SUMMARY
α- and β-neurexins (NRXNs) are transmembrane cell adhesion proteins that localize to pre-synaptic membranes in neurons and interact with the post-synaptic neuroligins (NLGNs). Their gene mutations are associated with the autistic spectrum disorders. The extracellular region of α-NRXNs, containing nine independently folded domains, has structural complexity and unique functional characteristics, distinguishing it from the smaller β-NRXNs. We have solved the X-ray crystal structure of seven contiguous domains of the α-NRXN-1 extracellular region at 3.0 Å resolution. The structure reveals an arrangement where the N-terminal five domains adopt a more rigid linear conformation and the two C-terminal domains form a separate arm connected by a flexible hinge. In an extended conformation the molecule is suitably configured to accommodate a bound NLGN molecule, as supported by structural comparison and surface plasmon resonance. These studies provide the structural basis for a multi-functional synaptic adhesion complex mediated by α-NRXN-1.
INTRODUCTION
Specialized synaptic adhesion proteins play an important role in synapse specification, development and maintenance. Genetic evidence implicates several synaptic adhesion proteins in the onset of neuro-developmental disorders, such as the autism spectrum disorders (ASD) (Abrahams and Geschwind, 2008). The NRXN-1 gene has been identified as one candidate for susceptibility to ASD in multiple genetic linkage studies, ranging from whole-genome scans to single-nucleotide polymorphism (SNP) screens (Glessner et al., 2009; Kim et al., 2008; Sebat et al., 2007; Szatmari et al., 2007; Yan et al., 2008). The neurexins (NRXNs) constitute a family of pre-synaptic transmembrane proteins, which are developmentally and spatially expressed at neuronal GABAergic and glutamatergic synapses (Ullrich et al., 1995).
In vertebrates there are three NRXN genes (NRXN 1-3), each encoding either the longer α-NRXN or the shorter β-NRXN via alternative promoter regions. For each gene, the α- and β-NRXNs yield class I transmembrane proteins that share the same C-terminal sequence and differ only in the extent of their N-terminal extracellular region. After post-translational cleavage of an α-specific signal peptide, the extracellular region of the mature α-NRXN contains nine independently folded domains, including six laminin-neurexin-sex hormone-binding globulin (LNS) domains separated by three epidermal growth factor (EGF)-like domains, and a C-terminal O-glycosylated stalk domain (~100 amino acids) that forms an extended linker to the transmembrane domain. The much smaller extracellular region of β-NRXNs contains a β-specific signal peptide (cleaved in the mature protein) and a short β-specific N-terminal sequence followed by a single LNS domain, which is identical to the LNS 6 domain of α-NRXN, and the same O-glycosylated stalk domain (Figure 1A). NRXN mRNAs are subject to extensive alternative splicing. There are five alternative splice sites in total named SS#1 through 5. α-NRXNs are subject to splicing at all five splice sites, and β-NRXNs only at SS#4 and #5. The diversity of NRXNs, which is achieved by different genes, alternative promoters and extensive splicing, results in the potential for thousands of isoforms (Tabuchi and Sudhof, 2002). Alternative splicing is thought to function in the regulation of protein-protein interactions and is potentially governing synapse specificity(Ullrich et al., 1995).
Figure 1. The structure of the α-NRXN-1 LNS 2-6 extracellular region.
