Abstract
OBJECTIVE
Genome-wide association studies have begun to elucidate the genetic architecture of type 2 diabetes. We examined whether single nucleotide polymorphisms (SNPs) identified through targeted complementary approaches affect diabetes incidence in the at-risk population of the Diabetes Prevention Program (DPP) and whether they influence a response to preventive interventions.
RESEARCH DESIGN AND METHODS
We selected SNPs identified by prior genome-wide association studies for type 2 diabetes and related traits, or capturing common variation in 40 candidate genes previously associated with type 2 diabetes, implicated in monogenic diabetes, encoding type 2 diabetes drug targets or drug-metabolizing/transporting enzymes, or involved in relevant physiological processes. We analyzed 1,590 SNPs for association with incident diabetes and their interaction with response to metformin or lifestyle interventions in 2,994 DPP participants. We controlled for multiple hypothesis testing by assessing false discovery rates.
RESULTS
We replicated the association of variants in the metformin transporter gene SLC47A1 with metformin response and detected nominal interactions in the AMP kinase (AMPK) gene STK11, the AMPK subunit genes PRKAA1 and PRKAA2, and a missense SNP in SLC22A1, which encodes another metformin transporter. The most significant association with diabetes incidence occurred in the AMPK subunit gene PRKAG2 (hazard ratio 1.24, 95% CI 1.09–1.40, P = 7 × 10−4). Overall, there were nominal associations with diabetes incidence at 85 SNPs and nominal interactions with the metformin and lifestyle interventions at 91 and 69 mostly nonoverlapping SNPs, respectively. The lowest P values were consistent with experiment-wide 33% false discovery rates.
CONCLUSIONS
We have identified potential genetic determinants of metformin response. These results merit confirmation in independent samples.
The number of common genetic variants reproducibly associated with type 2 diabetes is growing (1). Well-powered candidate gene association studies and, more recently, genome-wide association studies (GWASs) have identified over two dozen loci robustly and reproducibly associated with type 2 diabetes or related quantitative glycemic traits. While these discoveries have advanced our understanding of the genetics of type 2 diabetes, they only explain a small fraction of the overall genetic contribution to the disease. Furthermore, in most cases, the genes involved in type 2 diabetes risk have not yet been identified: the majority of the associations detected thus far merely mark genomic regions where a certain variant is overrepresented in diseased cases versus unaffected controls, and subsequent fine-mapping and functional studies are necessary before a molecular mechanism can be ascribed to each locus.
Nevertheless, progress in the translation of genetic discoveries to clinical practice can advance along parallel paths. On the one hand, knowledge of the specific gene or variant causing the molecular phenotype is not needed to determine whether the associated region can aid prediction or affect the response to therapy; and on the other hand, targeted approaches can be applied in vivo in humans that shed light on the function of genes of interest, as a way to narrow the regions to study. These two objectives can be achieved by controlled interventions in randomized clinical trials. Such pharmacogenetic or gene-environmental strategies can fulfill both complementary roles, testing whether genetic variants predict response to therapy and whether a particular pharmacologic or lifestyle intervention affects the mode of action of specific risk loci.
In type 2 diabetes, the field of pharmacogenetics remains in its infancy. Although pharmacogenetic investigation has yielded clinically actionable results in neonatal diabetes and maturity-onset diabetes of the young, extending these studies to common type 2 diabetes has been more arduous (2). With regard to metformin, intriguing results were obtained by Shu et al. (3) in their investigation of common variants in SLC22A1, which encodes the liver-specific organic cation transporter 1 responsible for the absorption of metformin into hepatocytes: in a study of 20 human participants, carriers of reduced-function polymorphisms of SLC22A1 had a 17% higher area under the glucose curve after an oral glucose tolerance test when treated with metformin, indicating decreased responsiveness. Unfortunately, these results have not been confirmed in the long-term follow-up of a large observational cohort of patients treated with metformin monotherapy (4). Recently, a preliminary association was discovered between a variant in SLC47A1, which encodes the multidrug and toxin extrusion protein 1 (involved in the excretion of metformin into the bile and urine) and the glucose-lowering effect of metformin (5).
The Diabetes Prevention Program (DPP) can help answer some of these questions (6). The strengths of this randomized clinical trial include its enrollment of participants at high risk of developing diabetes, multiethnic composition, comprehensive longitudinal measures, and standardized behavioral and pharmacologic interventions. Extensive in-depth phenotyping and the use of behavioral and pharmacologic interventions allow characterization of the effects of known type 2 diabetes variants on diabetes incidence and response to therapy. We therefore designed a large-scale genotyping study by which we tested 1,590 variants identified through prior genetic studies of type 2 diabetes or related traits, as well as those capturing all common variation in 40 biological candidate genes, for association with diabetes incidence or response to preventive interventions (lifestyle modification or metformin) in the DPP.
RESEARCH DESIGN AND METHODS
The DPP.
The DPP was a 27-center randomized clinical trial in the U.S. that assessed whether metformin or lifestyle interventions prevent or delay development of diabetes in high-risk individuals. The DPP enrolled 3,234 overweight or obese people without diabetes but with impaired glucose tolerance and elevated fasting glucose and randomized them to placebo, metformin (850 mg twice daily), or a lifestyle intervention program consisting of individual and group counseling sessions conducted by dietary and exercise professionals aimed at ≥7% weight loss and ≥150 min of physical activity per week. A fourth arm of 585 subjects assigned to troglitazone (400 mg daily) was stopped because of hepatotoxicity (7). The primary end point was development of diabetes, ascertained by semi-annual measurement of fasting glucose or an annual 75-g oral glucose tolerance test, either of which was confirmed on a second occasion. The metformin and lifestyle interventions reduced the incidence of diabetes by 31% (95% CI 17–43) and 58% (95% CI 48–66), respectively, versus placebo (6). The 2,994 participants in the placebo, metformin, and lifestyle arms who gave informed consent for genetic investigation are the subjects of this study, which was approved by institutional review boards at each of the 27 participating sites. Their demographic characteristics are shown in Table 1.
TABLE 1.
