Abstract
Simian Virus 40 usesthe large T antigen (Tag) to bind and inactivate retinoblastoma tumor suppressor proteins (Rb), which can result in cellular transformation. Tag is a modular protein with four domains connected by flexible linkers. The N-terminal J domain of Tag is necessary for Rb inactivation. Binding of Rb is mediated by an LXCXE consensus motif immediately C-terminal to the J domain. Nuclear magnetic resonance (NMR) and small angle X-ray scattering (SAXS) were used to study the structural dynamics and interaction of Rb with the LXCXE motif, the J domain and a construct (N260) extending from the J domain through the origin binding domain (OBD).NMR and SAXS data revealed substantial flexibility between the domains in N260. Binding of pRb to a construct containing the LXCXE motif and the J domainrevealed weak interactions between pRb and the J domain. Analysis of the complex of pRb and N260indicated that the OBD is not involved and retains its dynamic independencefrom the remainder of Tag. These results support a ‘chaperone’ model in which the J domain of Tag changes its orientation as it acts upon different protein complexes.
Keywords: SV40 T antigen, Retinoblastoma tumor suppressor, J domain, SAXS, NMR
Simian Virus 40 (SV40), a double stranded DNA polyomavirus, encodes the multi-functional multi-domain large T antigen (Tag) protein. Tag is required at several stages of viral productive infection, and is necessary and sometimes sufficient, to induce cellular transformation [1]. During viral infection, Tag is required for the initiation and elongation steps of viral DNA replication, transcriptional repressionof the late viral promoter, regulating cellular transcription, and virion assembly. SV40 transforms cells by evasion, inactivation, and deregulation of the growth pathways of the host [2]. It performs these actions primarily through the action of Tag.
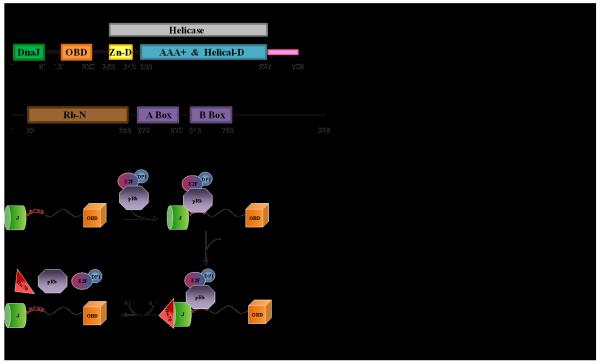
The amino-terminus of Tag is a J domain (Fig. 1A), and genetic and biochemical studies have established that Tag is a DnaJ molecular chaperone [3]. DnaJ chaperones work in concert with DnaK family chaperones. Tag binds the mammalian DnaK homologue Hsc70, and like other DnaJ-DnaK interactions, the binding of Tag to Hsc70 leads to activation of the Hsc70 ATPase activity. The J domain and its interaction with Hsc70 are essential for Tag mediated cellular transformation [4].
Figure 1. Domain structure of SV40 Tag (A) and pRb (B), and an overview of the chaperone model(C).
Tag bindsRb/E2F complexes primarily via the LXCXE motif, recruits its co-chaperone Hsc70, ATP hydrolysis occurs and the complex is disassembled.
One well defined role for the J domain in transformation is the disruption of the interaction between retinoblastoma (Rb) tumor suppressor proteins and transcriptional activator E2F proteins [5]. The Rb family contains pRb, p107, and p130 proteins. These multi-domain proteins (Fig. 1B) have been implicated in transcriptional activation and several other cellular processes [6]. One function of Rb proteins is to regulate the cell cycle by inhibiting proliferation. Rb is bound to the E2F/DP1 in G1 phase and represses its activity [7]. In late G1 phase, Rb is inactivated via phosphorylation by cyclin dependent kinases, which in turn releases E2F/DP1 and enables entry into the S phase and progression of the cell cycle [8]. Inactivation of Rb causes deregulated E2F activity, inappropriate cellular proliferation and may potentiate progression to tumorigenesis [9].
During both productive infection and cell transformation Tag induces the release of E2F transcription factors from the three retinoblastoma proteins, pRb, p107, and p130 [5, 10]. The release of E2F from Rb is dependent on a functional J domain and Hsc70-mediated ATP hydrolysis [3]. These observations led to a “chaperone” model in which Tag first binds to Rb-E2F complexes, and then energy derived from ATP hydrolysis by Hsc70, recruited by the J domain, is used to liberate E2F from Rb (Fig. 1C) [11].
The chaperone model involves Tag recruiting Hsc70 and multi-protein complexes so that the chaperone machinery can alter specific protein-protein interactions. In this model, Tag can alter the target complex by: (1) dislodging specific proteins from the complex; (2) altering the conformation and thus the activity of proteins in the complex; (3) targeting specific proteins in the complex for post-translational modification and/or degradation. This model is consistent with studies of Tag action on pRb/E2F complexes, in which it has been shown that energy derived from Hsc70-mediated ATP hydrolysis is used to release E2F4 from its association with p130 [5]. In the absence of ATP hydrolysis, Tag associates with both Hsc70 and p130/E2F4 complexes [5]. Hence, the assembly of Tag and Rb is a biologically important intermediate of the chaperone reaction.