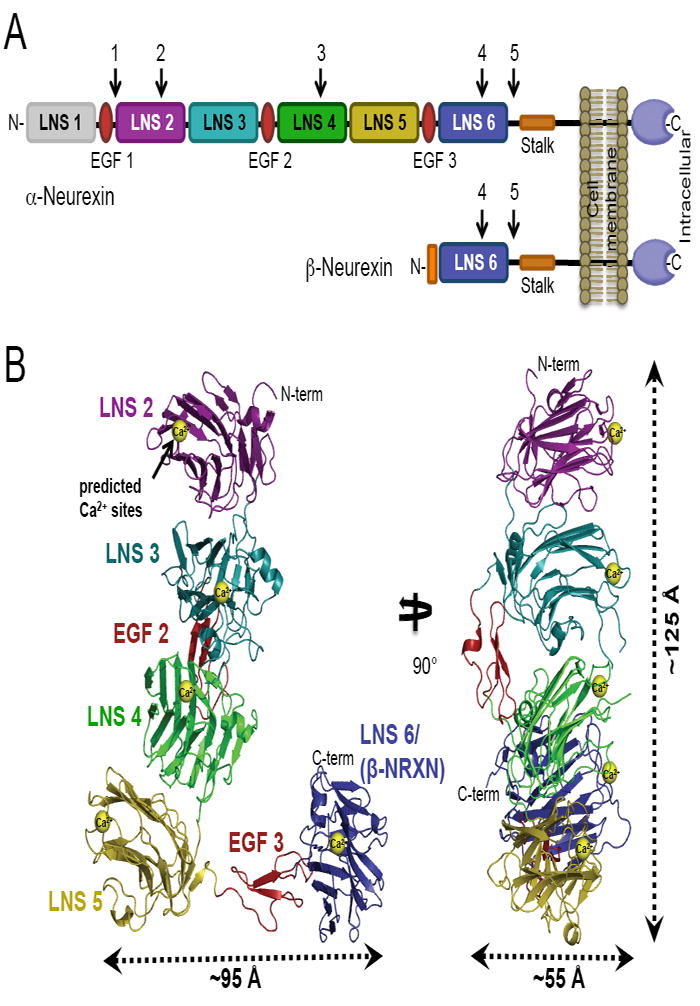
(A) Schematic representation of the mature α- and β-NRXN class I transmembrane proteins. The larger α-NRXNs differ from the shorter β-NRXNs in the extent of their N-terminal extracellular region, with α-NRXNs having six LNS domains interspersed by three EGF domains and β-NRXNs having only one LNS domain, identical to LNS 6 in α-NRXN. The positions for alternative splicing inserts are numbered and indicated with arrows.
(B) Ribbon representation of the crystal structure of α-NRXN-1 LNS 2-6 in two orthogonal orientations. The predicted Ca2+-binding sites, displayed as yellow spheres, were modeled from the Ca2+-bound structures of LNS 2, 4 and 6. Approximate dimensions of the asymmetric molecule are indicated.
See also Figure S1.
α- and β-neurexins are thought to function primarily as cell-adhesion molecules through their extracellular interaction with the post-synaptic neuroligins (NLGNs) (Ichtchenko et al., 1995; Ichtchenko et al., 1996). More recently they have been found to also associate with the post-synaptic leucine-rich repeat transmembrane proteins (LRRTMs) (de Wit et al., 2009; Ko et al., 2009), the α4β2 nicotinic acetylcholine receptor (Cheng et al., 2009), and the GABAA receptor (Zhang et al., 2010). α-NRXN-1 was originally identified as a high-affinity receptor for a spider neurotoxin, α-latrotoxin, which binds to pre-synaptic receptors causing massive neurotransmitter release (Davletov et al., 1995). Studies in mice identified the extracellular region of α-NRXN-1 to be essential for the regulation of Ca2+-dependent exocytosis in neurons (Missler et al., 2003; Zhang et al., 2005). In addition to the post-synaptic partner proteins described above, earlier studies showed that α-NRXNs interact with α-dystroglycan through the LNS 2 and LNS 6/β-NRXN domains (Sugita et al., 2001) and to the soluble neurexophilin proteins through the LNS 2 domain (Missler et al., 1998). The precise roles of each of these interactions in governing synaptic assemblies remain to be investigated.
The NLGN molecules constitute another family of class I transmembrane cell-adhesion proteins that are expressed post-synaptically at either glutamatergic (NLGN-1, -3, -4) or GABAergic (NLGN-2) synapses (Song et al., 1999; Varoqueaux et al., 2004). Structurally, the interaction between the NLGN and β-NRXNs has been extensively characterized. Two β-NRXN molecules bind in a Ca2+-dependent manner on opposing sides of the long axis of the dimeric NLGN molecule. Crystal structures of the complexes between NLGN-1 or NLGN-4 and β-NRXN-1 establish Ca2+-coordination at the binding interfaces and support a regulatory mechanism at the alternative splice site B of NLGN-1 (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007; Leone et al., 2010).
Compared to β-NRXN, the extracellular region of α-NRXN is structurally far more complex. It presents multiple independent folding domains including two known functional protein-binding domains, LNS 2 and LNS 6, which bind to multiple endogenous partner proteins (described above). However, questions persist: what is the synaptic disposition of the extracellular region of α-NRXN in the limited space of the synaptic cleft, and how does the association of the NRXNs with partners mediate synaptic function through protein-protein interactions? We recently demonstrated by single particle electron microscopy (EM) and small angle X-ray scattering (SAXS) that the extracellular region of α-NRXN-1, comprised of LNS 1 through LNS 6, adopts a semi-elongated shape with varying degrees of flexibility between the independent LNS domains, of which the LNS 1 domain displayed the most pronounced flexibility (Comoletti et al., 2010). In fact, removal of the LNS 1 and EGF 1 domains permitted the crystallization of the remainder of the structured extracellular domain.