Baseline characteristics of the DPP participants enrolled in the genetic study
| Overall | Placebo | Metformin | Lifestyle | |
|---|---|---|---|---|
| n | 2,994 | 1,000 | 990 | 1,004 |
| Sex | ||||
| Male | 982 (32.8) | 311 (31.1) | 344 (34.7) | 327 (32.6) |
| Female | 2,012 (67.2) | 689 (68.9) | 646 (65.3) | 677 (67.4) |
| Race or ethnic group | ||||
| White | 1,674 (55.9) | 557 (55.7) | 570 (57.6) | 547 (54.5) |
| African American | 612 (20.4) | 210 (21.0) | 206 (20.8) | 196 (19.5) |
| Hispanic | 498 (16.6) | 164 (16.4) | 158 (16.0) | 176 (17.5) |
| Asian/Pacific Islander | 128 (4.3) | 39 (3.9) | 33 (3.3) | 56 (5.6) |
| American Indian | 82 (2.7) | 30 (3.0) | 23 (2.3) | 29 (2.9) |
| Family history of diabetes | 2,089 (69.8) | 703 (70.4) | 683 (69.0) | 703 (70.1) |
| History of gestational diabetes mellitus | 321 (16.0) | 105 (15.2) | 104 (16.1) | 112 (16.5) |
| Quantitative traits | ||||
| Age (years) | 50.8 ± 10.7 | 50.5 ± 10.5 | 51.0 ± 10.4 | 50.7 ± 11.4 |
| Weight (kg) | 94.6 ± 20.3 | 94.8 ± 20.2 | 94.6 ± 20.0 | 94.5 ± 20.8 |
| BMI (kg/m2) | 34.1 ± 6.7 | 34.3 ± 6.7 | 34.0 ± 6.7 | 34.0 ± 6.8 |
| Waist circumference (cm) | 105.2 ± 14.6 | 105.3 ± 14.5 | 105.0 ± 14.6 | 105.3 ± 14.9 |
| Waist-to-hip ratio | 0.92 ± 0.09 | 0.92 ± 0.08 | 0.93 ± 0.09 | 0.93 ± 0.09 |
| Plasma glucose (mg/dl) | ||||
| In the fasting state | 106.7 ± 8.2 | 107.0 ± 8.4 | 106.7 ± 8.4 | 106.5 ± 7.9 |
| 2 h after an oral glucose load | 164.7 ± 17.1 | 164.6 ± 17.2 | 165.1 ± 17.2 | 164.4 ± 16.9 |
| Glycated hemoglobin (%) | 5.91 ± 0.51 | 5.91 ± 0.50 | 5.91 ± 0.51 | 5.91 ± 0.51 |
| Leisure physical activitiy (MET h/week) | 16.1 ± 25.9 | 16.6 ± 29.1 | 16.4 ± 25.9 | 15.4 ± 22.4 |
Data are n (%) or means ± SD.
SNP selection.
We selected SNPs in two ways: 1) SNPs in high-likelihood candidate genes and 2) SNPs identified by ongoing GWASs for type 2 diabetes or related metabolic traits. The 40 candidate genes were tentatively associated with type 2 diabetes, implicated in monogenic forms of diabetes, known to encode type 2 diabetes drug targets or drug-metabolizing/transporting enzymes, or involved in cellular metabolism, hormonal regulation, or response to exercise (Table 2). We used Tagger (8) to capture (at r2 ≥ 0.8) all common (minor allele frequency >5%) variations in European (CEU) and African (YRI) HapMap populations in these candidate genes. For seven additional genes (ACE, CASQ1, GCKR, IRS1, KCNQ1, LIPC, and NOS3), rather than attempting full coverage of genetic variation, we selected a limited number of SNPs previously associated with the phenotypes of interest. As the study evolved, it became obvious that previous reports of genetic association provided an equally compelling—or perhaps even higher—prior probability of true association with type 2 diabetes traits than biological function alone; thus, we also focused on GWASs whose results were available at the time this custom-made genotyping array was designed: SNPs associated with type 2 diabetes in the Diabetes Genetics Initiative (9), DIAGRAM (10), or three smaller 100K SNP GWASs in which we participated (11–13); SNPs tentatively associated with quantitative glycemic traits (fasting glucose, the insulinogenic index, and insulin resistance by homeostasis model assessment) in the Diabetes Genetics Initiative; or SNPs associated with obesity (14,15) or lipid traits (16–18). For quality control and analytical reasons, we also included some SNPs previously genotyped in these samples, as well as ancestry-informative markers to derive a global proportion of geographic ancestry in African American (19) or Hispanic (20) participants. Finally, we included a small number of SNPs provided by investigators leading ancillary studies approved by the DPP ancillary studies and genetics subcommittees. The total number of SNPs analyzed for each category is shown in Table 2.
TABLE 2.
Number of SNPs analyzed per selection category
| Category | n |
| Candidate genes (n SNPs) | 1,256 |
| Comprehensive coverage | 1,241 |
| Monogenic diabetes | 317 |
| ABCC8 (84), GCK (37), HNF1A (20), HNF1B (74), HNF4A (66), KCNJ11 (9), NEUROD1 (14), PDX1 (13) | |
| Monogenic obesity | 21 |
| MC4R (21) | |
| Previously associated with type 2 diabetes | 60 |
| CAPN10 (33), PTPN1 (27) | |
| Drug targets | 190 |
| PPARG (59), PRKAA1 (9), PRKAA2 (18), PRKAB1 (10), PRKAB2 (13), PRKAG1 (7), PRKAG2 (53), PRKAG3 (11), STK11 (10) | |
| Drug-metabolizing enzymes and transporters | 135 |
| CYP3A4 (15), SLC22A1 (47), SLC22A2 (44), SLC47A1 (29) | |
| Hormonal regulation | 117 |
| ADIPOQ (25), ADIPOR1 (22), ADIPOR2 (31), GCG (13), ITLN1 (11), ITLN2 (15) | |
| Cellular energy | 334 |
| FOXO1A (35), PCK1 (37), PCK2 (14), PKLR (10), PPARA (63), PPARGC1A (79), PPARGC1B (96) | |
| Response to exercise | 67 |
| CREB1 (13), MEF2A (38), MEF2D (16) | |
| Select SNPs in candidate genes | 15 |
| ACE (1), CASQ1 (2), GCKR (2), IRS1 (2), KCNQ1 (1), LIPC (1), NOS3 (6) | |
| Ancillary studies | 26 |
| ALOX5 (15), IL6 (1), renin-angiotensin system (7), USF1 (2), TNF (1) | |
| GWASs | 248 |
| Amish 100K GWAS (13): Type 2 diabetes | 46 |
| Framingham Heart Study 100K GWAS (11): Type 2 diabetes | 38 |
| Pima 100K GWAS (12): Type 2 diabetes | 12 |
| Obesity (14,15) | 11 |
| Diabetes Genetics Initiative (9) | |
| Fasting glucose | 37 |
| Insulin resistance by homeostasis model assessment | 16 |
| Insulinogenic index | 17 |
| Type 2 diabetes | 30 |
| DIAGRAM (10): Type 2 diabetes | 6 |
| Lipids (16–18) | 35 |
| Quality control | 60 |
| African American ancestry informative markers (19) | 29 |
| Hispanic ancestry informative markers (20) | 29 |
| Previously genotyped in DPP | 2 |
| Total | 1,590 |
Genotyping.
We initially designed a 1,536-SNP oligonucleotide pool array for the Illumina BeadArray platform (Illumina, San Diego, CA). In the 1,445 SNPs that passed quality control metrics, the sample pass rate was 99.8% and the average genotyping call rate per SNP was 98.5%. Because 91 SNPs failed genotyping on the oligonucleotide pool array, we assessed the adequacy of the coverage afforded by the successfully genotyped SNPs in each region. To rescue relevant SNPs, we used linkage disequilibrium (LD) to select proxy SNPs highly correlated to those that had failed and genotyped them on a Sequenom iPLEX platform. After quality control, 1,590 SNPs were available for analysis.
Statistical analysis.