If this model is correct, Hsc70 must have the ability to act on different cellular complexes, each containing a unique set of proteins. For example, genetic studies indicate that, in addition to targeting Rb-E2F complexes bound near the amino-terminus, the J domain acts on targets bound in the carboxy-terminal portion of Tag [12]. Thus, in each case the J domain must be in position so that Hsc70 is correctly oriented relative to the target complex. This suggests that the J domain must be able to adopt multiple orientations relative to the other Tag domains (Figure 1). This hypothesis is supported by cryo-EM studies of Tag [13], in which the absence of the J domain in the structure is presumed to arise from it having different orientations in different molecules on the grid.
Tag bindsRbproteins primarily through a consensus LXCXE binding motif (Tag 103-107) that resides in the linker between the OBD and the J domain (Fig. 1) [14]. However, the details of how Tag coordinates the protein interactions involved in inactivation of Rb remains poorly understood. A crystal structure has been determined of a complex of the N-terminal region of Tag, including the J domain (N117) and the LXCXE motif, in complex with the pocket domain of Rb (pRbA/B) [15]. The interaction interface of Tag in this structure primarily involves the LXCXE motif, but a few contacts between the J domain and pRbA/B were also observed. The chaperone model requires that the J domain be able to adopt different orientations in order to correctly position Hsc70 for action on its multiple targets. In contrast, the crystal structure suggests the J domain and pRb are in a fixed orientation. We propose that flexibility must exist in the linker between the J and OB domains of Tag to provide the variation in positioning required for function. To test this hypothesis we used a combination of NMR and small angle X-ray scattering (SAXS) experiments to characterize the relative flexibility of the linker between J and OB domains, and to determine if this flexibility is restricted upon binding to the Rb substrate.
MATERIALS AND METHODS
DNA constructs
A plasmid expressing SV40 Tag construct N260 (4-260) obtained from Xiaojiang Chen (University of Southern California) was subcloned into an in-house kanamycin resistant pbG101 vector (L. Mizoue, Center for Structural Biology) containing an H3C cleavable N-terminal 6xHis-GST tag. The Tag constructs N117 (4-117) and N102 (4-102) were subclonedfrom N260 into the ampicillin resistant pET15b vector containing an N-terminal 6xHis tag. pRbA/B (379-570/645-772)in an ampicillin resistant pGex-kg vector containing a thrombin cleavable GST tag was used in these studies because it corresponds to that used in the crystal structure of the pRb-Tag complex [15].
Protein Expression
The constructs of SV40 Tag (N260, N117, N102) were expressed in BL21 (DE3) Escherichia coli (E. coli) competent cells grown in LB or M9 minimal media containing 15NH4Cl and/or 13C glucose as needed. The cells containing N260 were grown at 37 °C and induced with 0.25 mM isopropyl -D-thiogalactopyranoside (IPTG) overnight at 18° C. Expression of N117 or N102 was induced with 1 mM IPTG at 37 °C for 5 hours. The pRbA/B protein was expressed in E.coli XA90 cells. The cells were grown in Terrific Broth at 37 °C, protein was induced with 0.4 mM IPTG and expressed overnight at 18 °C. All cells were harvested using a JLA 8.1 Beckman rotor at 7000 rpm and 4 °C for 20 minutes. Pellets were stored at −20 °C.
Protein Purification
The Tag protein constructs were purified using Nickel affinity chromatography (NiNTA) in 25 mMTris (pH 8.0) and 300 mMNaCl. Cleavage of the His tag on N102 and N117 was performed with thrombin at room temperature for 3 hrs while the protein remained bound to the resin. The cleaved protein was eluted by washing the resin with the purification buffer and followed by size exclusion chromatography (SEC) using a Superdex S75 column equilibrated with 20 mM NaH2PO4 (pH 7.2), 100 mMNaCl, and 5 mMdithiothreotol (DTT). Cleavage of the His-GST tag from N260 with H3C protease was performed via overnight dialysis in buffer containing 25 mMTris (pH 8.0), 200 mMNaCl and 2 mM DTT. A second NiNTA purification step was used to remove the His-GST tag and the H3C protease. An elution gradient of 5-20 mM imidazole was used to elute the cleaved N260 protein, followed by SEC over a Superdex S200 column equilibrated with 20 mM NaH2PO4 (pH 7.2), 100 mMNaCl and 5 mM DTT. The pRbA/Bprotein was purified using glutathione affinity chromatography in 1X PBS, 2 mM DTT, pH 7.2 and 10 mM imidazole for the elution. The fusion protein was dialyzed into 20 mM NaH2PO4 (pH 6.0), 50 mMNaCl, and 5 mM -mercapto ethanol (BME). The GST tag was cleaved at room temperature without agitation in the presence of 10% glycerol via addition of thrombin (100 units) every two hours for eight hours. Then the cleavage reaction remained at room temperature overnight. The pRbA/B was further purified with ion exchange chromatography (SourceS) using 20 mm NaH2PO4 (pH 6.0), 5 mM DTT with a 50 mM - 1 M NaCl elution gradient. The final purification step was SEC using a Superdex S200 column equilibrated with 20 mM NaH2PO4 (pH 7.2), 100 mMNaCl, and 5 mM DTT.
