Abstract
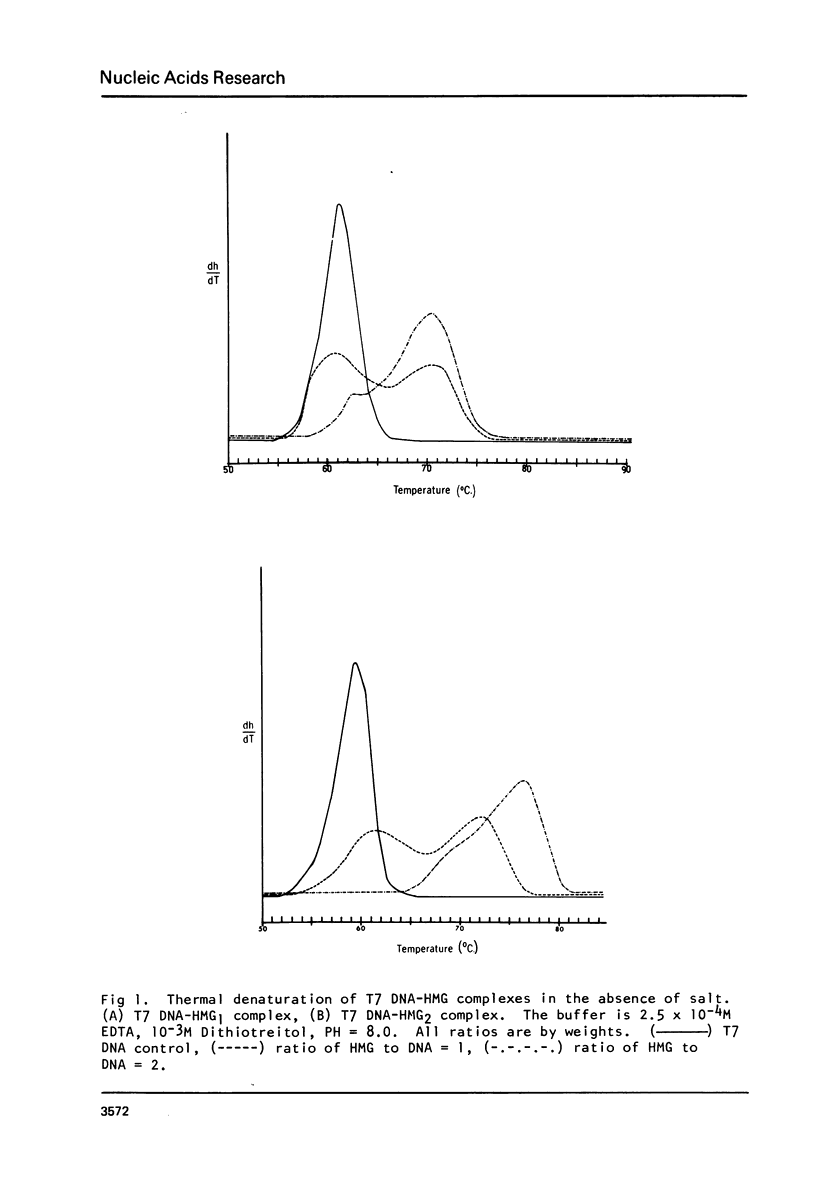
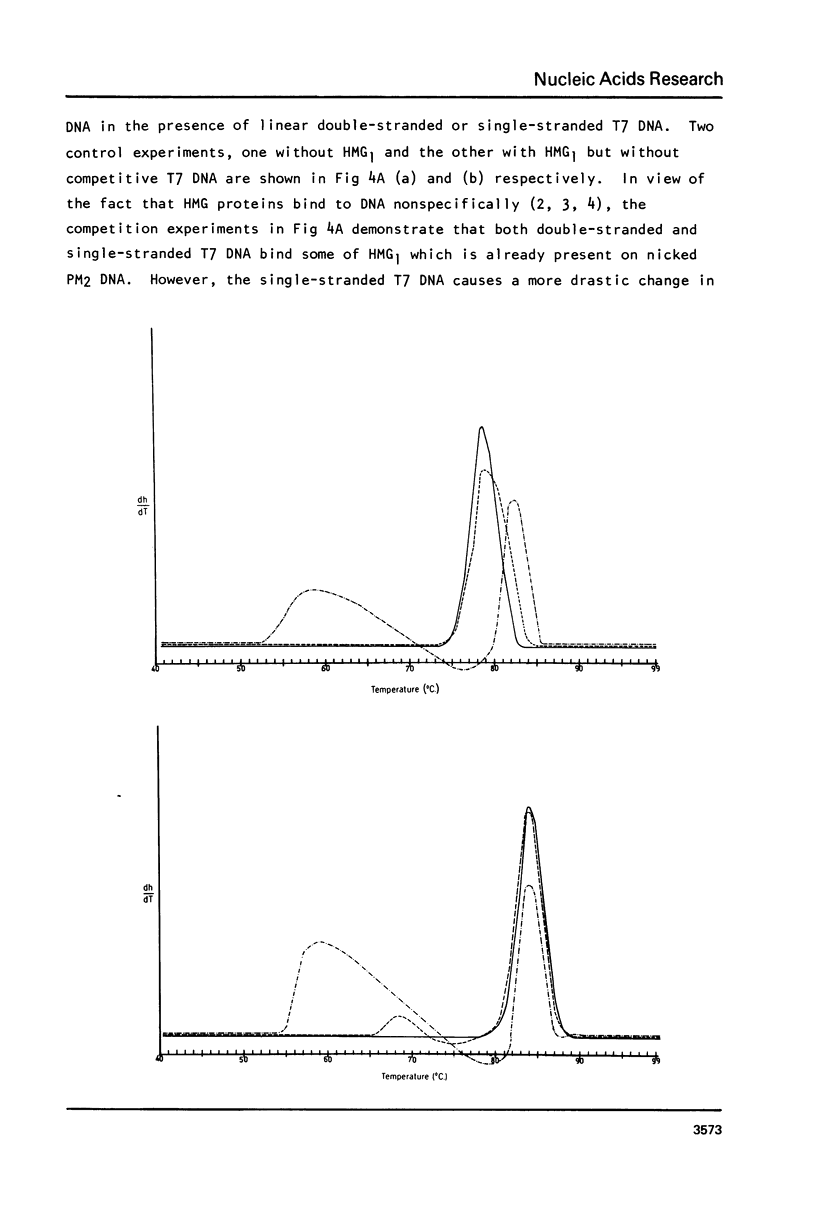
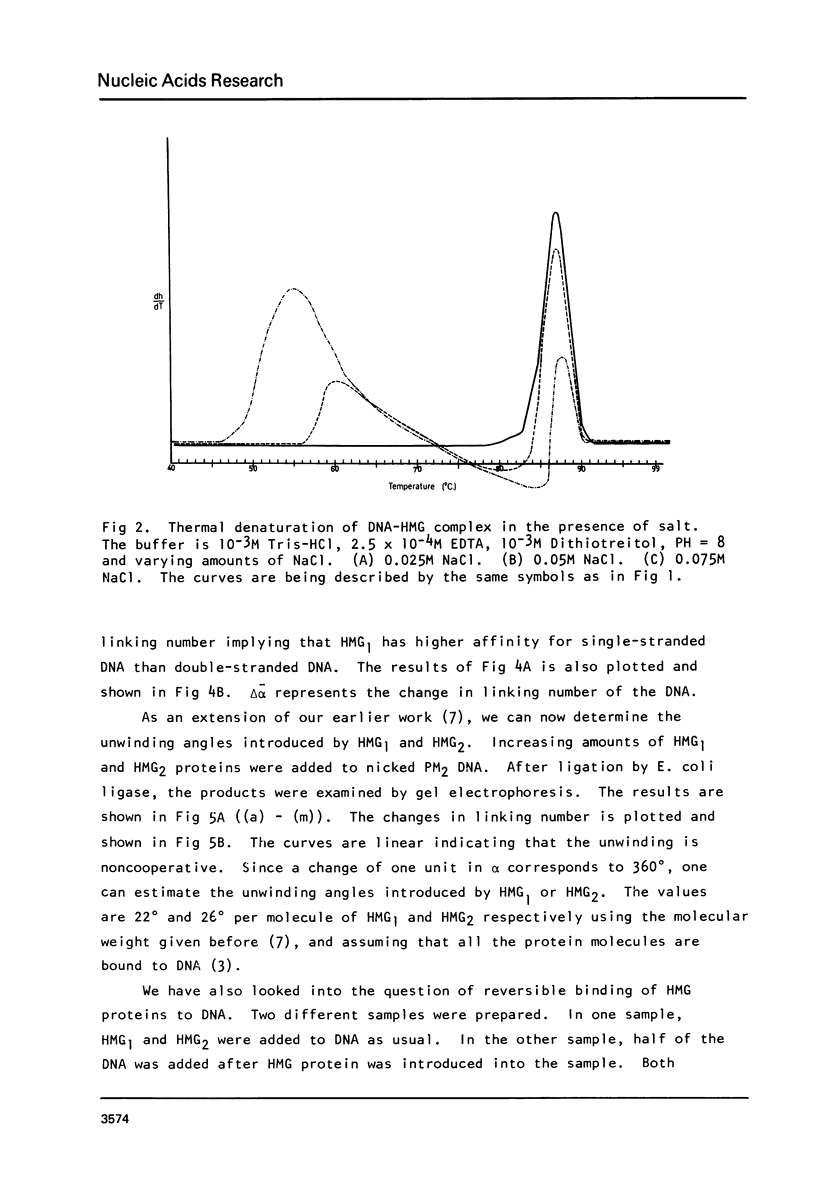
In a previous communication we have shown that both HMG1 and HMG2 nonhistone proteins change the DNA helical structure and the binding of HMG1 and HMG2 to DNA induces a net unwinding equivalent of DNA double helix (Javaherian, K., Liu, L. F. and Wang, J. C. (1978) Science, 199, 1345-1346). Employing melting absorption technique, we now show that in the presence of salt HMG1 and HMG2 destabilize DNA whereas in the absence of salt, they both stabilize DNA molecules. Consequently the folded structure of HMG must play an important role in melting DNA. Furthermore, by measuring topological winding number using competition unwinding