We tested the effect of each SNP on diabetes incidence under an additive genetic model by Cox proportional hazards models, using age, sex, ethnicity, and treatment arm as covariates and including treatment (metformin or lifestyle) × genotype interaction terms. In secondary analyses, we stratified participants by treatment arm; if the interaction P value was nominally significant, only stratified analyses were considered. We used the MACH software (21) and the HapMap CEU population to impute allelic calls at SNPs not directly genotyped in the DPP. Because of concerns regarding the accuracy of imputation methods in admixed populations, we restricted this procedure to individuals of self-described non-Hispanic white ethnicity. Genotype-phenotype correlations on imputed data were considered confirmatory of prior associations, as well as an initial fine-mapping exploration. Using the program STRUCTURE (22), we applied these markers trained on the HapMap populations to assign a proportion of global European ancestry to each DPP participant.
We considered two sequential approaches to correct for multiple hypothesis testing based on the number of SNPs examined (23). We first ran 1,000 permutations in which diabetes outcome was randomly assigned to an individual's genotype within each ethnicity and treatment group (keeping sex and age together with genotype, and BMI with diabetes outcome). The P value for the overall null hypothesis is the fraction of permutations (n/1,000) for which the scalar statistic is at least as extreme as that observed for the data (24). To estimate the expected proportion of type I errors among the rejected hypotheses, we also computed false discovery rates (FDRs) as in Benjamini and Hochberg (25).
RESULTS
Supplementary Table 1 (available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db10-0543/DC1) shows that we achieved adequate coverage of all 40 genes in the two targeted populations, with 37 genes reaching at least 80% of common variants captured at r2 ≥ 0.8 in Europeans and all 40 reaching at least 70% of common variants captured at that level (comparable numbers were obtained in Africans). The average proportion of European ancestry among the DPP self-described white participants, as determined by ancestry-informative markers, was 98.9%, and the average proportion of West-African ancestry among DPP self-described African American participants was 89.3%. Given these results, we used self-described ethnicity as a covariate for these analyses. The full set of results is available in supplementary Table 2.
Table 3 shows the candidate gene regions harboring variants nominally associated with diabetes incidence in the treatment-adjusted models for the full study (i.e., there was no evidence for interaction with either intervention); only the top SNP within each gene region (out of 85 nominal associations) is given. The most significant associations occurred at SNPs in the AMP kinase (AMPK) subunit gene PRKAG2 (hazard ratio [HR] 1.24, 95% CI 1.09–1.40, P = 7.0 × 10−4 for the top SNP rs5017427, which is consistent with an experiment-wide 34% FDR). Twelve other PRKAG2 SNPs were nominally associated with diabetes (five in the top ten). Although most of them are in moderate to high LD with the index SNP (r2 ranging from 0.49 to 1.0 in HapMap CEU), at least two of them (rs954482 and rs2727537) are only weakly correlated with rs5017427 (r2 0.07 and 0.05, respectively). Nevertheless, the consistency of the association signal in this region provides reassurance with regard to the absence of genotyping artifacts in our dataset. Of SNPs previously associated with type 2 diabetes in the 100K Amish, Framingham, or Pima GWASs, three (rs1422930 in ODZ2, rs1859441 near COL2A1 and SENP1, and rs385909 near SH3YL1) had consistent nominal associations with diabetes incidence in the DPP, and two had nominally significant associations (rs10520926 and rs3136279) in the opposite direction. On the other hand, none of the six SNPs selected from the DIAGRAM meta-analysis (original odds ratio [OR] ranging from 1.05 to 1.15) were nominally significant in the DPP. Fifteen SNPs in genes that cause either maturity-onset diabetes of the young or neonatal diabetes were nominally associated with diabetes incidence; one of them, rs11868513 in HNF1B (not in LD with the previously type 2 diabetes–associated SNP rs757210), was strongly associated with diabetes incidence in the placebo arm (HR 1.69, 95% CI 1.36–2.10, P = 2 × 10−6). Finally, 14 SNPs in genes that encode metformin transporters (SLC22A1, SLC22A2, and SLC47A1) were nominally associated with diabetes incidence. Of the 85 nominal associations with diabetes incidence in DPP, only two SNPs (rs651164 in SLC22A1 and rs3736265 in PPARGC1A) were nominally associated with type 2 diabetes in DIAGRAM in a consistent direction (OR 1.08, 95% CI 1.02–1.16, P = 0.01, and OR 1.15, 95% CI 1.01–1.31, P = 0.04, respectively), with 60 other SNPs not being nominally significantly associated in DIAGRAM and 23 SNPs not captured in that dataset.
TABLE 3.
Candidate gene variants nominally associated with diabetes incidence in the DPP
| SNP | Chromosome | Position (NCBI 36) | Gene | Alleles (m/M) | Overall, treatment-adjusted |
Placebo |
Metformin |
Lifestyle |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | Observed P | Permuted P | FDR Q | HR (95% CI) | Observed P | HR (95% CI) | Observed P | HR (95% CI) | Observed P | |||||
| rs5017427 | 7 | 150886136 | PRKAG2 | T/C | 1.24 (1.09–1.40) | 0.0007 | 0.001 | 0.34 | 1.26 (1.04–1.53) | 0.02 | 1.15 (0.93–1.42) | 0.20 | 1.34 (1.05–1.71) | 0.02 |