Nuclear Magnetic Resonance Spectroscopy
NMR experiments were conducted using Bruker DRX 600 and 800 MHz spectrometers equipped with z-axis gradient TXI cryoprobes. All experiments were collected at 25 °C. To obtain backbone resonance assignments, 800 μM13C,15N-enriched N117 was prepared in 1X PBS (pH 6.5), 2mM DTT and 10% D2O. The experiments acquired were 2D 15N-1H HSQC and13C-HSQC, and 3D HNCACB, CBCA(CO)NH, HN(CO)CA, HNCA, HNCO, 13C-NOESY, 3D 15N-NOESY, and 15N-TOCSY. Chemical shift perturbations induced by binding of pRb/AB were elucidated by comparing spectra of free 15N-enriched N117 and N260with the corresponding 1:1 complex with unlabeled pRb/AB in a buffer containing 20 mM NaH2PO4(pH 7.2), 100 mMNaCl, 5mM DTT and 5% D2O. 15N-1H HSQC was used for N117 and 15N-1H TROSY HSQC for N260. All NMR data were processed using Bruker software TopSpin [16] and analyzed using Sparky [17].
Small angle x-ray scattering
SAXS data were collected at the ALS beamline 12.3.1 (SIBYLS) LBNL Berkeley, California [18]. The wavelength of the incident beam λ was 1.0 Å and the sample-to-detector distances set to 1.5 m resulting in scattering vectors, q, ranging from 0.001 Å−1 to 0.32 Å−1. The scattering vector is defined as q = 4πsinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C and data were processed as described [18]. The experimental SAXS data for different protein concentrations were investigated for aggregation using Guinier plots [19]. The radius of gyration Rg was derived by the Guinier approximation I(q) = I(0) exp(−q2R 2g/3) with the limits qRg<1.3. The program GNOM [20] was used to compute the pair-distance distribution functions, P(r). This approach also provided the maximum dimension of the macromolecule, Dmax. In order to determine Rg and Dmaxfor the OBD and J domain,the FoXS server [21] was used to back-calculate SAXS from coordinates for OBD(PDBid: 2tbd) and for J domain (N102)(PDBid: 1gh6:A) and this data was then processed using GNOM.
The recently reported approach based on the Porod–Debye law was used to qualitatively assess the presence of structural disorder in the proteins [24]. The Porod-Debye law describes a fourth power approximation to the relationship between q and the observed intensities, I(q) [25, 26]. Transformation of scattering data as q4•I(q) vs. q4 (Porod-Debye plot) should display a curve asymptotically approaching a constant value as q approaches infinity. In practice, the plot for a highly globular protein will plateau rapidly at low q values, whereas the plot for a fully unfolded protein would be devoid of any discernible plateau [24].
Molecular graphics
Allimages were generated using PYMOL (DeLano Scientific, Palo Alto, CA, USA).
RESULTS
SV40 Tag is a modular protein containing three globular domains (J, origin binding, helicase) and a disordered C-terminal ‘host-range’ domain (Fig. 1). A segment of 29 residues links the J domain to the origin binding domain (OBD). Previous studies have shown the OBD aligns with the helicase domain (HD) [13]. A crystal structure of the J domain and a portion of the linker bound to pRb was reported and identified the residues involved in this interaction [15]. Potentialmodels for how pRb binds to Tag range from interaction exclusively with the LXCXE motif to formation of a tri-partate complex with the J domain and the OBD. Differences in these models impact the Tag mechanism of action on pRb function, in particular, how the J domain can adjust the orientation to correctly position Hsc70 for action on its targets. Our experiments were directed toward investigating changes in the relative flexibilityof the J domain upon pRb binding. Since the OBD is structurally aligned with the core of Tag [13], we have selected for our studies the N260 construct containing the residues spanning the J domain through the OBD.
The J and OB domains of Tag are structurally independent
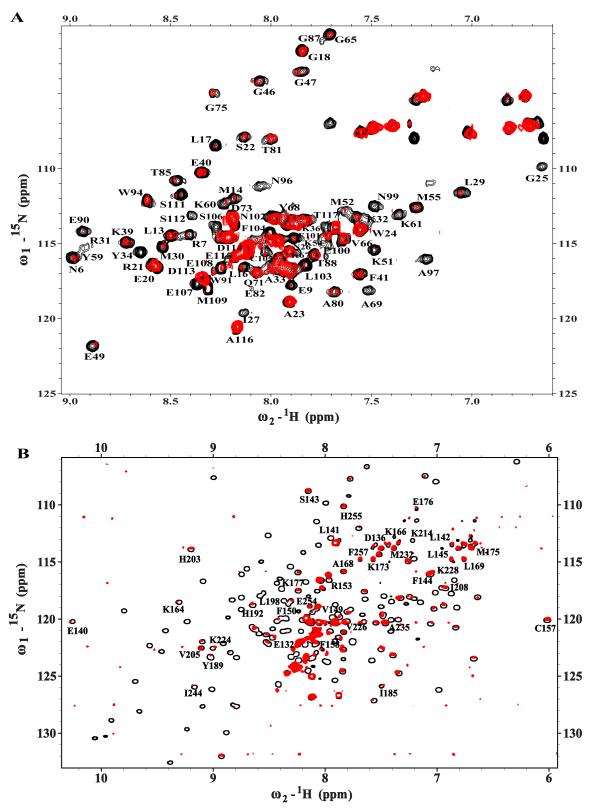
Our approach involved characterizing the structural and dynamic properties of the Tag N260 construct first, using heteronuclear NMR spectroscopy (Figure 2). Note that TROSY-HSQC was required for the larger N260construct to obtain sufficient resolution of the peaks. The spectra for all three constructs show well dispersed signals, which is indicative of the presence of folded globular domains. Inspection of these spectra reveals that N260 has many signals that reflect the presence of the J domain and OBD. In addition, a number of signals in the crowded central region of the spectrum spanning 7.8-8.2 ppm in the 1H dimension, which is typically associated with residues in segments lacking regular structure, were attributed to unstructured regions of N260including the long linker between the two folded domains.