Herein we present the crystal structure, determined at 3.0 Å resolution, of seven contiguous domains of the α-NRXN-1 extracellular region, including the LNS 2-6 domains and intervening EGF 2 and 3 domains. The domains are organized in an L-shaped conformation with LNS 2-5 forming the longer arm, and EGF 3 and LNS 6 making up the short arm. The achievement of a high-resolution picture of the domain arrangement and distinguishing binding interfaces provides novel insights into the structural basis for biological function. By a comparison with the structure of the β-NRXN:NLGN complex, the α-NRXN structure is suitably shaped to accommodate a NLGN molecule bound to LNS 6. The structural analysis is supported by surface plasmon resonance (SPR) studies in which we quantitatively compare the binding affinities of β-NRXN-1 and various α-NRXN-1 truncations for NLGN-1. As previously seen from affinity pull-down assays (Boucard et al., 2005), we demonstrate exclusive binding of NLGN-1 to the LNS 6/β-NRXN domain. Furthermore, using a series of α-NRXN constructs with incremental truncations of each LNS domain, we show that the addition of LNS 2-5 domains results relatively small changes to the binding affinity for NLGN-1. These data confirm the accessibility of the LNS 6 binding site in solution and support the argument for a high affinity binding interaction between NLGN-1 and the full-length α-NRXN molecule.
RESULTS
Three Dimensional Structure of α-NRXN-1
The crystal structure of the glycosylated α-NRXN-1 extracellular region, LNS 2 through LNS 6 (α-NRXN_2-6) containing SS#3, was solved at 3.0 Å resolution by molecular replacement and refined (Table 1). It reveals a unique arrangement of domains, which form an L-shape with the longer arm composed of the LNS 2-5 and EGF 2 domains and the short arm formed by the EGF 3 and LNS 6 domains (Figure 1B). The LNS domains of the long arm are stacked in a similar orientation such that the concave β-sheet face of the N-terminal LNS domain faces the convex β-sheet face of the following C-terminal LNS domain, making extensive interfacial contacts through abundant hydrophobic and electrostatic interactions, and organizing all of the predicted Ca2+-binding sites on the same side of the molecule with similar inter-Ca2+ site distances (26-42 Å) (Figure 1B and Supplemental Figure S1A). Each of the LNS domains, 2-5, has a Cys bond that anchors the C-terminal loop to the convex β-sheet face on the opposite side of the predicted Ca2+-binding region. Furthermore, they all anchor to the same β-sheet (β12), with the exception of LNS 4, which anchors to β13. This linker arrangement may help maintain the overall architecture by directing the assembly of each of the following C-terminal LNS domains. Accordingly, the last LNS domain (LNS 6) is devoid of the Cys bond and sequence homology indicates that the LNS 1 domain has the same Cys connectivity.
Table 1.
Data Collection and Refinement Statistics
| X-ray source | SSRL BL11-1 |
|---|---|
| Crystal data | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 199.74, 61.24, 155.58 |
| α, β, γ (°) | 90.00, 121.21, 90.00 |
| Data collection | |
| Processing software | XDS package |
| Wavelength (Å) | 0.97945 |
| Resolution (Å) | 30.0-3.02(3.13-3.02) |
| Rsym (%) | 14.0(62.8) |
| I / σI | 8.7(2.0) |
| Completeness (%) | 97.9(99.5) |
| No. of unique reflections | 31504(3223) |
| Redundancy | 3.4(3.4) |
| Refinement | |
| Resolution (Å) | 30.0-3.02(3.10-3.02) |
| Rwork / Rfree (%) | 21.4(31.2)/26.9(39.4) |
| No. atoms | 7878 |
| Protein | 7795 |
| Carbohydrate | 50 |
| Water | 33 |
| Average B overall (Å2) | 47.6 |
| R.m.s.d. bond length (Å) | 0.013 |
| R.m.s.d. bond angle (°) | 1.369 |
| Ramachandran Plot | |
| Preferred (%) | 93.6 |
| Allowed (%) | 6.1 |
| Outliers (%) | 0.3 |
Values in parentheses are for the highest-resolution shell
The relatively small EGF-like domains (~35 residues) maintain a compact structural fold through a conserved cysteine sequence pattern that results in the formation of three disulfide bonds. The EGF 2 domain is located in proximal apposition with the LNS 3-LNS 4 interface, opposite the predicted Ca2+-binding sites of both LNS domains. It further stabilizes the LNS 3-LNS 4 interface by forming hydrogen bonds at either of its termini with the LNS 3 and 4 domains. At the junction between LNS 5 and EGF 3 there are no stabilizing inter-domain contacts, making an apparent hinge point where the C-terminal EGF 3 and LNS 6 domains form a distinct arm of the protein. The EGF 3 domain separates LNS 5 and LNS 6 by ~35 Å, displacing LNS 6 from the linear arrangement of LNS 2-5 (Figure 1B). The extended conformation of the EGF 3-LNS 6 arm is stabilized by abundant interfacial contacts with an LNS 5 domain belonging to a symmetry related molecule (Supplemental Figure S1B). The presence of a flexible hinge between LNS 5 and EGF 3 suggests that the arm can assume multiple conformations, as supported by a previous study using independent techniques (Comoletti et al., 2010). The crystal structure likely represents one of these conformations.