experiments, we conclude that HMG1 has a higher affinity for a single-stranded DNA relative to double-stranded DNA. These results together suggest that HMG1 and HMG2 unwind DNA double helix by local denaturation of the DNA base pairs. The net unwinding angles have been measured to be 22 degrees and 26 degrees per molecule of HMG1 and HMG2 respectively.
Full text
PDF


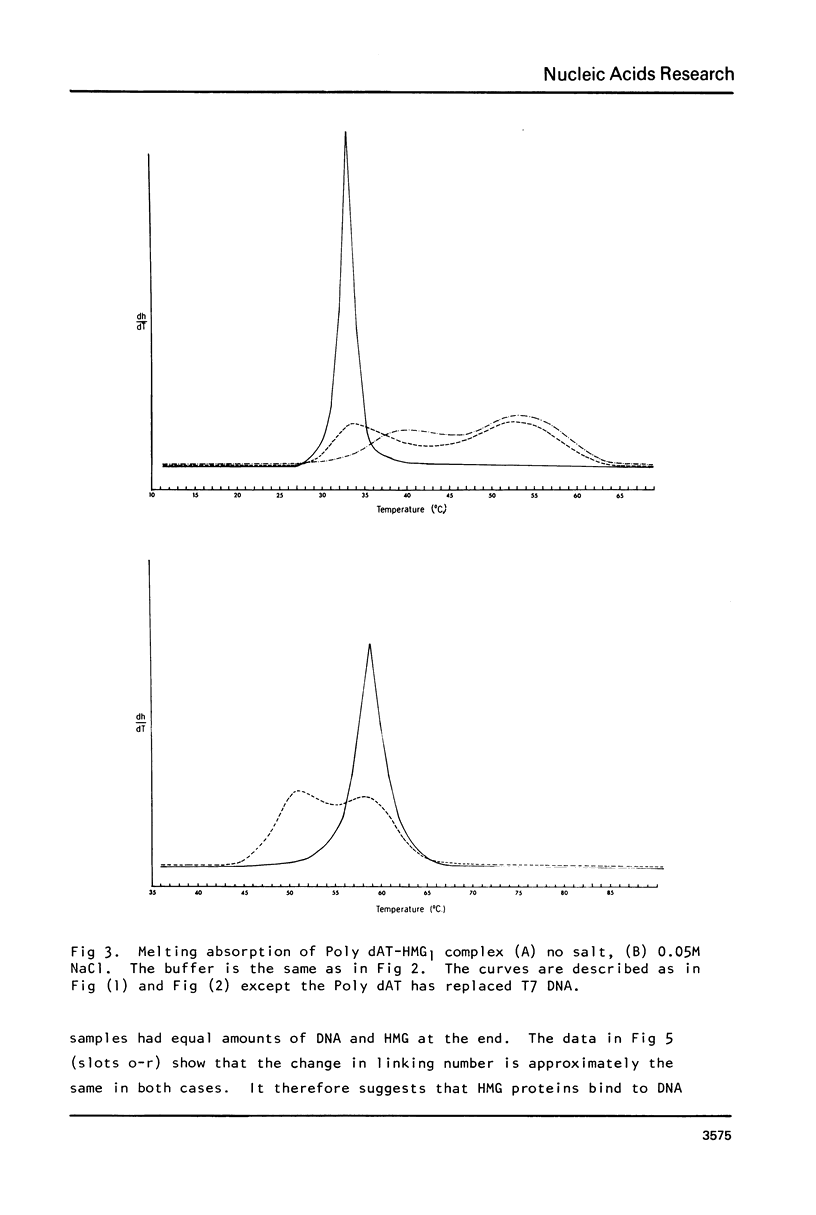
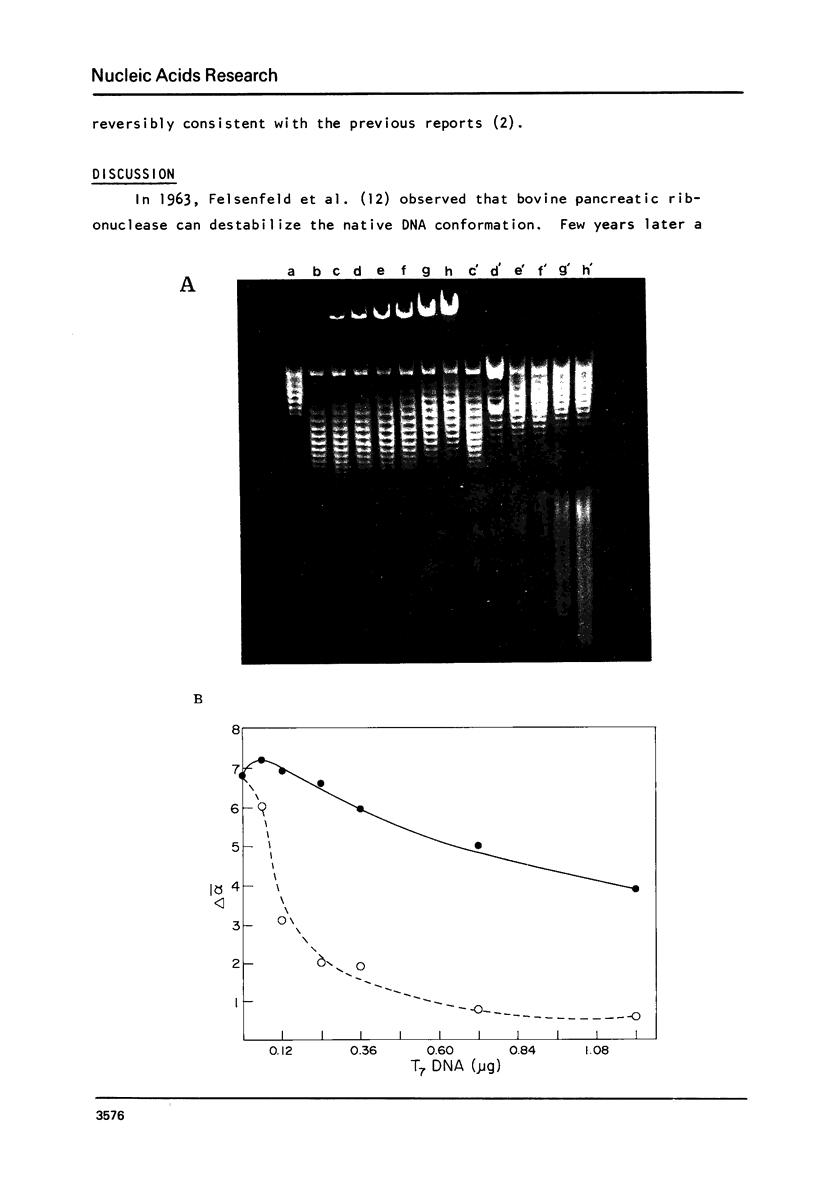

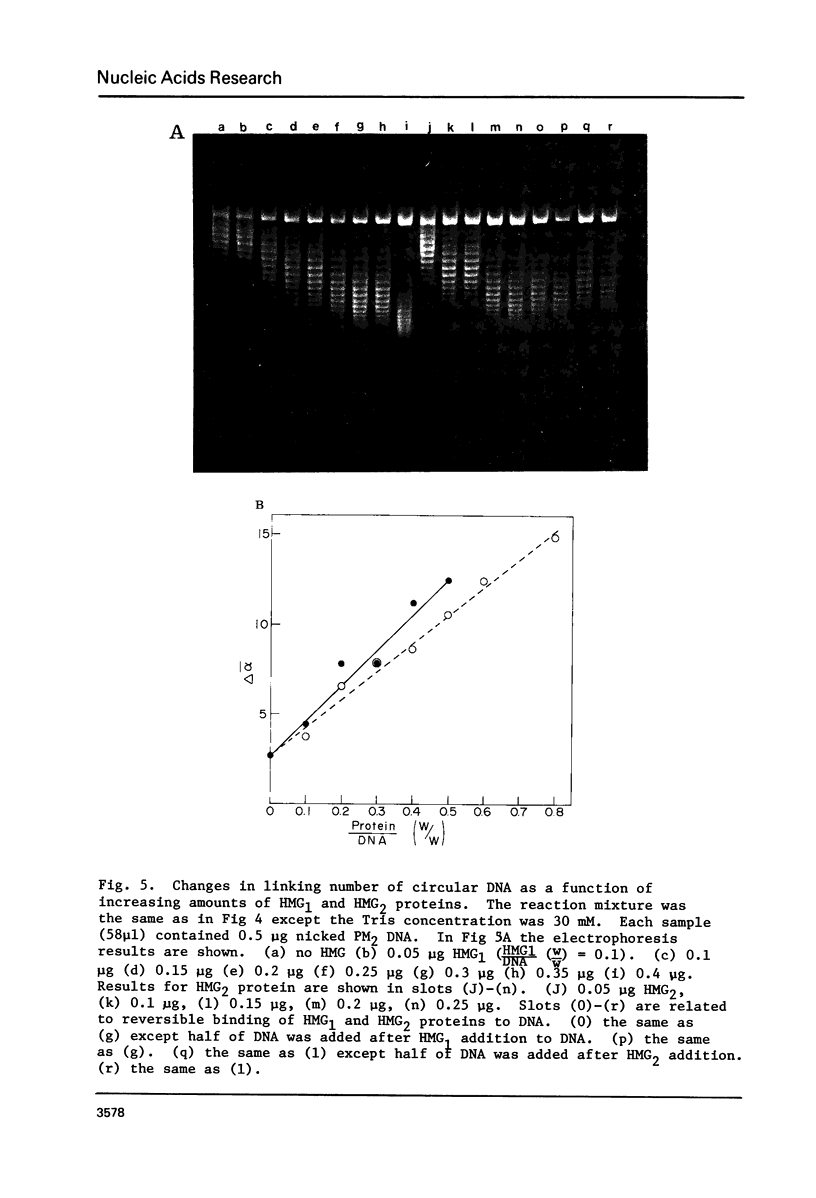


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Baker C., Isenberg I., Goodwin G. H., Johns E. W. Physical studies of the nonhistone chromosomal proteins HMG-U and HMG-2. Biochemistry. 1976 Apr 20;15(8):1645–1649. doi: 10.1021/bi00653a009. [DOI] [PubMed] [Google Scholar]