Figure 2. The J and OB domains of SV40 Large T antigen are structurally independent.
Overlays of 15N-1H HSQC NMR spectra of N117(red) and OBD (blue) on the 15N-1H TROSY-HSQC spectrum of N260 (black). Resonances arising from each domain correspond closely to those observed in the larger N260 construct.
To accurately identify the signals observed in the spectrum of N260, chemical shift assignments for the J domain and OBD are required. The corresponding NMR signal assignments have been reported for OBD [28] but not for the J domain. Consequently, 15N- and 13C,15N-enriched samples of the J domain N102 and N117 constructs were prepared and a series of standard multi-dimensional heteronuclear NMR experiments were recorded (See Materials and Methods). The assignments for the backbone amide resonances of N117 are provided in Figure S1 in the Supplementary Materials. Note that these include the resonances of residues in the linker, including the key LXCXE motif spanning residues 103-107, which as anticipated appear in the crowdedcentral region of the spectrum.
Overlays of the individual J domain and OBD spectra onto the N260 spectrum are shown in Figure 2. It is clear from these comparisons that many distinct resonances corresponding to the J domain and OBD appear in identical locations in the spectrum of N260 and resonances for a number of additional residues are found close by.We also note some additional signals are present in the central crowded region of the spectrum that arise from residues 118-130, which are not present in either the OBD or J domain construct. The observation that the large majority of the residues in the J domain and OBD have the same or very similar NMR chemical shifts, whether alone or tethered together in the context of the N260 construct, indicates that the J domain and OBD are structurally independent of each other and implies that the linker between them is flexible.
The J domain is flexibly tethered to the OBD
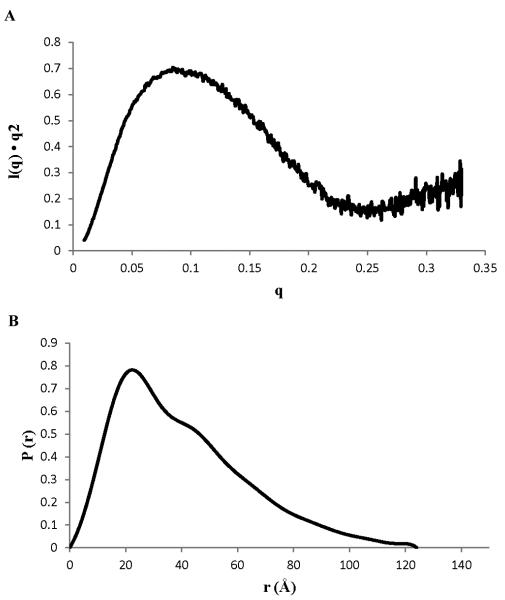
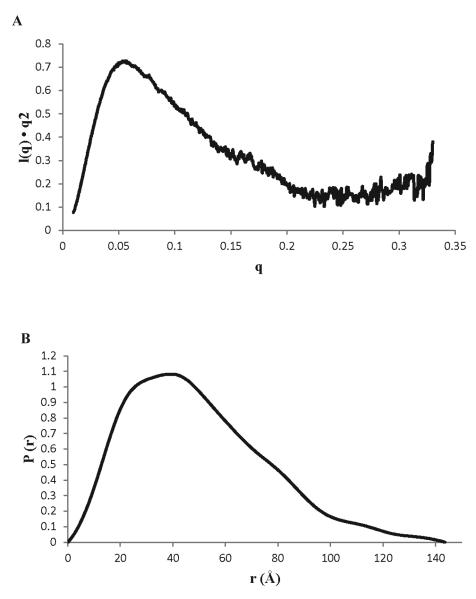
To test if the two domains of N260 are linked by a flexible tether, we turned to small angle X-ray scattering (SAXS) experiments. SAXS is well suited for characterizing the structural architecture of multi-domain proteins in solution and determining whether or not the domains are arranged in a fixed orientation [18]. Analysis of the SAXS data for N260 shows it has amaximal protein dimension (Dmax) and a radius of gyration (Rg) (froma Guinier plot, Figure S2) of 124Å and 30.6 Å, respectively. These values are significantly larger than the sum of Rg (14.75 + 14.58 = 29.33 Å) and Dmax (44.0 + 42.5 = 86.5 Å) values calculated from the x-ray crystal structure of the J domain [15] and the representative NMR structure of the OBD [28]. More significantly, the broadened P(r) function and elongation of the P(r) tail (Fig. 3B) are characteristic of population of highly extended conformations where the J domain and OBD are far from each other. In addition, the upward slope in the Kratky plot at high q values (Fig. 3A) indicates the presence of disordered polypeptide in the protein. In summary, the ensemble of the SAXS dataindicated that the J domain of N260 is flexibly attached to the OBD.
Figure 3. SAXS analysis for SV40 Tag N260.
