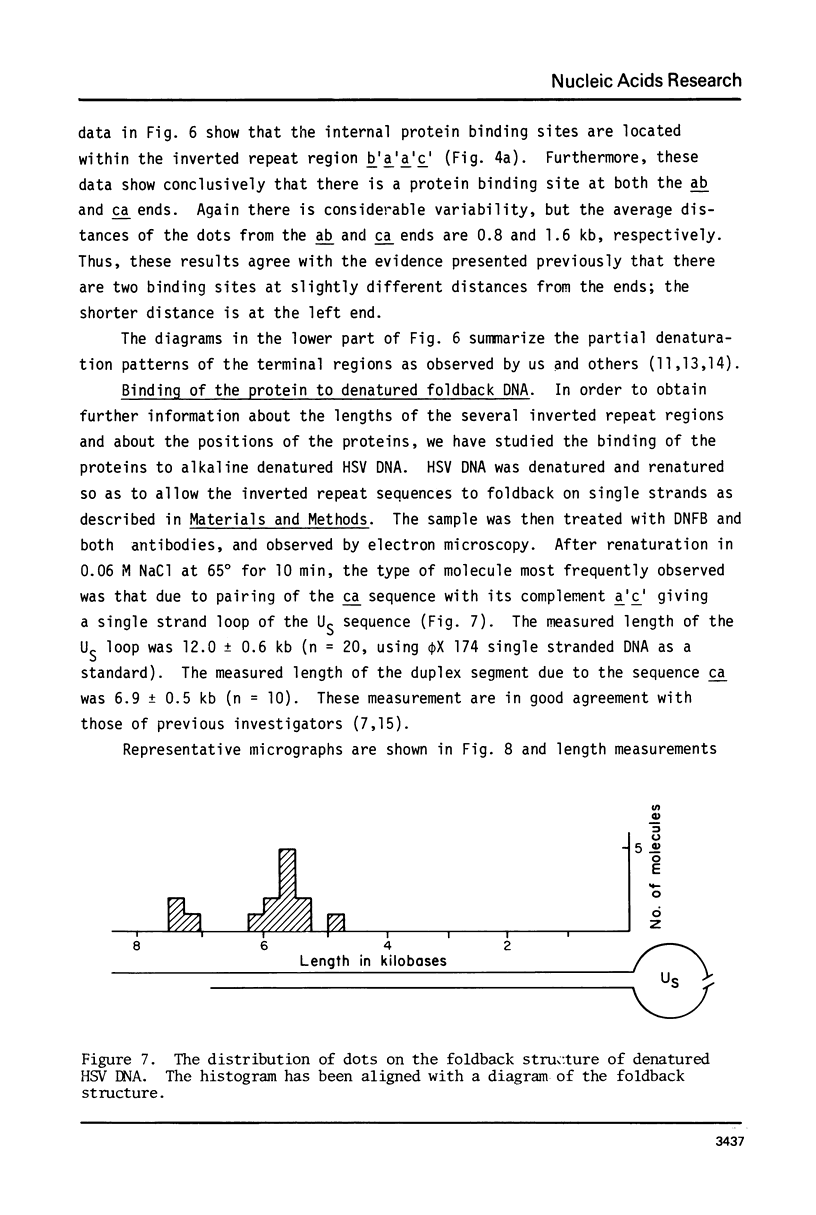
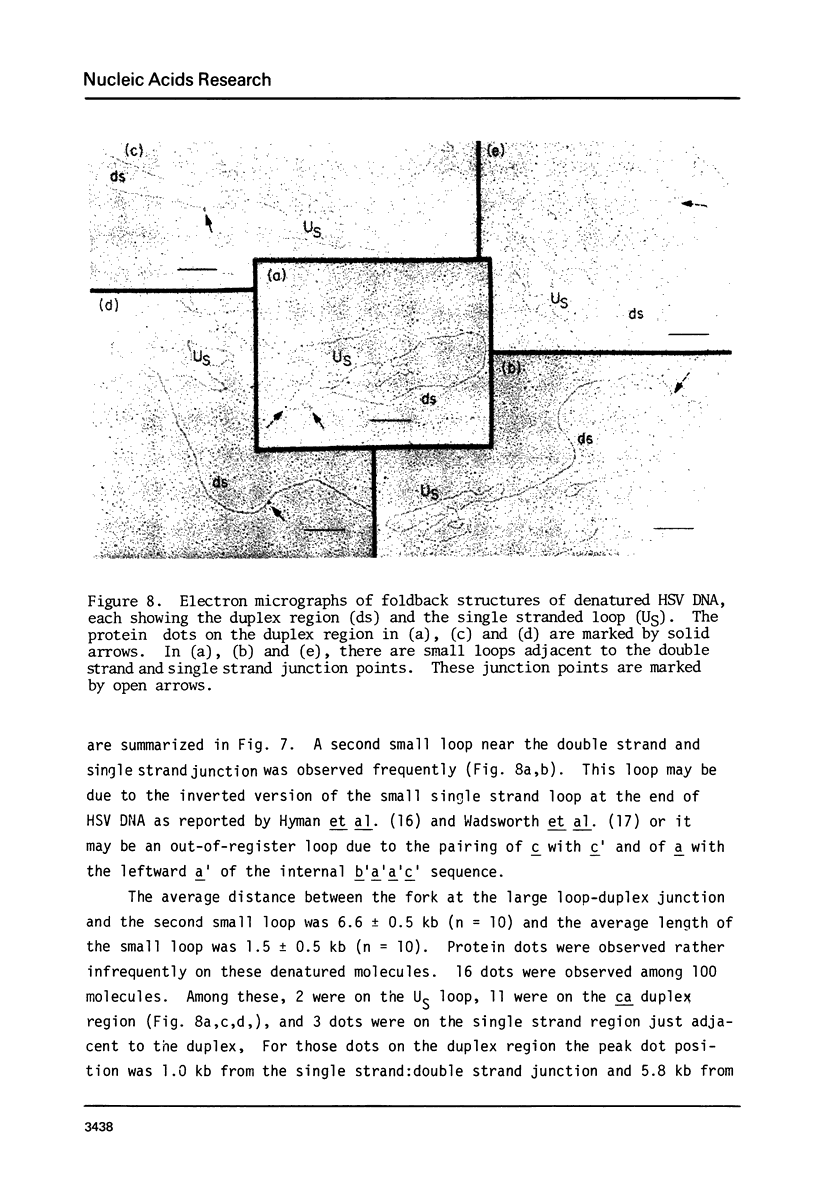
Exonuclease digestion experiments have suggested that there is a protein(s) bound close to one or both ends of herpes simplex virus-1 (HSV) DNA. The existence of such bound proteins has been positively demonstrated and their positions on the HSV genome determined by application of a newly developed method for electron microscopic mapping of proteins bound to nucleic acids. Purified HSV DNA was treated with dinitrofluorobenzene under conditions that covalently attach the dinitrophenyl (DNP) group to the proteins in a protein-nucleic acid complex. The HSV DNA-protein-(DNP)n complex was treated with rabbit anti-DNP IgG, and, in some cases, additionally treated with monovalent Fab fragments of goat anti-rabbit IgG, and mounted for examination in the electron microscope. Electron opaque dots representing the protein-(DNP)n-(IgG)m complex were seen on the HSV DNA. Direct measurements of the positions of the protein, as well as partial denaturation mapping, indicate that there are four positions for protein bound to HSV DNA: two near but not at the two ends and two at sites corresponding to the internal inverted repeats of the ends. These results suggest that there is a specific protein binding sequence within the direct terminal repeat of HSV DNA. The