- Bidney D. L., Reeck G. R. Purification from cultured hepatoma cells of two nonhistone chromatin proteins with preferential affinity for single-stranded DNA: apparent analogy with calf thymus HMG proteins. Biochem Biophys Res Commun. 1978 Dec 14;85(3):1211–1218. doi: 10.1016/0006-291x(78)90671-x. [DOI] [PubMed] [Google Scholar]
- Cary P. D., Crane-Robinson C., Bradbury E. M., Javaherian K., Goodwin G. H., Johns E. W. Conformational studies of two non-histone chromosomal proteins and their interactions with DNA. Eur J Biochem. 1976 Mar 1;62(3):583–590. doi: 10.1111/j.1432-1033.1976.tb10193.x. [DOI] [PubMed] [Google Scholar]
- FELSENFELD G., SANDEEN G., VONHIPPEL P. H. THE DESTABILIZING EFFECT OF RIBONUCLEASE ON THE HELICAL DNA STRUCTURE. Proc Natl Acad Sci U S A. 1963 Oct;50:644–651. doi: 10.1073/pnas.50.4.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H., Sanders C., Johns E. W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973 Sep 21;38(1):14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Shooter K. V., Johns E. W. Interaction of a non-histone chromatin protein (high-mobility group protein 2) with DNA. Eur J Biochem. 1975 Jun;54(2):427–433. doi: 10.1111/j.1432-1033.1975.tb04153.x. [DOI] [PubMed] [Google Scholar]
- Herrick G., Alberts B. Nucleic acid helix-coil transitions mediated by helix-unwinding proteins from calf thymus. J Biol Chem. 1976 Apr 10;251(7):2133–2141. [PubMed] [Google Scholar]
- Javaherian K., Amini S. Conformational study of calf thymus HMG14 nonhistone protein. Biochem Biophys Res Commun. 