(A)Kratky plot and (B) P(r) function.
RbA/B interacts with the Tag LXCXE motif and the J domain but not the OBD
Binding of pRb to consensus LXCXE motifs, including that of SV40 Tag, is well established[29, 30, 31]. The crystal structure of the complex of the Tag J domain extended to the LXCXE motif (N117 construct) with pRbA/B revealed a few additional contacts to the J domain[15], although the importance of these contacts to the interaction of Tag and pRb has not been directly investigated. Of importance here, if the contacts with the J domain are significant, the binding of pRb may fix the orientation of pRb and J, which has implications for the chaperone model. Following the crystal structure and previous studies of Rb binding to LXCXE motif proteins [31], we utilized the pRbA/B construct that contains the core A and B structured domains with a truncated linker.
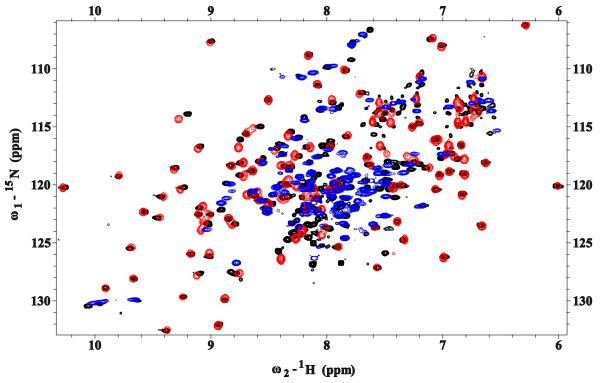
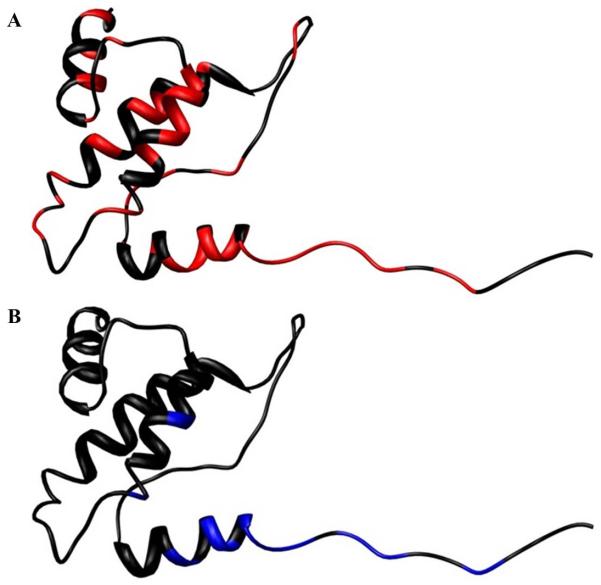
An NMR approach was used first to characterize contacts between Tag and pRb. Figure 4A shows an overlay of the 15N-1H HSQC spectrum of 15N-enriched N117 alone (black) and with a 1:1 ratio of pRbA/B (red). The comparison of these data revealed that binding of pRbA/B causes a combination of resonance disappearance, line broadening, perturbation of chemical shifts, and some unchanged peaks. These observations are indicative of a direct interaction with pRbA/B that extends beyond the LXCXE motif to residues in the J domain. The perturbation of some but not all signals in the J domain is consistent with a specific interaction. The pronounced broadening and disappearance of J domain peaks in the presence of Rb might be interpreted as due to the close proximity of the J domain to pRbA/B. However, given the large size of the N117-pRbA/B complex (52 kDa) and the fact that a substantial number of J domain signals are present in a basic15N-1H HSQC spectrum the most likely explanation is that the J domain does not tumble in synchrony with pRbA/B, which implies that the interaction with the J domain is transient.To obtain further insights into the interaction with pRbA/B, we took advantage of our NMR chemical shift assignments for the J domain and identified the residues whose resonances are perturbed by the addition of pRbA/B (Table 2). The disappearance of all of the resonancesfrom the LXCXE motif is consistent with it serving as an essential component of the binding interface and with the high affinity (Kd?? M) binding of the isolated motif to pRbA/B [REF]. We note that the residues whose resonances are perturbed include all of the J domain residues that are reported to be in contact with pRbA/B in the crystal structure [15] as well as a number of additional residues. Figure 5 shows a ribbon diagram of J domain with these perturbed residues mapped onto the structure. Since residues whose signals are perturbed are scattered throughout the J domain and are not clustered near the C-terminus close to the LXCXE motif, then it is possible to rule out that the perturbations are due to simple proximity effects. Chemical shift perturbations may arise from direct inter-molecular contacts or from structural changes induced upon binding of the ligand. Hence, it is not possible to determine if the binding site in solution is identical to that observed in the crystal structure, but the available evidence suggests this is likely to be so. Regardless, as noted above, the very observation of NMR signals from the J domain in this 52 kDa complex strongly implies the interaction is more dynamic than implied by the crystal structure.
Figure 4. Interaction of pRbA/B with Tag.
(A)Region from the 600 MHz15N-1H HSQC spectra for 15N-enriched N117acquired in the absence (black) and presence (red) of a 1.0 molar equivalent of pRbA/B. Sequence-specific assignments are provided for the resolved peaks in the spectrum. (B) 800-MHz 15N-1H TROSY-HSQC spectra for 15N-enriched N260acquired in the absence (black) and presence (red) of pRbA/B.Sequence-specific assignments are provided for resolved peaks that are present in the spectrum of the complex, all of which arise from OBD residues.
