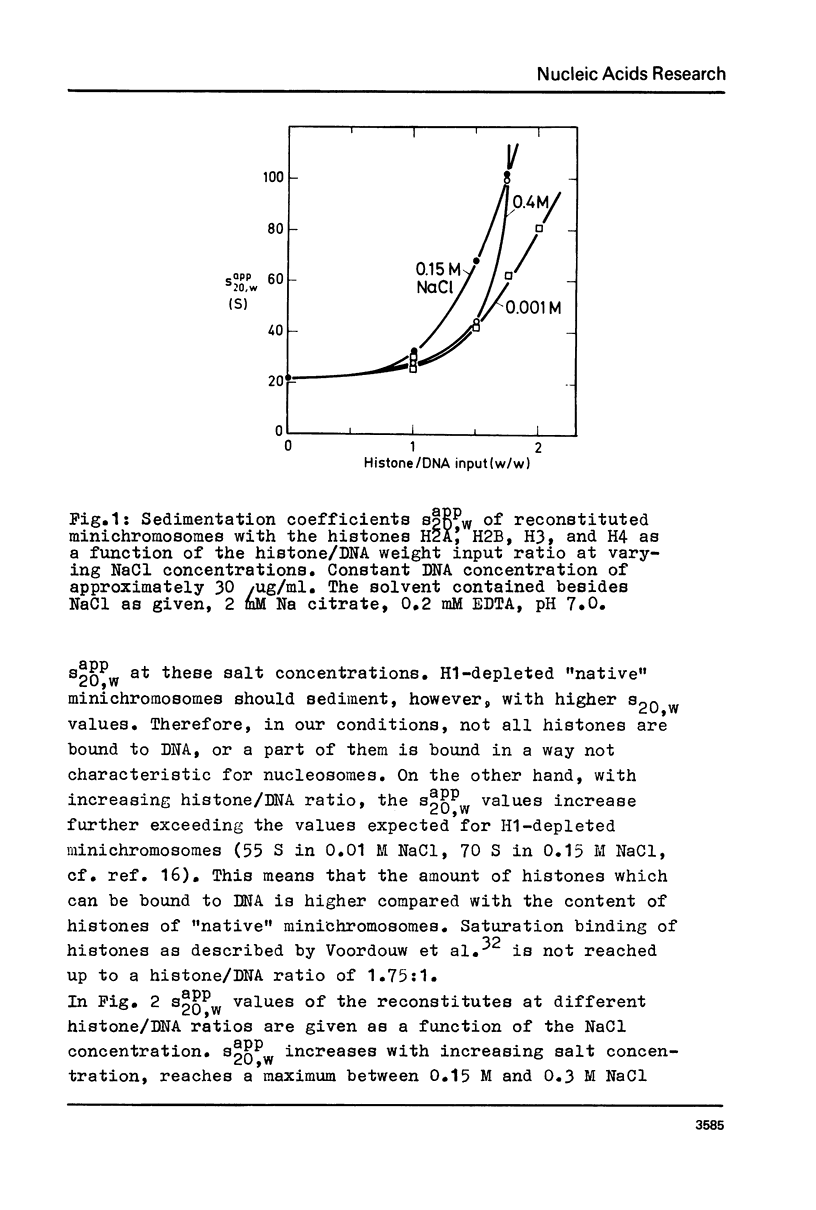
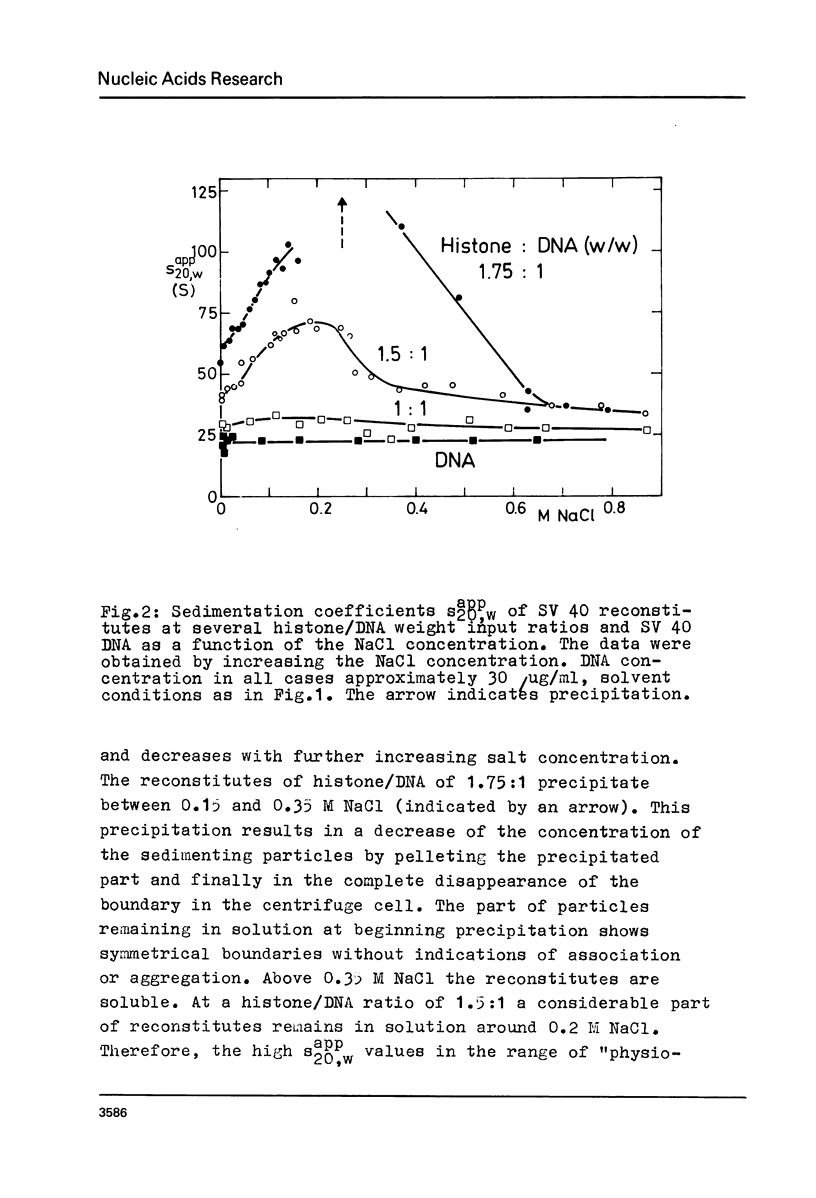
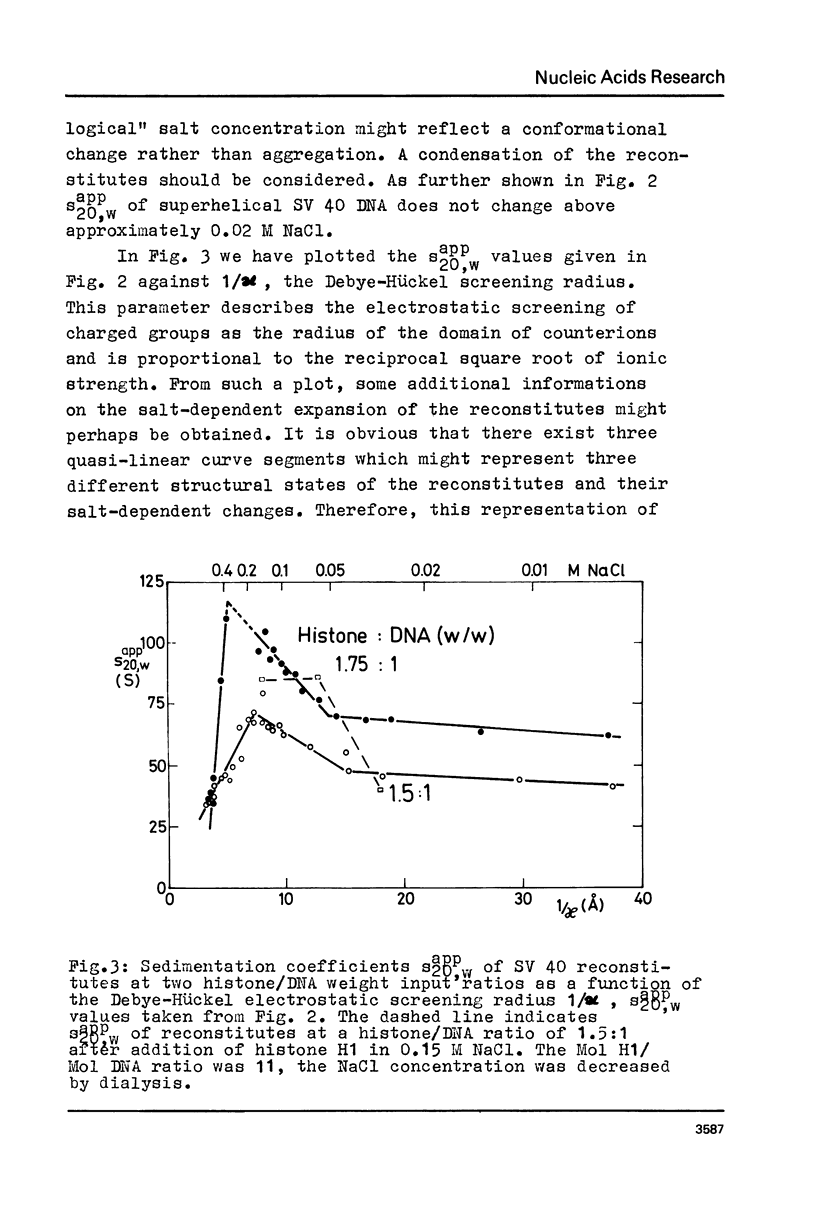
Abstract
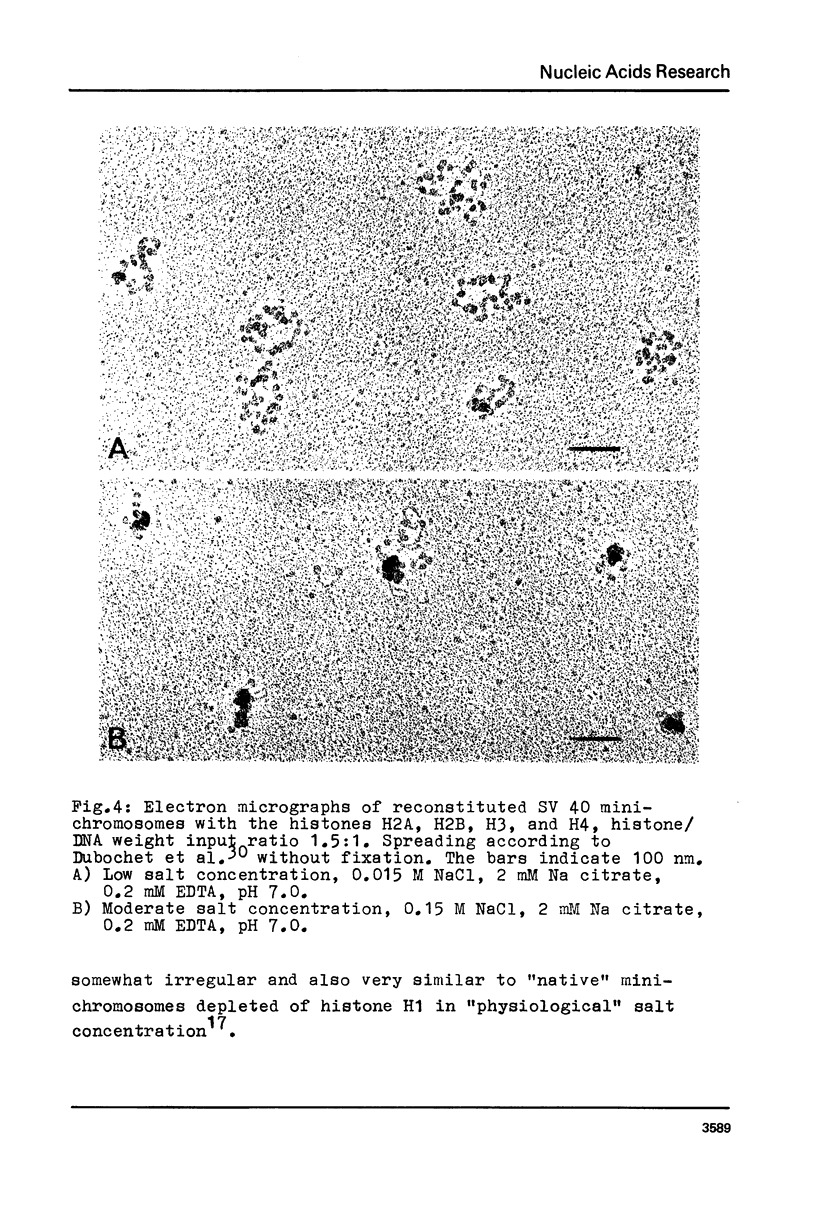
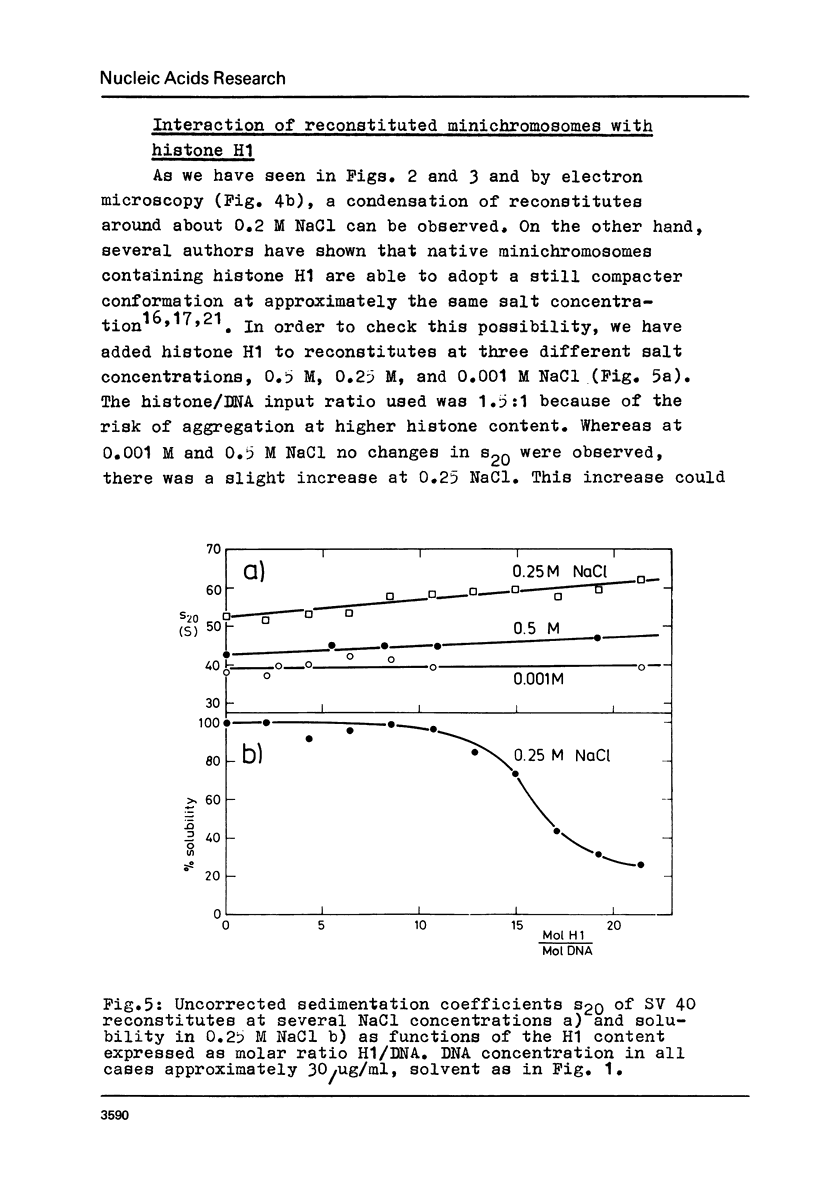
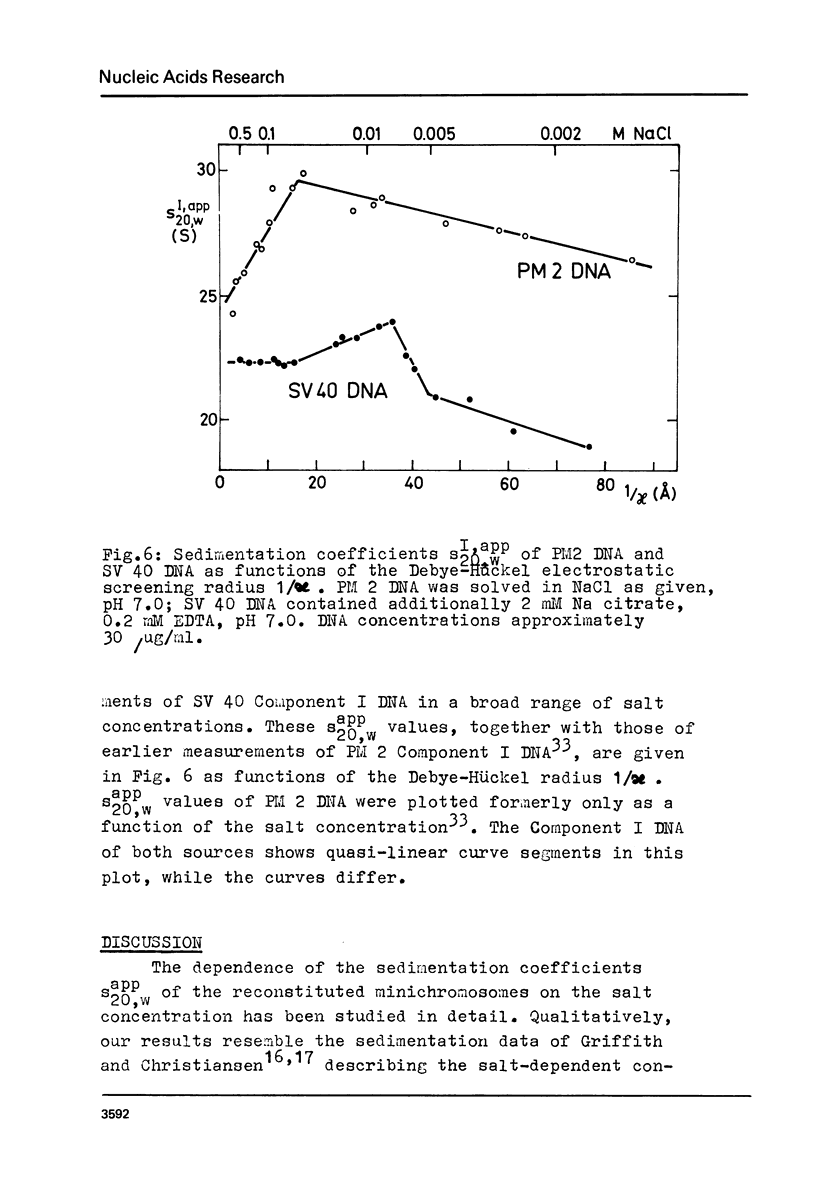
Structural changes of reconstituted SV 40 minichromosomes have been studied in relation to the salt concentration and addition of histone H1 by sedimentation and electron microscopy. Sedimentation data are represented as functions of the NaCl concentration and the Debye-Hückel electrostatic screening radius 1/alpha. The latter representation which proved to provide more information revealed three structural states of the SV 40 reconstitutes which can be additionally characterized by electron microscopy as follows: Expanded or relaxed conformation including free DNA spacers between the nucleosomes at low salt concentration (approx. 