Abstract
OBJECTIVE
Tissue-specific self-antigens are ectopically expressed within the thymus and play an important role in the induction of central tolerance. Insulin is expressed in both pancreatic islets and the thymus and is considered to be the primary antigen for type 1 diabetes. Here, we report the role of the insulin transactivator MafA in the expression of insulin in the thymus and susceptibility to type 1 diabetes.
RESEARCH DESIGN AND METHODS
The expression profiles of transcriptional factors (Pdx1, NeuroD, Mafa, and Aire) in pancreatic islets and the thymus were examined in nonobese diabetic (NOD) and control mice. Thymic Ins2 expression and serum autoantibodies were examined in Mafa knockout mice. Luciferase reporter assay was performed for newly identified polymorphisms of mouse Mafa and human MAFA. A case-control study was applied for human MAFA polymorphisms.
RESULTS
Mafa, Ins2, and Aire expression was detected in the thymus. Mafa expression was lower in NOD thymus than in the control and was correlated with Ins2 expression. Targeted disruption of MafA reduced thymic Ins2 expression and induced autoantibodies against pancreatic islets. Functional polymorphisms of MafA were newly identified in NOD mice and humans, and polymorphisms of human MAFA were associated with susceptibility to type 1 diabetes but not to autoimmune thyroid disease.
CONCLUSIONS
These data indicate that functional polymorphisms of MafA are associated with reduced expression of insulin in the thymus and susceptibility to type 1 diabetes in the NOD mouse as well as human type 1 diabetes.
Type 1 diabetes is caused by autoimmune destruction of insulin-producing β-cells of the pancreas in genetically susceptible individuals (1,2). Susceptibility to type 1 diabetes is under polygenic control, with IDDM1 in the major histocompatibility complex (MHC) showing the strongest effect (3). In addition to MHC-linked susceptibility, the contribution of several non-MHC genes has been reported (3–6). Most of the non-MHC genes identified to date are immune-regulating genes, which are considered to contribute to type 1 diabetes susceptibility through impaired regulation of autoimmune T-cell activation. Among these are genes encoding cytotoxic T-lymphocyte antigen 4 (CTLA4) in humans and mice (7), lymphoid tyrosine phosphatase (PTPN22) (8) in humans, and Cblb (9) and Ian4 (10) in rats. Most of these genes are therefore expected to confer susceptibility to autoimmune diseases in general but not to an autoimmune disease in a specific organ, as evidenced by the association of these genes with not only type 1 diabetes but also other autoimmune diseases, such as autoimmune thyroid diseases, rheumatoid arthritis, and/or systemic lupus erythematosus (7,11–13).
In contrast to immune-regulating genes conferring susceptibility to autoimmune diseases through dysregulation of T-cell activation, genes leading to organ specificity are largely unknown, with the only exception being IDDM2 located in the promoter region of the insulin gene (INS). IDDM2 is most likely to be encoded by a variable-number tandem repeat (VNTR) polymorphism in the cis-regulatory region of the insulin gene, which is associated with type 1 diabetes susceptibility through reduced expression of the insulin gene in the thymus (14–17). Accumulating lines of evidence indicate that tissue-specific self-antigens, including insulin, are also expressed within the thymus and play an important role in the induction of central tolerance (18–20). Abnormality in the regulation of intrathymic expression of self-antigen is therefore expected to cause autoimmune disease through impaired negative selection of antigen-specific autoreactive T-cells. Since expression of autoantigens in the thymus is regulated not only by cis-regulatory elements in the genes encoding self-antigens, as in the case of IDDM2, but also by trans-acting factors, tissue-specific transactivators regulating the expression of self-antigens are important candidate genes for organ-specific autoimmune diseases.
MafA has been identified as an islet-enriched transcriptional activator that binds to the RIPE3b1 element in the promoter of the insulin gene and has been postulated to regulate insulin transcription in response to serum glucose level in β-cells of the pancreas (21,22). Unlike previously known islet-enriched transcriptional factors, such as Pdx1 and NeuroD/BETA2, which are expressed in non–β-cells as well as in β-cells, the expression of MafA is restricted to only β-cells, suggesting that MafA is responsible for tissue-specific expression of insulin (23). MafA is therefore an important candidate among these transcriptional activators of insulin, leading to organ-specific autoimmunity to pancreatic β-cells by dysregulation of transcription of insulin in the thymus as an organ-specific self-antigen. Here, we report that MafA is expressed in the thymus and colocalizes with and modulates the expression of insulin in the thymus. Functional variants of mouse Mafa and human MAFA were identified and found to be associated with the expression level of insulin in the thymus and susceptibility to type 1 diabetes.
RESEARCH DESIGN AND METHODS
Female and male nonobese diabetic (NOD)/shi, NOD.nonobese nondiabetic (NON)-Mhc(H2) congenic (NOD.NON-H2), C3H/He, and NSY mice (24) were housed under specific pathogen-free conditions. All experiments were conducted in accordance with the Osaka University Guidelines, which are based on the National Institutes of Health's Guide for the Care and Use of Laboratory Animals. ICR mice with targeted disruption of Mafa (Mafa knockout mice) were provided by S.T. (25).
Semiquantitative RT-PCR.