| rs2453583 | 17 | 19382628 | SLC47A1 | T/A | 0.81 (0.72–0.92) | 0.001 | 0.003 | 0.34 | 0.84 (0.70–1.02) | 0.08 | 0.68 (0.54–0.86) | 0.001 | 0.94 (0.73–1.21) | 0.65 |
| rs315978 | 6 | 160572848 | SLC22A2 | T/C | 0.73 (0.60–0.89) | 0.002 | 0.002 | 0.34 | 0.75 (0.57–1.00) | 0.05 | 0.67 (0.47–0.96) | 0.03 | 0.74 (0.49–1.11) | 0.14 |
| rs4273018 | 15 | 97910079 | MEF2A | T/C | 1.20 (1.07–1.35) | 0.002 | 0.005 | 0.50 | 1.20 (1.00–1.44) | 0.05 | 1.37 (1.11–1.67) | 0.003 | 0.96 (0.76–1.23) | 0.76 |
| rs9551419 | 13 | 27378356 | PDX1 | T/C | 0.77 (0.65–0.92) | 0.004 | 0.003 | 0.34 | 0.77 (0.59–1.00) | 0.05 | 0.85 (0.63–1.14) | 0.27 | 0.67 (0.45–1.00) | 0.05 |
| rs1042531 | 20 | 55574386 | PCK1 | C/A | 0.84 (0.74–0.94) | 0.004 | 0.002 | 0.34 | 0.83 (0.69–1.00) | 0.05 | 0.98 (0.80–1.20) | 0.85 | 0.64 (0.49–0.84) | 0.001 |
| rs1342387 | 1 | 201180979 | ADIPOR1 | A/G | 1.17 (1.05–1.31) | 0.006 | 0.009 | 0.55 | 1.22 (1.02–1.45) | 0.03 | 1.07 (0.88–1.30) | 0.51 | 1.24 (0.99–1.56) | 0.07 |
| rs1388332 | 4 | 23438412 | PPARGC1A | G/A | 1.32 (1.08–1.61) | 0.006 | 0.007 | 0.53 | 1.17 (0.85–1.60) | 0.33 | 1.33 (0.94–1.88) | 0.10 | 1.51 (1.03–2.21) | 0.03 |
| rs6093976 | 20 | 42469194 | HNF4A | A/G | 0.78 (0.65–0.94) | 0.008 | 0.008 | 0.53 | 0.76 (0.57–1.00) | 0.05 | 0.85 (0.62–1.16) | 0.31 | 0.75 (0.51–1.10) | 0.14 |
| rs651164 | 6 | 160501364 | SLC22A1 | T/C | 1.18 (1.04–1.33) | 0.008 | 0.007 | 0.53 | 1.08 (0.90–1.30) | 0.39 | 1.14 (0.92–1.42) | 0.23 | 1.40 (1.09–1.78) | 0.01 |
| rs12330015 | 22 | 44968942 | PPARA | C/T | 0.79 (0.66–0.95) | 0.01 | 0.01 | 0.58 | 0.82 (0.63–1.07) | 0.15 | 0.78 (0.57–1.08) | 0.14 | 0.72 (0.49–1.06) | 0.10 |
| rs2755209 | 13 | 40035804 | FOXO1 | C/A | 1.16 (1.04–1.31) | 0.01 | 0.01 | 0.58 | 1.05 (0.89–1.25) | 0.56 | 1.29 (1.05–1.58) | 0.02 | 1.25 (0.97–1.60) | 0.08 |
| rs10875552 | 5 | 149169682 | PPARGC1B | C/T | 0.84 (0.74–0.96) | 0.01 | 0.008 | 0.53 | 0.91 (0.75–1.12) | 0.37 | 0.89 (0.72–1.11) | 0.32 | 0.65 (0.48–0.86) | 0.003 |
| rs739690 | 11 | 17373454 | KCNJ11 | C/G | 1.87 (1.15–3.05) | 0.01 | 0.01 | 0.66 | 1.16 (0.48–2.79) | 0.74 | 2.66 (1.33–5.31) | 0.01 | 2.23 (0.76–6.56) | 0.15 |
| rs7811022 | 7 | 99223619 | CYP3A4 | G/C | 1.33 (1.06–1.67) | 0.01 | 0.02 | 0.67 | 1.16 (0.81–1.65) | 0.42 | 1.99 (1.38–2.87) | 0.0002 | 0.95 (0.56–1.60) | 0.84 |
| rs832646 | 2 | 182266649 | NEUROD1 | C/T | 0.57 (0.35–0.91) | 0.02 | 0.02 | 0.68 | 0.59 (0.31–1.13) | 0.11 | 0.79 (0.37–1.72) | 0.56 | 0.24 (0.06–1.01) | 0.05 |
| rs11836547 | 12 | 1756708 | ADIPOR2 | C/G | 1.37 (1.05–1.78) | 0.02 | 0.02 | 0.67 | 1.60 (1.09–2.34) | 0.02 | 0.92 (0.56–1.52) | 0.75 | 1.72 (1.00–2.95) | 0.05 |
| rs916829 | 11 | 17397049 | ABCC8 | A/G | 0.80 (0.66–0.97) | 0.03 | 0.04 | 0.78 | 0.76 (0.56–1.02) | 0.06 | 0.89 (0.63–1.25) | 0.50 | 0.78 (0.52–1.16) | 0.21 |
| rs12951345 | 17 | 33151976 | HNF1B | G/T | 1.17 (1.02–1.34) | 0.03 | 0.03 | 0.73 | 1.13 (0.91–1.40) | 0.27 | 1.20 (0.95–1.52) | 0.12 | 1.20 (0.91–1.59) | 0.20 |
| rs709159 | 3 | 12456203 | PPARG | C/A | 1.16 (1.01–1.33) | 0.04 | 0.04 | 0.82 | 1.06 (0.86–1.31) | 0.56 | 1.21 (0.95–1.56) | 0.13 | 1.27 (0.96–1.68) | 0.09 |
| rs12058717 | 1 | 159108473 | ITLN1 | T/C | 1.28 (1.01–1.63) | 0.04 | 0.049 | 0.87 | 1.24 (0.89–1.74) | 0.20 | 1.35 (0.87–2.09) | 0.18 | 1.32 (0.77–2.28) | 0.31 |
SNPs in or near biological candidate genes showing nominal association with diabetes incidence in the DPP are shown. HRs are estimated for the minor allele (m) vs. the major allele (M) under an additive genetic model. Only the top SNP within each gene region is shown; the full set of results (including allele frequencies) is available in supplementary Table 2.
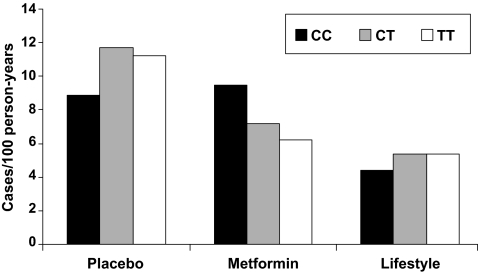
Table 4 shows the candidate gene regions harboring variants that have a nominally significant genotype × metformin interaction; only the top SNP within each gene region is given (out of 91 nominal associations). The best result was consistent with a study-wide 33% FDR. At rs8065082 in SLC47A1, there was a nominal interaction with metformin (P = 0.006), with the minor allele associated with lower diabetes incidence in the metformin arm (HR 0.78, 95% CI 0.64–0.96, P = 0.02) but not in the placebo arm (1.15, 0.97–1.37, P = 0.11). At this locus, major allele homozygotes did not benefit from metformin with regard to diabetes prevention (HR 1.07, 95% CI 0.77–1.50, vs. placebo, P = 0.68), whereas minor allele carriers did (0.58, 0.46–0.73, vs. placebo, P < 0.001; Fig. 1). We also noted a nominally significant interaction of a missense SNP in SLC22A1 (rs683369, encoding L160F) with metformin, with the major allele protecting from diabetes in the metformin arm (HR 0.69, 95% CI 0.53–0.89, P = 0.004) but not the placebo arm (1.01, 0.79–1.30, P = 0.91); the major allele is therefore associated with 31% risk reduction in diabetes incidence but only under the action of metformin. In this arm, the likelihood of developing diabetes depended on the number of phenylalanine alleles (HR 0.72, 95% CI 0.59–0.88, vs. placebo for LL homozygotes; 0.92, 0.66–1.28, for heterozygotes; and 1.44, 0.56–3.67, for FF homozygotes). There were five nominally significant interactions at SNPs encoding putative drug targets for metformin, in the gene encoding the AMPK kinase STK11 and the AMPK subunit genes PRKAA1, PRKAA2, and PRKAB2, respectively. A total of 22 SNPs in the ABCC8-KCNJ11 region also had nominally significant interactions with metformin, including rs5215, which is tightly linked to the widely replicated type 2 diabetes–associated missense SNP rs5219 (E23K) in KCNJ11.