There are two predicted N-linked glycosylation sites in the sequence of α-NRXN_2-6, one on SS#3 in LNS 4 at N813 and one at N1246 on LNS 6. The Fourier difference maps revealed part of an N-linked glycan bound to N1246 on LNS 6, fitting the electron density of two GlcNAc and one Man moieties. Density is missing for most of SS#3 including the N813 glycosylation site. In addition to the N-linked glycosylation, the Fourier difference maps revealed extra density extending away from the hydroxyl group on S705 in EGF 2. This serine residue belongs to a unique O-glycosylation consensus motif (C1-X- S/T-X-P-C2) found between the first and second conserved cysteines in EGF-like domains in several other extracellular proteins, including the coagulation factor IX and the Notch cell surface receptor signaling protein (Haines and Irvine, 2003; Harris and Spellman, 1993). The complete form of the carbohydrate structure found on these proteins is reported as Xylα1-3Xylα1-3Glcβ1-O-Ser (Okajima et al., 2008). Based on the conservation of the consensus motif in the α-NRXN EGF 2 domain, we have modeled a Glcβ1 monosaccharide linked to S705, which is supported by the present density although the resolution limits absolute identification of the sugar (Figure 2).
Figure 2. α-NRXN N- and O-Glycosylation.
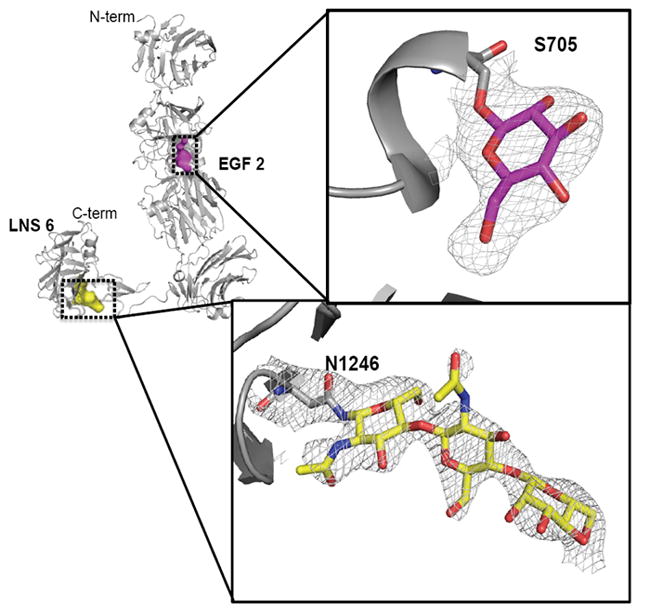
The α-NRXN_2-6 molecule with a β-Glc monosaccharide O-linked to S705 in EGF 2 and a β-GlcNAcβ1,4GlcNAcβ1,4Man trisaccharide N-linked to N1246 in LNS 6 shown modeled into the density. Evidence for a β-Glc modification at S705 comes from the conservation of an O-linked glycosylation motif found in EGF domains of other extracellular proteins, including Notch and Factor IX. The N-linked glycosylation on N1246 matches the consensus glycosylation site found in β-NRXN (PDB:3mw2). The framed figures show the 2Fo-Fc map (gray) for both sugars, contoured at 1.2 σ around S705 and at 1.0 σ around N1246.