Pancreatic islets were isolated from four or five female NOD.NON-H2 or C3H mice by collagenase digestion, as described previously (26). To collect insulitis-free pancreatic islets, NOD.NON-H2 mice, instead of NOD mice, were used for isolation of pancreatic islets for RT-PCR analysis. Total RNA was isolated from mouse pancreatic islets (at 14–16 weeks old) and thymus (at 3–25 days old) using Isogen (Nippon Gene, Toyama, Japan) and treated with 10 units RNase-free DNase I (Takara, Shiga, Japan) to remove genomic DNA. Then, 800 ng total RNA from each sample was subjected to cDNA synthesis using oligo-dT primers (ReverTra Aceα; Toyobo, Tokyo, Japan) (additional supplementary materials and methods for this article can be found in an online appendix, available at http://diabetes.diabetesjournals.org/cgi/content/full/db10-0476/DC1). PCR was performed using Ex Taq (Takara) within the log phase of the reaction (24–32 cycles). mRNA levels were measured by nonradioactive RT-PCR and charged-coupled device imaging, as described previously (27).
Immunohistochemical staining.
The thymus was embedded in OCT compound (Tissue-TEC; Miles, Elkhart, IN) and frozen by an acetone–dry-ice method. Then, 6-μm-thick frozen sections were cut with a cryostat, placed on slides, and fixed in cold acetone for 10 min. The sections were then rinsed in PBS, incubated for 5 min in 1% Triton X-100, and, after a second rinse, incubated in diluted serum derived from the same animal as the blocking serum. The sections were preincubated with 2.4G2 to block the Fcγ receptor and incubated for 60 min at room temperature with the first antibodies, washed with PBS, and incubated for 60 min on ice with fluorescein-conjugated second antibodies. The first antibodies were guinea pig anti-porcine insulin antibody (Dako Japan, Kyoto, Japan), rabbit anti-mouse MafA antibody (Bethyl Laboratories, Montgomery, TX), and 1:100 diluted sera from Mafa+/+ or Mafa−/− mice. Alexa Fluor 488–conjugated anti-rabbit IgG, Alexa Fluor 594–conjugated anti-guinea pig IgG, and Alexa Fluor 488–conjugated anti-mouse IgG were used as second antibodies. The sections were examined using a Provis AX80 (Olympus, Tokyo, Japan).
Genomic sequences.
Genomic DNA was amplified by PCR using Takara LA Taq (Takara). The refined products were subjected to direct sequencing, utilizing dye termination chemistry with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Tokyo, Japan). To minimize sequencing artifacts, sequencing was performed by direct sequencing of the PCR products, with both strands sequenced at least twice in each strain. The sequences were compared with the sequence of Mafa from the BALB/c mouse, which was provided by K.K.
Subjects for molecular scanning and case-control study.
A total of 96 unrelated Japanese subjects (16 control subjects, 16 individuals with type 1 diabetes, and 64 individuals with type 2 diabetes) were analyzed for de novo polymorphisms. One hundred and thirty-eight unrelated Japanese individuals (53 male and 85 female subjects) with type 1 diabetes, 347 unrelated Japanese individuals with type 2 diabetes (173 male and 174 female subjects), 190 unrelated Japanese individuals with autoimmune thyroid disease (AITD), and 348 unrelated healthy control subjects were subjected to genotyping. Means ± SD age at onset of type 1 diabetes was 17.4 ± 12.4 years (range 3–59). This study was approved by the appropriate ethical committees, and informed consent was obtained from all participants.
Statistical analysis.
Allele frequency was estimated by direct counting. The significance of the difference in distribution of alleles was determined by a Fisher direct probability test. Haplotypes were estimated by the expectation-maximization algorithm (Haploview version 3.32, four gamete rule [28]). Statistical analysis for semiquantitative RT-PCR was performed by unpaired t test. Statistical significance was defined as P < 0.05.
RESULTS
Putative transcriptional factors of Ins2 in thymus.
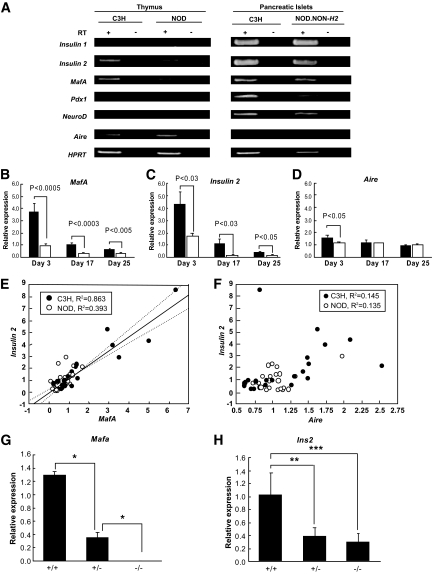
Since insulin, a target gene of MafA, is known to be expressed in both pancreatic islets and the thymus as a primary antigen in type 1 diabetes (29,30), the expression profiles of Ins1 and Ins2 in mouse pancreatic islets and neonatal thymus were determined by RT-PCR analysis in control C3H and NOD mice, a model of type 1 diabetes. RNA preparations from neonatal thymus contained the transcript of Ins2 but not Ins1, whereas transcripts of both Ins1 and 2 were detected in pancreatic islets (Fig. 1A), as shown in previous studies (31,32). To search for putative transcriptional regulators of Ins2 in the thymus, several transcription factors that have been reported to regulate insulin transcription in pancreatic islets and/or the thymus were also examined by RT-PCR analysis. Transcripts of Mafa and Aire, but not Pdx1 and Neurod1, were detected in the thymus, whereas Mafa, Pdx1, and Neurod1, but not Aire, were detected in the pancreatic islets (Fig. 1A), indicating a difference in the expression of insulin transactivators between the thymus and pancreatic islets.