TABLE 4.
Candidate gene variants showing a nominally significant interaction with the metformin intervention in the DPP
| SNP | Chromosome | Position (NCBI 36) | Gene | Alleles (m/M) | Interaction genotype * metformin |
Placebo |
Metformin |
Lifestyle |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed P | Permuted P | FDR Q | HR (95% CI) | Observed P | HR (95% CI) | Observed P | HR (95% CI) | Observed P | |||||
| rs11868513 | 17 | 33126805 | HNF1B | A/G | 0.0007 | 0.001 | 0.33 | 1.69 (1.36–2.10) | 0.000002 | 0.87 (0.65–1.16) | 0.33 | 1.08 (0.79–1.47) | 0.64 |
| rs4148609 | 11 | 17441307 | ABCC8 | A/G | 0.002 | 0.002 | 0.33 | 1.24 (1.04–1.48) | 0.02 | 0.79 (0.63–0.98) | 0.03 | 1.06 (0.83–1.37) | 0.64 |
| rs11086926 | 20 | 42492111 | HNF4A | G/T | 0.002 | 0.001 | 0.33 | 0.82 (0.61–1.11) | 0.20 | 1.81 (1.35–2.43) | 0.0001 | 1.29 (0.91–1.83) | 0.15 |
| rs10213440 | 4 | 23475437 | PPARGC1A | C/T | 0.002 | 0.002 | 0.33 | 0.76 (0.59–0.97) | 0.03 | 1.31 (1.03–1.66) | 0.03 | 1.07 (0.80–1.43) | 0.65 |
| rs4424892 | 15 | 97921908 | MEF2A | G/A | 0.003 | 0.006 | 0.38 | 0.88 (0.72–1.09) | 0.24 | 1.43 (1.14–1.80) | 0.002 | 0.78 (0.58–1.05) | 0.10 |
| rs8065082 | 17 | 19405783 | SLC47A1 | T/C | 0.006 | 0.008 | 0.41 | 1.15 (0.97–1.37) | 0.11 | 0.78 (0.64–0.96) | 0.02 | 1.09 (0.86–1.38) | 0.46 |
| rs6666307 | 1 | 154719358 | MEF2D | T/A | 0.009 | 0.005 | 0.38 | 0.54 (0.26–1.09) | 0.08 | 2.15 (1.22–3.80) | 0.01 | 1.11 (0.44–2.77) | 0.83 |
| rs3792269 | 2 | 241180152 | CAPN10 | G/A | 0.01 | 0.01 | 0.41 | 1.61 (1.28–2.02) | 0.00005 | 0.95 (0.70–1.28) | 0.73 | 0.81 (0.55–1.18) | 0.27 |
| rs758027 | 12 | 1662034 | ADIPOR2 | C/T | 0.01 | 0.002 | 0.33 | 0.34 (0.15–0.75) | 0.01 | 1.31 (0.76–2.29) | 0.33 | 0.75 (0.26–2.15) | 0.59 |
| rs7124355 | 11 | 17369536 | KCNJ11 | A/G | 0.01 | 0.01 | 0.41 | 0.84 (0.68–1.04) | 0.10 | 1.25 (0.99–1.59) | 0.06 | 0.89 (0.68–1.17) | 0.41 |
| rs662301 | 6 | 160616909 | SLC22A2 | T/C | 0.02 | 0.02 | 0.47 | 0.78 (0.49–1.22) | 0.27 | 1.57 (1.09–2.27) | 0.02 | 0.60 (0.31–1.17) | 0.13 |
| rs2908289 | 7 | 44190467 | GCK | A/G | 0.02 | 0.02 | 0.47 | 0.86 (0.69–1.08) | 0.19 | 1.29 (1.02–1.64) | 0.04 | 0.97 (0.73–1.31) | 0.86 |
| rs741765 | 19 | 1172545 | STK11 | T/C | 0.02 | 0.02 | 0.47 | 1.17 (0.96–1.43) | 0.13 | 0.82 (0.64–1.04) | 0.10 | 0.95 (0.72–1.26) | 0.73 |
| rs6701920 | 1 | 159181541 | ITLN2 | A/G | 0.02 | 0.01 | 0.41 | 1.86 (1.19–2.91) | 0.01 | 0.49 (0.17–1.40) | 0.19 | 0.69 (0.27–1.78) | 0.45 |
| rs741579 | 5 | 149165563 | PPARGC1B | G/A | 0.02 | 0.01 | 0.38 | 2.41 (1.08–5.37) | 0.03 | 0.23 (0.03–1.62) | 0.14 | 0.68 (0.17–2.71) | 0.58 |
| rs4810083 | 20 | 55553677 | PCK1 | T/C | 0.03 | 0.03 | 0.53 | 1.14 (0.95–1.37) | 0.15 | 0.84 (0.69–1.02) | 0.08 | 1.14 (0.90–1.44) | 0.27 |
| rs9803799 | 1 | 56952660 | PRKAA2 | G/T | 0.03 | 0.02 | 0.49 | 1.17 (0.85–1.62) | 0.34 | 0.66 (0.43–1.02) | 0.06 | 1.13 (0.72–1.78) | 0.59 |
| rs17367421 | 1 | 153553865 | PKLR | C/G | 0.03 | 0.03 | 0.53 | 0.47 (0.23–0.96) | 0.04 | 1.21 (0.73–1.99) | 0.46 | 1.36 (0.77–2.38) | 0.29 |
| rs683369 | 6 | 160471194 | SLC22A1 | G/C | 0.03 | 0.03 | 0.53 | 0.99 (0.77–1.27) | 0.91 | 1.45 (1.12–1.88) | 0.004 | 0.82 (0.58–1.17) | 0.27 |
| rs249429 | 5 | 40817996 | PRKAA1 | C/T | 0.04 | 0.03 | 0.53 | 1.22 (1.01–1.46) | 0.04 | 0.89 (0.71–1.13) | 0.34 | 1.24 (0.97–1.59) | 0.09 |
| rs6733736 | 2 | 162704406 | GCG | G/A | 0.04 | 0.01 | 0.43 | 0.65 (0.20–2.13) | 0.48 | 3.70 (1.56–8.80) | 0.003 | 0.59 (0.08–4.45) | 0.61 |
| rs4253652 | 22 | 44947503 | PPARA | G/A | 0.04 | 0.02 | 0.47 | 0.60 (0.27–1.33) | 0.20 | 2.29 (1.10–4.76) | 0.03 | 1.07 (0.44–2.65) | 0.88 |
| rs6690158 | 1 | 145123867 | PRKAB2 | T/C | 0.04 | 0.03 | 0.53 | 0.58 (0.33–1.03) | 0.06 | 1.42 (0.85–2.36) | 0.18 | 1.56 (0.87–2.81) | 0.14 |
SNPs in or near biological candidate genes showing a nominally significant interaction with the metformin intervention in the DPP are shown. HRs are estimated for the minor allele (m) vs. the major allele (M) under an additive genetic model. Only the top SNP within each gene region is shown; the full set of results (including allele frequencies) is available in supplementary Table 2.