Table 2.
T antigen residues perturbed upon interaction with RbA/B as observed by NMR (black) and X-ray crystallography (bold).
| T antigen Residues affected by RbA/B interaction |
|---|
| N6; R7; E9; L13; M14; L17; S22; I27; L29; M30; R31; K32 Y34; K39; E40; G46; G47; E49; K51; M55; Y59; K60; K61; G65; G75; G76; T81; E82; T85; G87; E90; Y94; N96; A97; N99; E100; E101; L103; F104; C105; S106; E107; E108; M109; S111; S112 |
Figure 5. Map of residues on the J domain whose resonances are altered upon binding of pRbA/B.
Ribbon diagrams of the J domain (PDBID: 1gh6:A),(A)colored red for residues whose resonances disappear or are broadened in the complex (Table 2) and (B) colored yellow for residues identified as contacting pRbA/B in the structure [15].
To determine whether the OBD is also involved in the interaction with pRbA/B, an NMR chemical shift perturbation assay was performed using the N260 construct. Figure 4B shows an overlay of the 15N-1H TROSY-HSQC spectrum of N260 alone (black) and with a 1:1 ratio of pRbA/B (red), along with circles that identify resonances that are identical to those found in the spectrum of the isolated OBD. As described above for the experiment with N117, there is clear evidence of interaction from the extensive degree of peak disappearance and increases in line widths as a result of formation of a large complex (69 kDa). The figure reveals a number of signals from the OBD but none from the J domain can still be observed.
This observation does not rule out that the more pronounced broadening and disappearance of J domainverus OBD signals in the presence of Rb is simply due to it being closer to the Rb than the OB domain. However, as for the N117-pRbA/B complex, the that fact that we see signals in a basic 15N-1H HSQC spectrum of this 69 kDa complex indicates that the OBD does not tumble in synchrony with pRbA/B. The most likely interpretation of these observations is that the OBD does not interact with pRbA/B, but given the limitations of the NMR analysis, it is not possible to rule out alternate explanations such as a highly transient weak interaction with a small OBD interface.
The Tag J domain remains flexibly attached to the OBD after pRbA/B is bound
To test the effect of pRbA/B binding on the flexibility of N260, we performed additional SAXS experiments on the purified pRbA/B and the pRbA/B-N260 complex (Figure 6). The data for pRbA/B matched well to that previously published [32]. The calculated P(r) function had a Dmax of ~145 Å (Table 1) about 20 Å greater than for N260 alone. In addition, a new feature in the curve was observed in the P(r) function at r ~80 Å, which corresponds to the average distance between the J-domain and pRbA/B domains (Fig. 6B).
Figure 6. SAXS data for the complex of pRbA/Band N260.
(A) Kratky plot and (B) P(r) function.
Table 1.
Comparison of SAXS-derived parameters for N260, pRb/AB, and the N260-pRb/AB complex.
| Proteins | N260 | pRbA/B | N260-pRbA/B Complex |
|---|---|---|---|
|
Rg (Å), Guinier Analysis |
30.6 | 28.1 | 41.6 |
|
Rg (Å), P(r) analysis |
32.9 | 27.1 | 42.24 |
|
Dmax (Å), P(r) analysis |
124 | 96.5 | 143.5 |
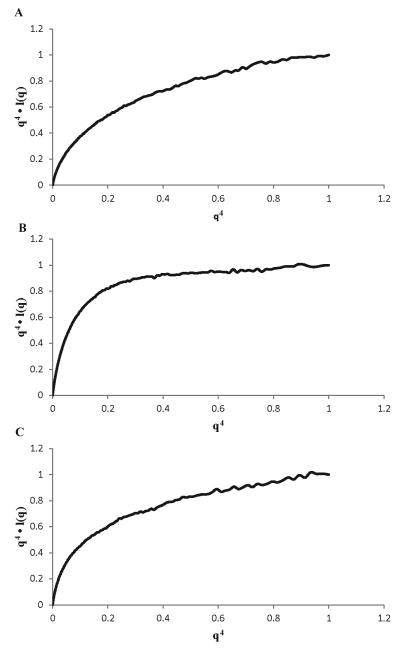
We performed a Porod-Debye analysis for pRbA/B, N260 and their complex to obtain direct insights into their flexibility. In plots of the normalized q4•I(q) vs. q4, we find the globular pRbA/B reaches a plateau at lower q than the flexible N260, and the complex is in between (Fig. 7). Table 3 lists the Porod volumes determined from the three SAXS data sets, along with the experimental packing densities. The ideal value for a compact globular protein is 1.37 g-cm3. The value of 0.8 g-cm3 for N260 is consistent with extensive inter-domain flexibility. Although a higher value of 0.9 g-cm3is found for the complex, this is still not close to the canonical value for a compact protein. This observation suggests there is little if any global compaction of N260 upon binding of pRb and that the OBD remains flexible, consistent with the NMR results and the upward slope in the Kratky plot at high q values for the complex (Fig. 6A). Together, the data indicate that the Tag J domain remains flexibly attached to the OBD when pRbA/B is bound.
Figure 7. Porod-Debye analysis of SAXS data for the N260-pRb/AB complex.