FIG. 1.
Regulation of Ins2 transcription in thymus by Mafa. A: Gene expression of insulin (Ins1 and Ins2) and putative transcriptional factors of insulin (Mafa, Pdx1, Neurod1, and Aire) in thymus and pancreatic islets by RT-PCR. B–D: Semiquantitative RT-PCR analysis of expression of Ins2 (B), Mafa (C), and Aire (D) in C3H (■) and NOD (□) thymus. Relative mRNA levels normalized to HPRT expression were determined. Data are expressed as means ± SE. E: Close correlation of relative expression levels between Ins2 and Mafa expression in thymus. F: No correlation between Ins2 and Aire expression in thymus. G and H: Mafa directly regulates insulin transcription in thymus. Semiquantitative RT-PCR of Mafa (G) and Ins2 (H) in Mafa+/+, +/−, and −/− mice. Relative mRNA levels normalized to HPRT expression were determined. Data are expressed as means ± SE. *P = 0.003; ** P = 0.001; ***P < 0.0001, ANOVA.
Reduced expression of Mafa and Ins2 in NOD thymus.
Semiquantitative RT-PCR analyses of Mafa, Ins2, and Aire were performed on thymic cDNA of NOD and control C3H mice aged 3–25 days. We found highest expression of Mafa and Ins2 in the neonatal thymus (3 days of age), and the expression of both transcripts gradually decreased with age (Fig. 1B and C). In contrast, Aire showed stable expression throughout the ages we examined (Fig. 1D). In mice of all ages (3–25 days) examined, the expression levels of Mafa and Ins2 were significantly lower in the thymus of NOD mice than in control mice. The largest differences between NOD mice and control mice were observed at 3 days of age, both in Mafa (73% reduction, P < 0.0005) and Ins2 (60% reduction, P < 0.03) (Fig. 1B and C). No significant difference was observed between male and female mice (data not shown). In contrast to the marked difference in Mafa transcript, Aire transcript in NOD mice was comparable with that in control mice, with only a slight difference in 3-day-old neonates (28% reduction, P < 0.05) (Fig. 1D). The expression level of Ins2 in the thymus was strongly correlated with that of Mafa (R2 = 0.809) (Fig. 1E). When the data were stratified by strain, the correlation was stronger in control (R2 = 0.863) than in NOD (R2 = 0.393) mice. No correlation was found between Ins2 and Aire in either control C3H (R2 = 0.145) (Fig. 1F) or NOD (R2 = 0.135) mice.
Mafa regulates Ins2 transcription in thymus.
To demonstrate the direct effect of MafA on insulin expression in the thymus, the expression of Mafa and Ins2 in the thymus was studied in Mafa knockout mice. Mafa transcript was absent in the thymus in homozygous knockout mice (Mafa−/−) and was significantly decreased in heterozygous knockout mice (Mafa+/−) compared with that in wild-type littermates (Mafa+/+) (P < 0.0001, ANOVA) (Fig. 1G). Ins2 transcript in the thymus was significantly lower in homozygous and heterozygous knockout mice than in wild-type littermates (P = 0.001, P = 0.003, respectively) (Fig. 1H). These data clearly indicate a direct link of Mafa with Ins2 transcription in the thymus.
Detection of insulin and MafA proteins by immunohistochemical analysis.
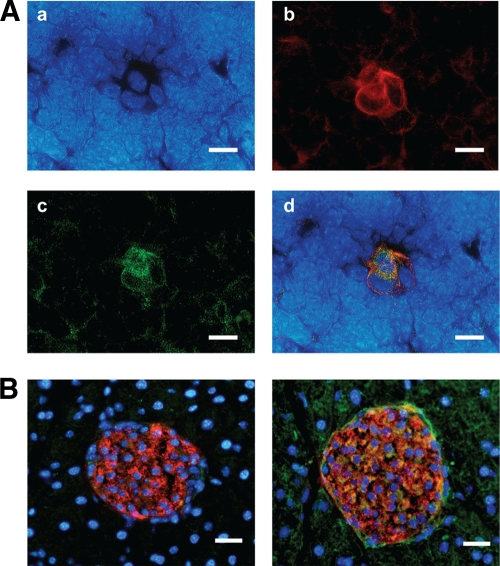
We next investigated the expression of insulin and MafA protein in the C3H thymus (7 days old) using immunohistochemical analysis. Insulin-positive cells were observed in the thymic medulla and were costained with anti-cytokeratin antibody, suggesting that the site of insulin expression is medullary thymic epithelial cells (supplementary Fig. 1A). The thymus was also costained by anti-MafA antibodies and insulin antibodies; MafA staining was observed in the cells positive for insulin, indicating the colocalization of MafA with insulin in the thymus (Fig. 2A, supplementary Fig. 1B).
FIG. 2.
Immunohistochemical staining of thymic medulla. A: Colocalization of insulin and Mafa in thymic medulla (C3H female, 7 days old). Nuclear staining by DAPI (4′,6-diamidino-2-phenylindole) (a), anti-insulin antibody (b), anti-Mafa antibody (c), and merged image (d). Bar scale represents 10 μm. B: Anti–β-cell antibodies in serum of Mafa−/− mouse. Mafa+/+ serum (left panel) and Mafa−/− serum (right panel) were used as first antibodies (Alexa Fluor 488, green). Nuclear staining (DAPI, blue). Anti-insulin antibody (Alexa Fluor 594, red). Bar scale represents 100 μm. (A high-quality digital representation of this figure is available in the online issue.)