FIG. 1.
Diabetes incidence in the DPP, by genotype at rs8065082 in the SLC47A1 gene. This SNP is in tight LD with rs2289669 (r2 ∼0.8), whose major allele predicts a poorer response to metformin (5). In the DPP, major allele homozygotes at rs8065082 did not benefit from metformin with regard to diabetes prevention, whereas minor allele carriers did (P < 0.001).
Table 5 shows the candidate gene regions harboring variants that have a nominally significant interaction with the lifestyle intervention; only the top SNP within each gene region is given (out of 69 nominal associations). The best result was consistent with an experiment-wide 84% FDR. Twelve of the top findings were in four AMPK subunit genes (PRKAA2, PRKAB2, PRKAG1, and PRKAG2), and 11 SNPs clustered around the peroxisome proliferator–associated receptor γ coactivators 1α and 1β (PPARGC1A and PPARGC1B, respectively).
TABLE 5.
Candidate gene variants showing a nominally significant interaction with the lifestyle intervention in the DPP
| SNP | Chromosome | Position (NCBI 36) | Gene | Alleles (m/M) | Interaction genotype * lifestyle |
Placebo |
Metformin |
Lifestyle |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed P | Permuted P | FDR Q | HR (95% CI) | Observed P | HR (95% CI) | Observed P | HR (95% CI) | Observed P | |||||
| rs3792269 | 2 | 241180152 | CAPN10 | G/A | 0.005 | 0.008 | 0.84 | 1.61 (1.28–2.02) | 0.00005 | 0.95 (0.70–1.28) | 0.73 | 0.81 (0.55–1.18) | 0.27 |
| rs2425640 | 20 | 42461451 | HNF4A | A/G | 0.01 | 0.01 | 0.84 | 0.83 (0.69–1.01) | 0.06 | 0.93 (0.75–1.16) | 0.51 | 1.23 (0.96–1.58) | 0.11 |
| rs1342514 | 1 | 56959948 | PRKAA2 | G/C | 0.01 | 0.005 | 0.84 | 1.07 (0.90–1.29) | 0.44 | 1.14 (0.92–1.41) | 0.25 | 0.71 (0.54–0.92) | 0.01 |
| rs4725408 | 7 | 150881542 | PRKAG2 | C/T | 0.02 | 0.005 | 0.84 | 0.82 (0.39–1.73) | 0.61 | 1.02 (0.41–2.55) | 0.97 | 2.90 (1.52–5.56) | 0.001 |
| rs7599142 | 2 | 162701737 | GCG | A/G | 0.02 | 0.01 | 0.84 | 1.02 (0.84–1.23) | 0.86 | 1.00 (0.81–1.22) | 0.97 | 0.66 (0.51–0.86) | 0.002 |
| rs17367421 | 1 | 153553865 | PKLR | C/G | 0.02 | 0.01 | 0.84 | 0.47 (0.23–0.96) | 0.04 | 1.21 (0.73–1.99) | 0.46 | 1.36 (0.77–2.38) | 0.29 |
| rs12374408 | 4 | 23463581 | PPARGC1A | T/C | 0.02 | 0.03 | 0.84 | 1.13 (0.94–1.36) | 0.20 | 0.94 (0.76–1.17) | 0.60 | 0.77 (0.59–1.02) | 0.07 |
| rs3751151 | 12 | 119926582 | HNF1A | T/A | 0.02 | 0.02 | 0.84 | 0.89 (0.73–1.09) | 0.26 | 0.82 (0.65–1.04) | 0.10 | 1.30 (1.01–1.67) | 0.04 |
| rs11024298 | 11 | 17448407 | ABCC8 | T/G | 0.02 | 0.01 | 0.84 | 1.17 (0.82–1.67) | 0.39 | 1.01 (0.65–1.58) | 0.95 | 0.44 (0.21–0.92) | 0.03 |
| rs6008306 | 22 | 45018756 | PPARA | T/C | 0.02 | 0.01 | 0.84 | 0.52 (0.25–1.08) | 0.08 | 0.96 (0.49–1.91) | 0.91 | 2.05 (0.97–4.33) | 0.06 |
| rs1422429 | 5 | 149146627 | PPARGC1B | A/G | 0.02 | 0.02 | 0.84 | 0.83 (0.69–0.99) | 0.04 | 0.96 (0.79–1.17) | 0.70 | 1.19 (0.94–1.50) | 0.15 |
| rs1054442 | 12 | 47675587 | PRKAG1 | C/A | 0.03 | 0.02 | 0.84 | 1.22 (1.02–1.45) | 0.03 | 1.08 (0.88–1.32) | 0.47 | 0.87 (0.68–1.12) | 0.27 |
| rs9965495 | 18 | 56184656 | MC4R | T/C | 0.03 | 0.02 | 0.84 | 1.09 (0.90–1.31) | 0.39 | 1.20 (0.97–1.49) | 0.09 | 0.75 (0.58–0.98) | 0.04 |
| rs1008284 | 17 | 33136571 | HNF1B | T/C | 0.03 | 0.03 | 0.84 | 1.30 (1.08–1.58) | 0.01 | 0.91 (0.71–1.15) | 0.43 | 0.89 (0.68–1.18) | 0.42 |
| rs2018675 | 17 | 19382123 | SLC47A1 | T/C | 0.03 | 0.02 | 0.84 | 1.18 (0.99–1.40) | 0.06 | 1.13 (0.92–1.38) | 0.24 | 0.88 (0.69–1.13) | 0.33 |
| rs8032587 | 15 | 98068933 | MEF2A | C/T | 0.03 | 0.02 | 0.84 | 1.19 (0.72–1.95) | 0.50 | 1.07 (0.56–2.05) | 0.83 | 0.31 (0.10–1.00) | 0.05 |
| rs7626560 | 3 | 12450088 | PPARG | A/G | 0.04 | 0.04 | 0.84 | 1.16 (0.93–1.45) | 0.19 | 1.12 (0.88–1.44) | 0.36 | 0.76 (0.55–1.06) | 0.10 |
| rs17161829 | 7 | 99187763 | CYP3A4 | A/G | 0.04 | 0.01 | 0.84 | 1.05 (0.69–1.60) | 0.82 | 1.24 (0.77–1.98) | 0.38 | 0.32 (0.11–0.93) | 0.04 |
| rs6701920 | 1 | 159181541 | ITLN2 | A/G | 0.04 | 0.02 | 0.84 | 1.86 (1.19–2.91) | 0.01 | 0.49 (0.17–1.40) | 0.19 | 0.69 (0.27–1.78) | 0.45 |
| rs461473 | 6 | 160463552 | SLC22A1 | T/C | 0.04 | 0.04 | 0.84 | 0.92 (0.64–1.33) | 0.67 | 0.93 (0.64–1.36) | 0.72 | 1.50 (1.01–2.22) | 0.04 |
| rs17159890 | 1 | 145094998 | PRKAB2 | C/A | 0.046 | 0.05 | 0.84 | 0.84 (0.56–1.26) | 0.40 | 1.01 (0.62–1.63) | 0.98 | 1.88 (1.13–3.14) | 0.02 |
| rs11904814 | 2 | 208135043 | CREB1 | C/A | 0.046 | 0.05 | 0.84 | 1.03 (0.86–1.23) | 0.78 | 1.15 (0.93–1.42) | 0.18 | 0.74 (0.57–0.96) | 0.03 |
| rs17373414 | 3 | 188068221 | ADIPOQ | T/C | 0.049 | 0.06 | 0.84 | 0.82 (0.57–1.18) | 0.28 | 0.90 (0.63–1.29) | 0.57 | 1.34 (0.90–1.98) | 0.15 |
SNPs in or near biological candidate genes showing a nominally significant interaction with the lifestyle intervention in the DPP are shown. HRs are estimated for the minor allele (m) vs. the major allele (M) under an additive genetic model. Only the top SNP within each gene region is shown; the full set of results (including allele frequencies) is available in supplementary Table 2.