previous observation that HSV DNA is more sensitive to digestion by a 3' than by a 5' exonuclease then indicates that the bound protein(s) is more intimately associated with one strand of the specific sequence than with the complementary strand.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann J. M. A model for replication of the ends of linear chromosomes. Nucleic Acids Res. 1976 Nov;3(11):3167–3171. doi: 10.1093/nar/3.11.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Kudler L., Hyman R. W. Exonuclease III digestion of herpes simplex virus DNA. Virology. 1979 Jan 15;92(1):68–81. doi: 10.1016/0042-6822(79)90215-0. [DOI] [PubMed] [Google Scholar]
- Pater M. M., Hyman R. W., Rapp F. Isolation of herpes simplex virus DNA from the "hirt supernatant". Virology. 1976 Dec;75(2):481–483. doi: 10.1016/0042-6822(76)90046-5. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Davidson N. An electron microscopic method for the mapping of proteins attached to nucleic acids. Nucleic Acids Res. 1978 Aug;5(8):2825–2846. doi: 10.1093/nar/5.8.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Davidson N., Wimmer E. An electron microscope study of the proteins attached to polio virus RNA and its replicative form (RF). Nucleic Acids Res. 1978 Dec;5(12):4711–4723. doi: 10.1093/nar/5.12.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]