Plots of q4•I(q) vs q4 plots for (A) N260, (B) pRbA/B, and (C) N260-pRbA/B complex.
Table 3.
Porod-Debye Parameters derived from the SAXS data for N260, pRb/AB, and the N260-pRb/AB complex.
| Porod Vol. (Å3) | dprotein(g•cm3) | |
|---|---|---|
| pRbA/B | 61803 | 1.07 |
| N260 | 63833 | 0.78 |
| N260-pRbA/B | 126380 | 0.92 |
DISCUSSION
SV40 Tag is a modular protein with multiple domains linked by flexible tethers. The dynamic nature of modular proteins presents challenges for characterization by traditional structural biology approaches. Here we applied a combination of NMR and SAXS to investigate how binding of Rb to Tag affects the structural dynamics of the Tag architecture and to determine implications of these data for the evolving chaperone model.
The chaperone model involves Tag recruitment of Hsc70 and multi-protein complexes involved in transcription and DNA replication that lead to alterations in specific protein-protein interactions. If this model is correct, the chaperone must have the ability to act on different cellular complexes, each containing a unique set of proteins. For example, genetic studies indicate that, in addition to targeting Rb-E2F complexes bound near the amino-terminus, the J domain acts on targets bound to the C-terminus of Tag [12]. In each case the J domain must be in position so that Hsc70 is correctly oriented relative to the target complex. This suggests that the J domain must be able to adopt multiple orientations relative to the other Tag domains. This hypothesis is supported by cryo-EM studies of Tag [13], in which the absence of the J domain in the structure is presumed to arise from it having different orientations in different molecules on the grid. One prediction of the chaperone model is that Hsc70 directly contacts Rb, E2F or both. Another prediction of the chaperone model is that to effect release of E2F from Rb, Hsc70 must be positioned to influence the Rb-E2F interaction.
A mechanism for release of E2F/DP1 from Rb has recently been proposed [33]. Rb has structured N terminal and central (pocket) domains as well as an unstructured C terminal domain (Fig. 1). The pocket domain is composed of two structured domains (A and B box) connected by a 64 residue unstructured linker. The transactivation domain of E2F binds to the unphosphorylated pocket domain, making critical interactions with both the A and B sub-domains. When phosphorylated, the Rb linker binds to pocket domain at the E2F binding site. The linker can be displaced from the pocket in the presence of the transactivation domain of E2F. This suggests that there is a binding competition between E2F and the pocket linker, which could explain the mechanism of Rb inactivation and E2F release.
Recently, evidence for the direct contact of Hsc70 with E2F has been reported [34]. Hsc70 is known to bind unfolded regions of proteins in a substrate-binding domain. An unstructured region was observed in E2F and shown to increase stimulation of ATP hydrolysis by Hsc70, but only in the presence of a J protein [34]. This suggests that the J domain is required to attain proper positioning of Hsc70 for contact with the E2F substrate.
These observations and our results support the chaperone model and highlight the intrinsically dynamic nature of the protein complexes. Our data has shown that the J domain is structurally and dynamically independent of the other Tag domains. This provides a required degree of flexibility for altering the spatial localization of the Hsc70-Tag-Rb complex to drive the dissociation of E2F transcription factors. In particular, despite binding of both Hsc70 and Rb, the residual dynamics can allow positioning Hsc70 for direct contact with E2F yielding an increased rate of ATP hydrolysis and disassembly of the entire complex, which would lead to improper progression of the cell cycle.
Supplementary Material
Figure S1. HSQC spectrum of Tag construct, N117, with assignments of resonances labeled.
Figure S2. Guinier analysis of SAXS data for N260, pRbA/B and the N260-pRb/AB complex.Gunier plots of(A) N260, (B) pRbA/B, and (C) N260-pRbA/B complex
Highlights.
The data reveal flexible linkage of origin binding and J domains of SV40 T antigen
Contacts between J domain and pRb in the crystal structure are transient in solution
The origin binding domain remains dynamically independent in the complex with pRb
The results support a ‘chaperone’ model for the action of the J domain
AKNOWLEDGEMENTS
We thank Paul Cantalupo and Xiaojiang Chen for assistance in expression and purification of the pRbA/B, and members of the Chazin laboratory for assistance in experimental procedures and many helpful discussions. This research wassupported an operating grantfromthe National Institutes of Health (R01 CA120997), as well as aRuth L. Kirschstein National Research Service Awardto CKW (T32 CA009582-18, 19) and funding for access to core facilities from the Vanderbilt-Ingram Cancer Center (P30 CA68485). The X-ray scattering technology and applications to the determination of macromolecular shapes and conformations at the SIBYLSbeamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, were supported in part by the U.S. Department of Energy program Integrated Diffraction Analysis Technologies (IDAT) and the DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) under Contract Number DE-AC02-05CH11231.
Abbreviations
- BME
beta-mercapto ethanol
- DTT
dithiothreotol
- IPTG
Isopropyl -D-1-thiogalactopyranoside
- OBD
Origin Binding Domain
- HD
Helicase Domain
- Hsc70
Heat shock cognate protein 70
- NMR
nuclear magnetic resonance
- P(r)
Probability of Distribution
- Rb
Retinoblastoma tumor suppressor protein
- Rg
Radius of Gyration
- SAXS
Small Angle X-ray Scattering
- SEC
Size Exclusion Chromatography
- SV40
Simian Virus 40
- Tag
Large T-antigen
APPENDIX
A figure showing NMR assignments for the backbone amide resonances of N117 labeled on an HSQC spectrum. Guinier analyses for pRbA/B, N260 and their complex.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Experimental and theoretical scattering profiles, and P(r) functions, will be deposited in the BIOISIS database (www.bioisis.net).