Review of 1,609 SNPs imputed in non-Hispanic white DPP participants (supplementary Table 3) revealed the nominal association of other PRKAG2 SNPs with diabetes incidence (best P = 5 × 10−5). Imputed SNPs in the PRKAA1, PRKAA2, and ABCC8-KCNJ11 regions also had nominally significant interactions with metformin.
DISCUSSION
We conducted a large-scale genotyping study in the DPP, with the aim to test whether common variants in candidate genes involved in major spheres of human physiology predict diabetes incidence or response to preventive interventions in a multiethnic at-risk population. Our secondary purpose was to characterize the mechanism of action of previously associated variants. We provide evidence supporting a previously reported association of variants in the metformin transporter gene SLC47A1 with weaker metformin response, here defined as the reduced ability of metformin to lower diabetes incidence (5). We identified a number of nominal associations with diabetes incidence or metformin response in several compelling candidate genes; however, none stand strict statistical correction for multiple hypothesis testing by FDRs.
Correction for multiple tests requires careful consideration in genetic association studies (26). When large numbers of SNPs are tested, methods that are valid in the presence of correlations due to LD, such as permutation methods or evaluation of FDR, are preferred over those that assume independence of SNPs. The scope of the present analysis is guided by technological convenience, and it might be argued that the number of distinct scientific hypotheses formulated, rather than the physical size of the genotyping array, is most relevant to the interpretation of results. However, what constitutes a single hypothesis (e.g., an SNP, a gene, an entire pathway, or a constellation of phenotypes) is subjective. On the other hand, correcting for the equivalent of the universe of independent common variants in the human genome (empirically estimated at ∼1 million [27]) is gaining increasing favor among genetic statisticians. In this context, the novel findings reported here should be viewed as hypothesis-generating.
We previously quantified the power of the DPP to detect modest genetic effects on diabetes incidence (28). Assuming there are no gene-treatment interactions, these calculations show that the overall DPP cohort has 83% power to detect a previously reported effect size of ∼20% for an SNP of 10% frequency at an α level of 0.05, while the placebo, lifestyle modification, and metformin arms have 53, 34, and 44% power, respectively. The DPP has inadequate power for detecting an effect size of <10%. Thus, it is not surprising that the DPP does not replicate all GWAS-derived findings in that range or that it fails to reach genome-wide significance in discovery efforts. Our null results on diabetes incidence for truly associated variants may be due to the high-risk population at baseline, the short time of follow-up (3.2 years on average), and/or the use of interventions effective in reducing diabetes incidence. On the other hand, considering the number of variants likely to influence the phenotypes under study, even submaximal power is likely to provide a number of true positive associations. In this context, genotyped and imputed SNPs in the gene encoding the AMPK γ2 subunit (PRKAG2) merit further consideration. While the association of SNPs in genes that encode metformin transporters with type 2 diabetes in the entire DPP cohort (if real) requires explanation, this could be due to a sufficiently strong effect in the metformin arm alone. Alternatively, SNPs in this region could be capturing variants in other nearby genes: for instance, immediately upstream of SLC22A1 and SLC22A2 in chromosome 6 lies the gene encoding the insulin-like growth factor 2 receptor (IGF2R), an excellent biological candidate.
This study constitutes the first large-scale prospective pharmacogenetic evaluation of metformin action in a controlled clinical trial. The UK Prospective Diabetes Study (29) and A Diabetes Outcome Progression Trial (ADOPT) (30) investigators independently showed that a substantial proportion of patients with type 2 diabetes eventually fail metformin therapy, defined by a need for additional pharmacotherapy to control hyperglycemia. Given the higher prior probability afforded by the known biological role of SLC47A1 in disposing of metformin and the previously reported genetic association of the major allele at SNP rs2289669 with poorer metformin response (5), validation in the DPP can be convincing without achieving the levels of statistical significance required for novel findings. Our index SNP (rs8065082) is in tight LD with rs2289669 (r2 ∼0.8 in HapMap CEU) and the direction of effect is consistent in DPP, a cohort nearly 10-fold larger than the one documented in the original report from Rotterdam (5). Thus, our findings confirm those of Becker et al. (5) and suggest that major allele homozygotes at this locus (∼30% of the European population) may experience suboptimal responses to metformin treatment.
Our findings on the SLC22A1 locus and metformin response are less robust. While our noted association with a missense SNP appears compelling, it is not among the most functional human variants described by Shu et al. (3), and it is in weak LD with rs622342 (r2 ∼0.14 in HapMap CEU), a SLC22A1 SNP associated with metformin response in another report from Rotterdam (31). SNP rs622342 was included among our tag SNPs but showed no evidence of an interaction with metformin (nominal P = 0.69) or an effect on diabetes incidence in any arm, raising the possibility that the original finding may have been spurious. Similarly, the SLC22A2 missense SNP rs316019 (A270S), reported to influence metformin renal excretion and affect its plasma concentrations (32), did not significantly interact with metformin in the DPP (nominal P = 0.35). Our novel findings in the putative metformin drug targets STK11 and AMPK require confirmation, as do those in MEF2A and MEF2D, themselves regulated by AMPK (33). One of the most significant interactions with metformin occurred at an SNP in HNF4A; given its role in hepatic gluconeogenesis (34), this intriguing result deserves further exploration. In contrast, the multiple interactions noted in the ABCC8-KCNJ11 locus reported previously (35) do not offer a clear mechanism of action. Finally, nominal associations with response to lifestyle modification should be replicated in cohorts that underwent a similar intervention.
In summary, we have conducted a large-scale genetic association study in the DPP and replicated the association of a polymorphism in a metformin transporter with metformin response. Other hypothesis-generating results require more detailed characterization in the DPP and follow-up in independent samples. A focus on likely functional variants may uncover loci with stronger effects.