1978 Dec 29;85(4):1385–1391. doi: 10.1016/0006-291x(78)91157-9. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Amini S. Physicochemical studies of non-histone protein HMG17 with DNA. Biochim Biophys Acta. 1977 Oct 4;478(3):295–304. doi: 10.1016/0005-2787(77)90147-2. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Liu J. F., Wang J. C. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science. 1978 Mar 24;199(4335):1345–1346. doi: 10.1126/science.628842. [DOI] [PubMed] [Google Scholar]
- Jensen D. E., von Hippel P. H. DNA "melting" proteins. I. Effects of bovine pancreatic ribonuclease binding on the conformation and stability of DNA. J Biol Chem. 1976 Nov 25;251(22):7198–7214. [PubMed] [Google Scholar]
- Levy W B., Wong N. C., Dixon G. H. Selective association of the trout-specific H6 protein with chromatin regions susceptible to DNase I and DNase II: possible location of HMG-T in the spacer region between core nucleosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2810–2814. doi: 10.1073/pnas.74.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shooter K. V., Goodwin G. H., Johns E. W. Interactions of a purified non-histone chromosomal protein with DNA and histone. Eur J Biochem. 1974 Sep 1;47(2):263–270. doi: 10.1111/j.1432-1033.1974.tb03690.x. [DOI] [PubMed] [Google Scholar]
- Yu S. S., Li H. J., Goodwin G. H., Johns E. W. Interaction of non-histone chromosomal proteins HMG1 and HMG2 with DNA. Eur J Biochem. 1977 Sep;78(2):497–502. doi: 10.1111/j.1432-1033.1977.tb11762.x. [DOI] [PubMed] [Google Scholar]