Disruption of Mafa induces autoantibodies to pancreatic β-cells.
To examine the direct link between Mafa expression and autoimmunity against the pancreas, mouse pancreas was stained with sera from Mafa−/− and Mafa+/+ mice. The cytoplasm of insulin-positive islet cells was costained with Mafa−/− serum but not with Mafa+/+ serum (Fig. 2B), suggesting that Mafa−/− serum contains autoantibodies against cytoplasmic proteins of pancreatic β-cells. Mouse IgG-positive cells were also detected in the cells around islets, suggesting that disruption of Mafa may induce autoantibodies against not only β-cells but also other cells in the pancreas. Autoantibodies against other endocrine organs, such as thyroid, adrenal, and salivary gland, were not detected in Mafa−/− serum (supplementary Fig. 1C).
Mafa variants were identified in NOD mouse but not other strains.
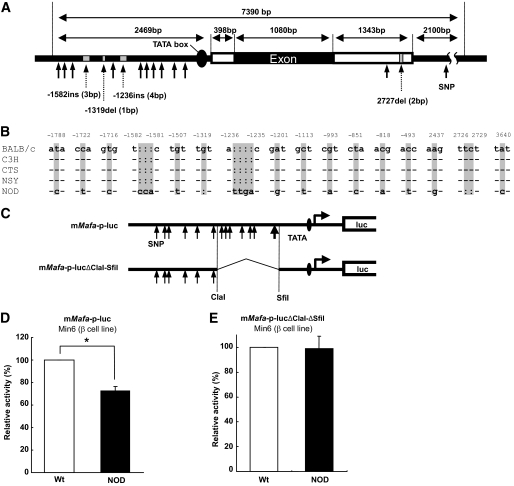
The entire nucleotide sequence of mouse Mafa was identical among the control strains (BALB/c, C3H, and CTS mice) and an animal model of type 2 diabetes (NSY mice) (Fig. 3A and B). Only NOD mice showed variation in the nucleotide sequence of Mafa compared with other strains (BALB/c, C3H, CTS, and NSY mice) (Fig. 3B). Ten single nucleotide polymorphisms (SNPs), a 1-bp deletion, and two insertions in the promoter region; one SNP and one 2-bp deletion in the 3′ untranslated region; and one SNP in the 3′ flanking region in the NOD mouse were newly identified (supplementary materials and methods).
FIG. 3.
Reduced promoter activity of Mafa in NOD mice. A: Localization of sequence variants of Mafa in NOD mice. B: Comparison of Mafa sequences in BALB/c, C3H, CTS, NSY, and NOD mice. Numbers represent position of each variant relative to transcription start site. C: Constructs of luciferase reporter plasmids. D: Comparison of promoter activity of mMafa-p-luc between BALB/c and NOD mice. Data are expressed as means ± SE. *P < 0.001, Student t test. E: Comparison of promoter activity of mMafa-p-lucΔClaI-ΔSfiI between BALB/c and NOD mice. Data are expressed as means ± SE.
Reduced transcriptional activity of Mafa promoter in NOD mouse.
NOD Mafa promoter activity was significantly lower than that of wild-type mice by 27% (P < 0.0001, Student t test) (Fig. 3C, upper panel, and D), indicating that these variants identified in the Mafa promoter could contribute to the reduced expression of Mafa in NOD mice. When an interval between ClaI (AT/CGAT, −1,263 bp upstream of the transcription start site) and SfiI (GGCCACTT/GGGCC, −399 bp) sites was removed from mMafa-p-luc (mMafa-p-lucΔClaI-SfiI) (Fig. 3C, lower panel), the promoter activity appeared to be comparable between NOD and control mice (Fig. 3E), indicating the importance of the sequence between the ClaI and SfiI sites in the regulation of Mafa transcription.
Identification of variants in human MafA gene (MAFA).
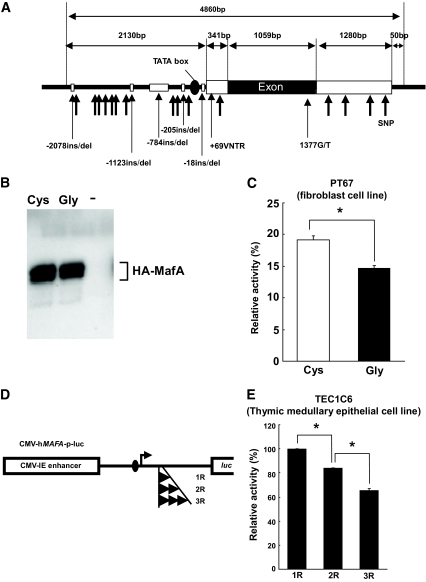
The complete MAFA gene in 16 unrelated patients with type 1 diabetes, 16 unrelated control subjects, and 64 unrelated patients with type 2 diabetes was sequenced. In total, 18 SNPs, five insertions/deletions, and VNTRs were newly identified (Genebank accession no. AB205138) (supplementary materials and methods Fig. 4A).
FIG. 4.