Supplementary Material
ACKNOWLEDGMENTS
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the coordinating center for the design and conduct of the study and collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the Office of Research on Minority Health, the National Institute of Child Health and Human Development, the National Institute on Aging, the Office of Research on Women's Health, the Centers for Disease Control and Prevention, and the American Diabetes Association. Bristol-Myers Squibb and Parke-Davis provided medication. This research was also supported in part by the intramural research program of the NIDDK. LifeScan, Health O Meter, Hoechst Marion Roussel, Merck-Medco Managed Care, Merck and Company, Nike Sports Marketing, Slim Fast Foods, and Quaker Oats donated materials, equipment, or medicines for concomitant conditions. McKesson BioServices, Matthews Media Group, and the Henry M. Jackson Foundation provided support services under subcontract with the coordinating center. A complete list of centers, investigators, and staff can be found in the online appendix.
This work was funded by R01 DK072041 to K.A.J., T.I.P., A.R.S., D.A., and J.C.F. J.C.F. is also supported by the Massachusetts General Hospital and a Clinical Scientist Development Award by the Doris Duke Charitable Foundation. This work was partially supported by a Doris Duke Charitable Foundation Distinguished Scientist Clinical Award to D.A. P.W.F. was supported in part by grants from Novo Nordisk, the Swedish Heart-Lung Foundation, the Swedish Diabetes Association, Påhlssons Foundation, the Swedish Research Council, and a Career Development Award from Umeå University. J.C.F. received consulting honoraria from Publicis Healthcare, Merck, bioStrategies, XOMA, and Daiichi-Sankyo and has been a paid invited speaker at internal scientific seminars hosted by Pfizer and Alnylam Pharmaceuticals.
No other potential conflicts of interest relevant to this article were reported.
R.L.H. and W.C.K. contributed to the participant recruitment, interventions, and outcomes assessment. P.W.F., T.I.P., R.L.H., R.S., A.R.S., W.C.K., D.A., and J.C.F. compiled the list of candidate genes. J.B.M. and P.I.W.d.B. performed the tagging procedure in European and African populations. P.W.F., T.I.P., R.L.H., R.S., A.R.S., and J.C.F. selected SNPs within those genes. J.B.M. directed the genotyping with supervision from J.C.F. K.A.J., P.I.W.d.B., T.I.P., R.L.H., and J.C.F. constructed the analytical pipeline. K.A.J. conducted all statistical analyses, with input from P.I.W.d.B., T.I.P., R.L.H., S.F., W.C.K., and D.A. K.A.J., R.L.H., and P.I.W.d.B. implemented the permutation procedure, with input from T.I.P., D.A., and J.C.F. J.B.M. and P.I.W.d.B. derived individual estimates of global ancestry and carried out SNP imputation. J.C.F. wrote the manuscript. All authors contributed to the discussion and reviewed and edited the manuscript.
The investigators gratefully acknowledge the commitment and dedication of the participants of the DPP.
Footnotes
Clinical trial reg. no. NCT00004992, clinicaltrials.gov.
The opinions expressed in this article are those of the investigators and do not necessarily reflect the views of the Indian Health Service or other funding agencies.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1. Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet 2008;24:613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pearson ER. Pharmacogenetics in diabetes. Curr Diab Rep 2009;9:172–181 [DOI] [PubMed] [Google Scholar]
- 3. Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 2007;117:1422–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou K, Donnelly LA, Kimber CH, Donnan PT, Doney AS, Leese G, Hattersley AT, McCarthy MI, Morris AD, Palmer CN, Pearson ER. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes 2009;58:1434–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes 2009;58:745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM. Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diabetes Prevention Program Research Group The Diabetes Prevention Program: Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care 1999;22:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005;37:1217–1223 [DOI] [PubMed] [Google Scholar]
- 9. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 10. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Boström KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jørgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjögren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ. Wellcome Trust Case Control Consortium Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Florez JC, Manning AK, Dupuis J, McAteer J, Irenze K, Gianniny L, Mirel DB, Fox CS, Cupples LA, Meigs JB. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes 2007;56:3063–3074 [DOI] [PubMed] [Google Scholar]
- 12. Hanson RL, Bogardus C, Duggan D, Kobes S, Knowlton M, Infante AM, Marovich L, Benitez D, Baier LJ, Knowler WC. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes 2007;56:3045–3052 [DOI] [PubMed] [Google Scholar]
- 13. Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, Zhang L, Zhao Y, Mitchell BD, O'Connell J, Shuldiner AR. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes 2007;56:3053–3062 [DOI] [PubMed] [Google Scholar]
- 14. Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O'Rahilly S, Purmann C, Rees MG, Ridderstråle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N. Wellcome Trust Case Control Consortium Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN. Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009;41:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009;41:18–24 [DOI] [PubMed] [Google Scholar]
- 16. Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 2008;40:189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 2009;41:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O'Brien SJ, Reich D. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 2004;74:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am J Hum Genet 2007;80:1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet S 2006;79:2290 [Google Scholar]
- 22. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dudoit S, Shaffer JP, Boldrick JC. Multiple hypothesis testing in microarray experiments. Statistical Science 2003;18:71–103 [Google Scholar]
- 24. Potter DM. Ominibus permutation tests of the association of an ensemble of genetic markers with disease in case-control studies. Genet Epidemiol 2006;30:438–446 [DOI] [PubMed] [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300 [Google Scholar]
- 26. Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet 2005;366:1315–1323 [DOI] [PubMed] [Google Scholar]
- 27. Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008;32:381–385 [DOI] [PubMed] [Google Scholar]
- 28. Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, Franks PW, Hanson RL, Shuldiner AR, Knowler WC, Altshuler D, Florez JC. Diabetes Prevention Program Research Group Extension of type 2 diabetes genome-wide association scan results in the Diabetes Prevention Program. Diabetes 2008;57:2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turner RC, Cull CA, Frighi V, Holman RR. Glycemic control with diet, sulfonylurea, metformin, or insulin in patients with type 2 diabetes mellitus: progressive requirement for multiple therapies (UKPDS 49): UK Prospective Diabetes Study (UKPDS) Group. JAMA 1999;281:2005–2012 [DOI] [PubMed] [Google Scholar]
- 30. Kahn SE, Haffner SM, Heise MA, Herman WH, Holman RR, Jones NP, Kravitz BG, Lachin JM, O'Neill MC, Zinman B, Viberti G. ADOPT Study Group Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N Engl J Med 2006;355:2427–2443 [DOI] [PubMed] [Google Scholar]
- 31. Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J 2009;9:242–247 [DOI] [PubMed] [Google Scholar]
- 32. Song IS, Shin HJ, Shim EJ, Jung IS, Kim WY, Shon JH, Shin JG. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin Pharmacol Ther 2008;84:559–562 [DOI] [PubMed] [Google Scholar]
- 33. Holmes BF, Sparling DP, Olson AL, Winder WW, Dohm GL. Regulation of muscle GLUT4 enhancer factor and myocyte enhancer factor 2 by AMP-activated protein kinase. Am J Physiol Endocrinol Metab 2005;289:E1071–E1076 [DOI] [PubMed] [Google Scholar]
- 34. Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci U S A 2003;100:4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Florez JC, Jablonski KA, Kahn SE, Franks PW, Dabelea D, Hamman RF, Knowler WC, Nathan DM, Altshuler D. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes 2007;56:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.