REFERENCES
- [1].Saenz-Robles MT, Sullivan CS, Pipas JM. Transforming functions of Simian Virus. Oncogene. 2001;20:7899–7907. doi: 10.1038/sj.onc.1204936. [DOI] [PubMed] [Google Scholar]
- [2].Pipas JM. SV40: Cell transformation and tumorigenesis. Virology. 2009;384(2):294–303. doi: 10.1016/j.virol.2008.11.024. 20. [DOI] [PubMed] [Google Scholar]
- [3].Sullivan CS, Pipas JM. T antigens of Simian Virus 40: molecular chaperones for viral replication and tumorigenesis. MicrobiolMolBiol Rev. 2002;66(2):179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kelley WL. The J-domain family and the recruitment of chaperone power. TIBS. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- [5].Sullivan CS, Cantalupo P, Pipas JM. The molecular chaperone activity of simian virus large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol. 2000;20(17):6233–4. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nature Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- [7].Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of pRb with E2F inhibits the transcriptional activity of E2F. Genes & Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- [8].Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification of retinoblastoma protein by at least two distinct cyclin-Cdk complexes. Mol. Cell. Biol. 1998;18:735–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hernando E, Nahle Z, Juan G, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430(7001):797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- [10].Helt AM, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24(2):159–69. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- [11].Brodsky JL, Pipas JM. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol. 1998;72(7):5329–34. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rushton JJ, Jiang D, Srinivasan A, Pipas JM, Robbins PD. Simian virus 40 T antigen can regulate p53-mediated transcription independent of binding p53. J Virol. 1997;71(7):5620–3. doi: 10.1128/jvi.71.7.5620-5623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gomez-Lorenzo MG, Valle M, Frank J, Gruss C, Sorzano CO, Chen XS, Donate LE, Carazo JM. Large T antigen on the simian virus 40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 2003;22(23):6205–13. doi: 10.1093/emboj/cdg612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zalvide J, Stubdal H, DeCaprio JA. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim HY, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 2001;20(1-2):295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. http://www.bruker-biospin.com/topspin-overview.html.
- [17].Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2006. [Google Scholar]
- [18].Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr., Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guinier A, Fournet G. Small angle scattering of X-rays. Wiley; New York: 1955. [Google Scholar]
- [20].Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992:495–503. [Google Scholar]
- [21].Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A Web server for Rapid Computation and Fitting of SAXS Profiles. NAR. 2010;38(Suppl):W540–4. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rambo RP, Tainer JA. Characterizing flexible and instrinsically unstructured biological macromolecules by SAS using the porod-debye law. Biopolymers. 2011;95(8):559–71. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Porod G. Die Röntgenkleinwinkelstreuung von dichtgepacktenkolloidenSystemen I. Teil. KolloidZeitschrift. 1951;124:83–115. [Google Scholar]
- [26].Debye P, Anderson HR, Brumberger H. Scattering by an Inhomogeneous Solid. II. The Correlation Function and Its Application. J. Appl. Phys. 1957;28:679–683. [Google Scholar]
- [27].Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80(6):2946–53. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Luo X, Sanford DG, Bullock PA, Bachovchin WW. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat Struct Bio. 1996;3(12):1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- [29].Singh M, Krajewski M, Mikolajka A, Holak TA. Molecular determinants for the complex formation between the retinoblastoma protein and LXCXE sequences. J Biol Chem. 2005;280(45):37868–76. doi: 10.1074/jbc.M504877200. [DOI] [PubMed] [Google Scholar]
- [30].Dong WL, Caldeira S, Sehr P, Pawlita M, Tommasino M. Determination of the binding affinity of different human papillomavirus E7 proteins for the tumour suppressor pRb by a plate-binding assay. J. Virol. Methods. 2001;98(1):91–8. doi: 10.1016/s0166-0934(01)00361-5. [DOI] [PubMed] [Google Scholar]
- [31].Lee C, Cho Y. Interactions of SV40 large T antigen and other viral proteins with retinoblastoma tumour suppressor. Rev Med Virol. 2002;12(2):81–92. doi: 10.1002/rmv.340. [DOI] [PubMed] [Google Scholar]
- [32].Balog ER, Burke JR, Hura GL, Rubin SM. Crystal structure of the unliganded retinoblastoma protein pocket domain. Proteins. 2011;79(6):2010–2014. doi: 10.1002/prot.23007. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-inducedconformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J.Biol Chem. 2010;285(21):16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Garimella R, Liu X, Qiao W, Liang X, Zuiderweg ER, Riley MI, Van Doren SR. Hsc70 contacts helix III of the J domain from polyomavirus T antigens: addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb. Biochemistry. 2006;45(22):6917–29. doi: 10.1021/bi060411d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. HSQC spectrum of Tag construct, N117, with assignments of resonances labeled.
Figure S2. Guinier analysis of SAXS data for N260, pRbA/B and the N260-pRb/AB complex.Gunier plots of(A) N260, (B) pRbA/B, and (C) N260-pRbA/B complex