Gly346Cys and +69VNTR polymorphisms in human MAFA are functional. A: Localization of sequence variants identified in hMAFA. B: Western blotting of HA-MafA-Gly346 and HA-MAFA-Cys346 using anti-HA antibody. C: MAFA-346Cys showed significantly higher promoter activity than MAFA-346Gly. Data are expressed as means ± SE. *P < 0.003, Student t test. D: Constructs of luciferase reporter plasmid containing MAFA promoter with each allele of +69VNTR (1R, 2R, and 3R). E: 1R allele had significantly higher promoter activity than the 2R or 3R alleles. Data are expressed as means ± SE. *P < 0.0001, ANOVA.
The effect of two polymorphisms with a potential functional alteration, an SNP with an amino acid substitution (1377G/T, Gly346Cys), and +69VNTR in the 5′untranslated region (UTR) on the expression and function of MAFA was studied. Luciferase reporter plasmid containing the MAFA response element (MARE) in the promoter region (3 × MARE/RBGP-luc [33]) was transfected into NIH3T3 fibroblast cells (PT67 cells), together with the expression plasmid for hemagglutinin (HA)-tagged MAFA-Gly346 (HA-MAFA-Gly346) or HA-tagged MAFA-Cys346 (HA-MAFA-Cys346). HA-MAFA-Gly346 and HA-MAFA-Cys346 were confirmed to be expressed and phosphorylated at similar levels in the cell lysates by Western blotting, using anti-HA antibody (Fig. 4B). HA-MAFA-Gly346 showed a 23% reduction of promoter activity compared with HA-MAFA-Cys346 (P < 0.003, Student t test) (Fig. 4C), indicating that the 346Cys allele has higher transcriptional activity. The effect of +69VNTR on MAFA transcription was also assessed by luciferase assay. Luciferase reporter plasmids containing the cytomegalovirus (CMV)-hMAFA promoter (CMV-hMAFA-p-luc) (Fig. 4D) with each allele of +69VNTR (1R, 2R, or 3R) were transfected into TEC1C6 cells (a thymic medullary epithelial cell line). The allele with one repeat of the VNTR motif (1R) showed significantly higher promoter activity than the allele with two repeats (2R allele) (P < 0.0001, ANOVA) (Fig. 4E). An allele with three repeats (3R allele) showed significantly lower promoter activity than the 2R allele (P < 0.0001). A similar tendency was observed in Min6 cells (a pancreatic β-cell line) with transfection of luciferase reporter plasmids containing hMAFA-p-luc (1R vs. 2R: P < 0.0001; 2R and 3R: P < 0.005) (supplementary Fig. 3).
Gly346Cys was significantly associated with susceptibility to type 1 diabetes, with a significantly lower frequency of the Cys allele in type 1 diabetic than in control subjects (1.8 vs. 5.1%; odds ratio [OR] 0.34 [95% CI 0.13–0.85], P = 0.0222) (supplementary Table 1), suggesting that Cys346 of MAFA, which showed higher transcriptional activity than Gly346 of MAFA, is protective against type 1 diabetes. Whereas homozygotes with Cys346 were absent from both case and control subjects, heterozygotes with Cys346 were negatively associated with type 1 diabetes (0.33 [0.13–0.83], P = 0.0196) (supplementary Table 1), suggesting a dominant mode of protection by Cys346. Given the well-known association of INS (IDDM2) polymorphisms with type 1 diabetes and insulin expression in the thymus, type 1 diabetic patients were stratified by INS polymorphism (patients with the −23HphI+/+ genotype, which is associated with lower transcriptional activity of insulin in the thymus as high-risk subjects and those with −23HphI+/− as low-risk case subjects). The protective effect of Cys346 was concentrated in patients with high-risk IDDM2 (high-risk patients versus control subjects, P = 0.0095, 0.23 [0.08–0.71]) (supplementary Table 2).
The Gly346Cys polymorphism and +69VNTR were in linkage disequilibrium (D′ = 1.0), giving rise to three haplotypes (MAFA-ht1–3) (Table 1 and supplementary Table 3). A haplotype association study revealed that the frequency of MAFA-ht3 (1R-Cys) was significantly lower in patients with type 1 diabetes than in control subjects (P < 0.03) (supplementary Table 3). When subjects with type 1 diabetes were limited to the high-risk IDDM2 genotype (−23HphI+/+ as high-risk case subjects), the association was further concentrated (OR 0.23 [95% CI 0.07–0.70], P = 0.0089) (Table 1). Stratification of cases by HLA haplotype (cases with high-risk IDDM1 versus control subjects) did not affect the association with type 1 diabetes (data not shown). No association with susceptibility to type 2 diabetes was found for MAFA (for Gly346Cys 1.52 [0.85–2.71], P = NS) (supplementary Table 4).
TABLE 1.
Haplotype association of MAFA with susceptibility to high-risk type 1 diabetes
| Haplotype | Polymorphism |
Frequency |
OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|
| +69 VNTR | +1,377 G/T | High-risk case subjects* | Control subjects | |||
| MAFA-ht1 | 1R | G (Gly) | 0.721 | 0.700 | 1.11 (0.78–1.53) | NS |
| MAFA-ht2 | Non-1R | G (Gly) | 0.266 | 0.249 | 1.10 (0.77–1.57) | NS |
| MAFA-ht3 | 1R | T (Cys) | 0.012 | 0.051 | 0.23 (0.07–0.70) | 0.0089 |
*High-risk case subjects: type 1 diabetic patients with the high-risk IDDM2 allele (−23Hph+/+). Fisher direct probability test. NS, not significant.