0.001 M-0.05 M NaCl), increasing condensation at moderate salt concentration (approx. 0.05 M-0.3 M NaCl) and expansion of this condensed state above approx. 0.3 M NaCl. The condensation of the reconstitutes at moderate salt concentration does not require the presence of histone H1. H1 seems to stabilize the condensed state against electrostatic expansion. The condensation might be promoted by salt-dependent conformational changes of naked superhelical DNA as revealed by sedimentation measurements.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Carpenter B. G., Rattle H. W. Magnetic resonance studies of deoxyribonucleoprotein. Nature. 1973 Jan 12;241(5385):123–126. doi: 10.1038/241123a0. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Inglis R. J., Matthews H. R. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974 Feb 1;247(5439):257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Böttger M., Kuhn W. Sedimentation analysis of conformation changes of circular PM 2 DNA in relation to the ionic strength. Biochim Biophys Acta. 1971 Dec 30;254(3):407–411. doi: 10.1016/0005-2787(71)90872-0. [DOI] [PubMed] [Google Scholar]
- Böttger M., Scherneck S., Fenske H. A sedimentation study of the interaction of superhelical SV40 DNA with H1 histone. Nucleic Acids Res. 1976 Feb;3(2):419–429. doi: 10.1093/nar/3.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M. Conformational variation in superhelical deoxyribonucleic acid. Biochem J. 1978 Apr 1;171(1):281–283. doi: 10.1042/bj1710281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I. Subunit associations among chromatin particles. Nucleic Acids Res. 1977 Nov;4(11):3877–3886. doi: 10.1093/nar/4.11.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B. G., Baldwin J. P., Bradbury E. M., Ibel K. Organisation of subunits in chromatin. Nucleic Acids Res. 1976 Jul;3(7):1739–1746. doi: 10.1093/nar/3.