To clarify whether the association of MAFA is limited to autoimmunity against β-cells, an association study with AITDs (Graves's disease and Hashimoto's thyroiditis) was performed. The Cys allele frequency of MAFA Gly346Cys was comparable between subjects with AITDs and control subjects (3.2 vs. 5.1%, P = NS) (Table 2). Among 190 subjects with AITDs, 18 were positive for GAD antibody, indicating autoimmunity against pancreatic β-cells as well as the thyroid gland in these patients. The frequency of the disease-protective Cys allele in AITD patients without anti-islet autoimmunity was similar to that in control subjects (3.5 vs. 5.1%, P = NS), whereas it was similar in AITD patients with anti-islet autoimmunity and type 1 diabetic patients (0.0 vs. 1.8%, P = NS).
TABLE 2.
Genotype and allele frequencies of MAFA +1,377 G/T in AITDs
| Control subjects | AITD subjects | OR (95% CI) | P value | |
|---|---|---|---|---|
| n | 263 | 190 | ||
| Genotype | ||||
| G/G | 236 (89.7) | 178 (93.7) | ||
| G/T | 27 (10.3) | 12 (6.3) | 0.59 (0.29–1.19) | NS |
| Allele | ||||
| G (Gly) | 499 (94.9) | 368 (96.8) | ||
| T (Cys) | 27 (5.1) | 12 (3.2) | 0.60 (0.30–1.20) | NS |
Data are n (%), unless otherwise indicated. Fisher exact probability test. NS, not significant.
DISCUSSION
The organ specificity of autoimmune disease is caused by impaired immunological tolerance to tissue-restricted peripheral antigens. Negative selection of autoreactive T-cells plays an important role in central tolerance to peripheral antigens, which are ectopically expressed in the thymus (34,35). Recent studies strongly suggest that insulin is the primary antigen for type 1 diabetes in humans (36) and the NOD mouse (37) and that the negative selection of insulin-specific T-cells could be impaired in individuals with type 1 diabetes. The expression level of insulin in the human thymus is known to be associated with a regulatory polymorphism in the promoter region of the INS gene, INS-VNTR (IDDM2) (14,15), whereas such variants have not been identified in the NOD mouse. The present study indicates the existence of an alternative mechanism regulating ectopic insulin transcription in the thymus other than Aire and INS-VNTR. Multiple transcription factors including Pdx-1, Neurod1, and Mafa are reported to be involved in β-cell–specific transcription of insulin (23). Of these transcription factors, only Mafa was shown to be expressed in the thymus and to be closely correlated with Ins2 expression, suggesting that Mafa plays a pivotal role in regulating Ins2 transcription in the thymus. Targeted disruption of Mafa resulted in a marked decrease in Ins2 transcription in the thymus, indicating a direct link of Mafa with Ins2 transcription in the thymus. MAFA expression was also confirmed in a thymic cDNA library in humans (supplementary Fig. 4). These results support our hypothesis that MafA is involved in the pathogenesis of type 1 diabetes through the expression of insulin in the thymus and the thymic deletion of insulin-specific autoreactive T-cells.
MafA has been shown to be a key molecule regulating the tissue-specific expression of insulin in β-cells, but its effect is weak unless it is coexpressed with Pdx1 and NeuroD/BETA2 (38). The expression level of insulin in the thymus is ∼100-fold lower than that in pancreatic islets, and it is thought to be sufficient to induce negative selection of autoreactive T-cells in the thymus. In this sense, it is reasonable to speculate that MafA, whose transcriptional activity is modest without coexpression of Pdx1 and NeuroD/Beta2, can control central tolerance to the insulin peptide by regulating insulin transcription in the thymus where Pdx1 and Neurod1 are not expressed. A series of studies showed that newly established medullary thymic epithelial cell (mTEC) clones spontaneously express Ins2 despite the absence of detectable Pdx1 (39), and the regulation of Ins2 transcription in mTEC clones was not coordinated with the expression level of Aire (40). Garcia et al. (41) also demonstrated that Ins2 expression was detected in the thymus of Pdx1−/− mice as well as wild-type controls, further supporting our hypothesis of a Pdx1-independent transcriptional mechanism of insulin in the thymus.
Several reports (42–45) have shown that the thymic expression of insulin is Aire-dependent, suggesting an essential role of Aire in insulin expression in the thymus. Based on the data of our semiquantitative RT-PCR analysis, the expression level of Aire was not correlated with that of Ins2 in either NOD (R2 = 0.135) or control (R2 = 0.145) C3H thymus, in contrast to the strong correlation between Mafa and Ins2 (NOD: R2 = 0.393, C3H: R2 = 0.863). When the data were restricted to 3-day-old neonatal animals, the expression of Aire in the NOD thymus was reduced (P < 0.05), and there was a weak correlation between the expression levels of In2 and Aire in the NOD thymus (R2 = 0.516) but not in the control thymus (R2 = 0.042, data not shown). The insight derived from these observations is that the expression level of Ins2 in the control thymus is precisely regulated by Mafa, rather than Aire, in normal conditions. Taking these results together, we speculate that insulin expression in the thymus is turned on by Aire and precisely regulated by MafA in normal conditions. In the present study, targeted disruption of Mafa led to reduced expression of Ins2 in the thymus and induced autoantibodies against the pancreas including cytoplasmic protein of pancreatic β-cells, indicating a direct link between Mafa and autoimmunity against the pancreas.
Molecular scanning of the human MAFA gene revealed several polymorphisms that may affect the function of the gene, including Gly346Cys in the vicinity of the dimerization domain (supplementary Fig. 5) and +69VNTR in the 5′UTR. In vitro studies revealed that the Gly346Cys polymorphism is functional, with the Gly allele showing a 23% reduction in transactivation activity compared with the Cys346 allele. +69VNTR in the 5′UTR was also functional, with the number of repeats of VNTRs being negatively correlated with the promoter activity. Based on a hypothesis-driven association study (46), MAFA-ht4 containing these two polymorphisms with higher transcriptional activity was significantly associated with protection against type 1 diabetes, and the association was especially concentrated in cases with the high-risk allele (−23HphI+/+; INS-VNTR class I/class I) at the IDDM2 locus. These data suggest the involvement of gene-gene interaction of MAFA with INS-VNTR (IDDM2) in type 1 diabetes susceptibility, possibly due to a common underlying mechanism for both genes in the disease pathogenesis of type 1 diabetes. Several non-HLA susceptibility loci for type 1 diabetes have been mapped in recent genome-wide association studies in Caucasian populations (47), but markers located near MAFA were not reported to be associated with type 1 diabetes. One possible explanation may be the difference in genetic background between Japanese and Caucasian populations, including IDDM2, the susceptibility allele of which shows large ethnic differences in frequency.
In the present study, an association of MAFA with type 1 diabetes, but not with AITDs, was observed. Further analysis revealed that the frequency of the disease-protective allele in AITD patients without anti-islet autoimmunity was similar to that in control subjects, whereas the frequency in AITD patients with anti-islet autoimmunity was similar to that in patients with type 1 diabetes, suggesting that MAFA is associated with autoimmunity against pancreatic β-cells but not other endocrine organs. These data support our hypothesis that MAFA is involved in autoimmunity specific to pancreatic β-cells but not other endocrine organs such as the thyroid gland.
In conclusion, the present study demonstrated that MafA expression in thymic medullary epithelial cells is closely correlated with that of Ins2, suggesting that the expression of peripheral-tissue antigens in the thymus is regulated by antigen-specific mechanisms. Reduced expression of Mafa and Ins2 in the NOD thymus could lead insulin-specific T-cells to escape negative selection, resulting in infiltration of insulin-specific T-cells into pancreatic islets and the development of type 1 diabetes. This hypothesis is further supported by the association of regulatory polymorphisms in human MAFA with susceptibility to type 1 diabetes. These data suggest a new concept that variation in antigen-specific transcriptional factors plays a critical role in induction of central tolerance to self-antigens, in addition to the more generic role played by Aire.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the Japan Diabetes Foundation (to S.N.); a grant from the Osaka Medical Research Foundation for Incurable Diseases (to S.N.); a grant-in-aid for young scientists (B) (to N.B.); grants-in-aid for scientific research (C) (to S.N. and H.I.); Health and Labor Sciences Research Grants (to H.I.); a grant from the Ministry of Health, Labor, and Welfare (to H.I.); a grant for research on intractable diseases (to H.I.); and a grant-in-aid for the Genome Network Project (to S.T.) from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
No potential conflicts of interest relevant to this article were reported.
S.N. designed this study, researched data, and wrote the manuscript. K.K. designed this study and researched data. Y.K. provided statistical support for human data. N.B. and Y.H. contributed to the animal studies. K.Y. contributed to the animal studies and researched data. T.F. contributed to the animal studies and provided statistical support. S.A. researched data. T.K. and S.T. provided knockout animals and researched data. H.I. reviewed and edited the manuscript and directed the overall project.
MIN6 cells and TEC1C6 cells were kindly provided by Dr. Junichi Miyazaki and Dr. Michiyuki Kasai, respectively. We thank M. Moritani, Y. Yoshizaki, S. Shimoyoshi, and S. Hayase for their skillful technical assistance; Dr. M. Kobayashi for islet isolation; and Dr. T. Ogihara for encouragement.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1. Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med 2009;360:1646–1654 [DOI] [PubMed] [Google Scholar]
- 2. Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 2001;358:221–229 [DOI] [PubMed] [Google Scholar]
- 3. Cox NJ, Wapelhorst B, Morrison VA, Johnson L, Pinchuk L, Spielman RS, Todd JA, Concannon P. Seven regions of the genome show evidence of linkage to type 1 diabetes in a consensus analysis of 767 multiplex families. Am J Hum Genet 2001;69:820–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 2007;448:591–594 [DOI] [PubMed] [Google Scholar]
- 5. Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet, 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511 [DOI] [PubMed] [Google Scholar]
- 8. Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337–338 [DOI] [PubMed] [Google Scholar]
- 9. Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, Saitoh Y, Seino Y, Yasuda K, Serikawa T, Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet 2002;31:391–394 [DOI] [PubMed] [Google Scholar]
- 10. Hornum L, Romer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes 2002;51:1972–1979 [DOI] [PubMed] [Google Scholar]