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R. S., Paul J. Role of non-histone components in determining organ specificity of rabbit chromatins. FEBS Lett. 1970 Aug 17;9(4):242–244. doi: 10.1016/0014-5793(70)80366-0. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. The multifunctional role of histone H1, probed with the SV40 minichromosome. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):215–226. doi: 10.1101/sqb.1978.042.01.024. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hartwig M. Organization of mammalian chromosomal DNA: supercoiled and folded circular DNA subunits from interphase cell nuclei. Acta Biol Med Ger. 1978;37(3):421–432. [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ide T., Nakane M., Anzai K., Ando T. Supercoiled DNA folded by non-histone proteins in cultured mammalian cells. Nature. 1975 Dec 4;258(5534):445–447. doi: 10.1038/258445a0. [DOI] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Littau V. C., Burdick C. J., Allfrey V. G., Mirsky S. A. The role of histones in the maintenance of chromatin structure. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1204–1212. doi: 10.1073/pnas.54.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B. R., Schmitz K. S. Quasielastic light scattering by biopolymers. Conformation of chromatin multimers. Biochem Biophys Res Commun. 1976 Nov 22;73(2):224–232. doi: 10.1016/0006-291x(76)90697-5. [DOI] [PubMed] [Google Scholar]
- Upholt W. B., Gray H. B., Jr, Vinograd J. Sedimentation velocity behavior of closed circular SV40 DNA as a function of superhelix density, ionic strength, counterion and temperature. J Mol Biol. 1971 Nov 28;62(1):21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Chumackov P. M., Georgiev G. P. Minichromosome of simian virus 40: presence of histone HI. Nucleic Acids Res. 1976 Aug;3(8):2101–2113. doi: 10.1093/nar/3.8.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Nedospasov S. A., Schmatchenko V. V., Bakayev V. V., Chumackov P. M., Georgiev G. P. Compact form of SV40 viral minichromosome is resistant to nuclease: possible implications for chromatin structure. Nucleic Acids Res. 1977 Oct;4(10):3303–3325. doi: 10.1093/nar/4.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Kalif D., Eisenberg H. Studies of ColE1-plasmid DNA and its interactions with histones: sedimentation velocity studies of monodisperse complexes reconstituted with calf-thymus histones. Nucleic Acids Res. 1977;4(5):1207–1223. doi: 10.1093/nar/4.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A., von Mickwitz C. U., Grade K., Lindigkeit R. Complexes of DNA with arginine-rich and slightly lysine-rich histones. Transcription and electron microscopy. Biochim Biophys Acta. 1978 Mar 29;518(1):172–176. doi: 10.1016/0005-2787(78)90126-0. [DOI] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]