- 11. Torres B, Aguilar F, Franco E, Sanchez E, Sanchez-Roman J, Jimenez Alonso J, Nunez-Roldan A, Martin J, Gonzalez-Escribano MF. Association of the CT60 marker of the CTLA4 gene with systemic lupus erythematosus. Arthritis Rheum 2004;50:2211–2215 [DOI] [PubMed] [Google Scholar]
- 12. Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VE, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet 2005;76:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet 2009;10:43–55 [DOI] [PubMed] [Google Scholar]
- 14. Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 1997;15:289–292 [DOI] [PubMed] [Google Scholar]
- 15. Pugliese A, Zeller M, Fernandez A, Jr, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 1997;15:293–297 [DOI] [PubMed] [Google Scholar]
- 16. Lucassen AM, Screaton GR, Julier C, Elliott TJ, Lathrop M, Bell JI. Regulation of insulin gene expression by the IDDM associated, insulin locus haplotype. Hum Mol Genet 1995;4:501–506 [DOI] [PubMed] [Google Scholar]
- 17. Kennedy GC, German MS, Rutter WJ. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat Genet 1995;9:293–298 [DOI] [PubMed] [Google Scholar]
- 18. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol 2001;2:1032–1039 [DOI] [PubMed] [Google Scholar]
- 19. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science 2002;298:1395–1401 [DOI] [PubMed] [Google Scholar]
- 20. Chentoufi AA, Palumbo M, Polychronakos C. Proinsulin expression by Hassall's corpuscles in the mouse thymus. Diabetes 2004;53:354–359 [DOI] [PubMed] [Google Scholar]
- 21. Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem 2002;277:49903–49910 [DOI] [PubMed] [Google Scholar]
- 22. Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci U S A 2002;99:6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A 2004;101:2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ueda H, Ikegami H, Yamato E, Fu J, Fukuda M, Shen G, Kawaguchi Y, Takekawa K, Fujioka Y, Fujisawa T, et al. The NSY mouse: a new animal model of spontaneous NIDDM with moderate obesity. Diabetologia 1995;38:503–508 [DOI] [PubMed] [Google Scholar]
- 25. Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol 2005;25:4969–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gotoh M, Maki T, Kiyoizumi T, Satomi S, Monaco AP. An improved method for isolation of mouse pancreatic islets. Transplantation 1985;40:437–438 [DOI] [PubMed] [Google Scholar]
- 27. Taniguchi H, Yamato E, Tashiro F, Ikegami H, Ogihara T, Miyazaki J. Beta-cell neogenesis induced by adenovirus-mediated gene delivery of transcription factor pdx-1 into mouse pancreas. Gene Ther 2003;10:15–23 [DOI] [PubMed] [Google Scholar]
- 28. Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 2002;71:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanahan D. Peripheral-antigen-expressing cells in thymic medulla: factors in self-tolerance and autoimmunity. Curr Opin Immunol 1998;10:656–662 [DOI] [PubMed] [Google Scholar]
- 30. Gianani R, Eisenbarth GS. The stages of type 1A diabetes: 2005. Immunol Rev 2005;204:232–249 [DOI] [PubMed] [Google Scholar]
- 31. Brimnes MK, Jensen T, Jorgensen TN, Michelsen BK, Troelsen J, Werdelin O. Low expression of insulin in the thymus of non-obese diabetic mice. J Autoimmun 2002;19:203–213 [DOI] [PubMed] [Google Scholar]
- 32. Heath VL, Moore NC, Parnell SM, Mason DW. Intrathymic expression of genes involved in organ specific autoimmune disease. J Autoimmun 1998;11:309–318 [DOI] [PubMed] [Google Scholar]
- 33. Kataoka K, Noda M, Nishizawa M. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene 1996;12:53–62 [PubMed] [Google Scholar]
- 34. Kronenberg M, Rudensky A. Regulation of immunity by self-reactive T cells. Nature 2005;435:598–604 [DOI] [PubMed] [Google Scholar]
- 35. Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 2005;435:590–597 [DOI] [PubMed] [Google Scholar]
- 36. Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 2005;435:224–228 [DOI] [PubMed] [Google Scholar]
- 37. Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005;435:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishizawa M, Kataoka K, Vogt PK. MafA has strong cell transforming ability but is a weak transactivator. Oncogene 2003;22:5938–5946 [DOI] [PubMed] [Google Scholar]
- 39. Palumbo MO, Levi D, Chentoufi AA, Polychronakos C. Isolation and characterization of proinsulin-producing medullary thymic epithelial cell clones. Diabetes 2006;55:2595–2601 [DOI] [PubMed] [Google Scholar]
- 40. Levi D, Polychronakos C. Regulation of insulin gene expression by cytokines and cell-cell interactions in mouse medullary thymic epithelial cells. Diabetologia 2009;52:2151–2158 [DOI] [PubMed] [Google Scholar]
- 41. Garcia CA, Prabakar KR, Diez J, Cao ZA, Allende G, Zeller M, Dogra R, Mendez A, Rosenkranz E, Dahl U, Ricordi C, Hanahan D, Pugliese A. Dendritic cells in human thymus and periphery display a proinsulin epitope in a transcription-dependent, capture-independent fashion. J Immunol 2005;175:2111–2122 [DOI] [PubMed] [Google Scholar]
- 42. Liston A, Gray DH, Lesage S, Fletcher AL, Wilson J, Webster KE, Scott HS, Boyd RL, Peltonen L, Goodnow CC. Gene dosage–limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med 2004;200:1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med 2005;202:33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A 2005;102:7233–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabater L, Ferrer-Francesch X, Sospedra M, Caro P, Juan M, Pujol-Borrell R. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J Autoimmun 2005;25:312–318 [DOI] [PubMed] [Google Scholar]
- 46. Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, Petros C, Rollins J, Bennet AM, Wiman B, de Faire U, Wennberg C, Olsson PG, Ishii N, Sugamura K, Hamsten A, Forsman-Semb K, Lagercrantz J, Paigen B. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet 2005;37:365–372 [DOI] [PubMed] [Google Scholar]
- 47. Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 2008;40:1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.