Comparison of LNS domains
The independently solved NRXN-1 LNS domains 2, 4 and 6/β-NRXN show high structural homology despite low sequence identity (~20%) (Koehnke et al., 2008; Koehnke et al., 2010; Rudenko et al., 1999; Sheckler et al., 2006; Shen et al., 2008). Similarly, a comparison of each of the individual domains with the corresponding domain in the α-NRXN_2-6 molecule shows high structural similarity (average rmsd of 0.5-0.9 Å for all atoms). The five LNS domains in α-NRXN_2-6, which include the previously unsolved LNS 3 and 5, all maintain the characteristic β-sheet sandwich containing 13 β strands making up the core of the domain(Sheckler et al., 2006) (Figure 3). The face containing the predicted Ca2+-binding site, previously coined the “hyper-variable” surface (Sheckler et al., 2006), also contains the sites for SS#2, 3 and 4 in LNS 2, 4, and 6, respectively, and a non-spliced insert that creates a distinct flexible loop at the surface in LNS 3 (Figure 3). In the structure, the LNS 3 loop shows high flexibility (average B-factor 88 Å2) and is oriented towards the LNS 4 domain with proximity to the LNS 4 Ca2+-binding site. Long-range electrostatic attractions between the positively charged loop and negatively charged LNS 4 surface are likely to influence the orientation of the flexible LNS 3 loop (Supplemental Figure S2A). The β11-β12 loop, which extends across the concave side of the β-sheet, diverges in structure for all of the LNS domains. Compared to the LNS 6 domain, the β11-β12 loop is significantly larger in the LNS 2-5 domains, covering a greater surface area of the concave side of the β-sheet (Supplemental Figure S2B). In LNS 2 and 3 the β11-β12 loop makes stabilizing contacts between the adjacent LNS domains, LNS 3 and 4, respectively. In LNS 4 the loop also makes extensive contacts, predominantly with the symmetry-related LNS 5 domain that participates in the organization of the EGF 3-LNS 6 arm. In LNS 5 the homologous loop is solvent accessible, as is the small β11-β12 loop in LNS 6. Similar to the plant lectins, which have a related fold and homologous β11-β12 loop involved in carbohydrate binding specificity, the variation in the β11-β12 loop in the different LNS domains of α-NRXN is likely to confer selectivity for the interacting surfaces (Rini, 1995; Sheckler et al., 2006). The structure demonstrates that this variable surface loop in each domain functions in distinctive ways in the assembly of the full-length α-NRXN protein.
Figure 3. Comparison of the α-NRXN_2-6 LNS domains.
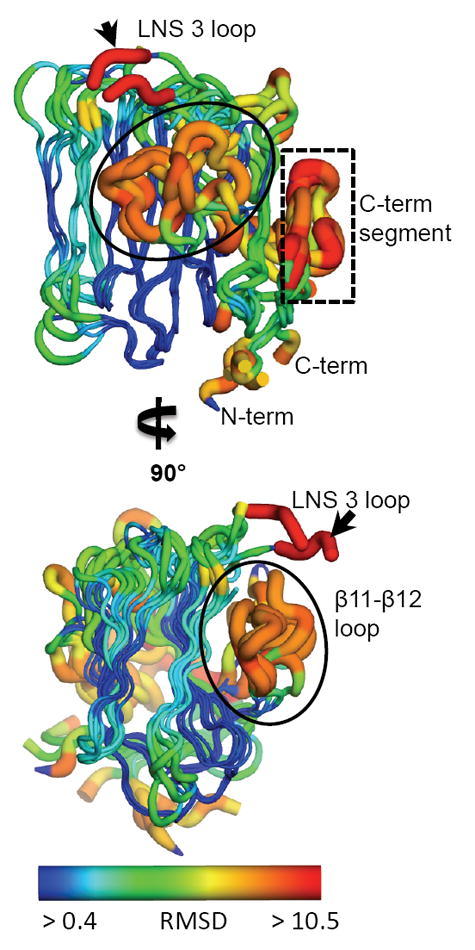
The five LNS domains in the α-NRXN_2-6 structure share a conserved conformation with a characteristic 13 β-sheet fold. Regions of high variation, measured by deviation in the Cα backbone, include the β11-β12 loop (circled), a loop at the Ca2+-binding face in LNS 3 (small black arrow), and the C-terminal segment including the α-helix and proceeding β-sheet (boxed). Relative variation is depicted by color and size of the tube, where thin blue tubes represent low variation and wide orange tubes show greater variation. Residues that do not align with any other residues in a sequence alignment of all five domains are colored in red. (also see Supplemental Figure S2).
Structural comparison with the β-NRXN/NLGN complex
α-NRXN-1 has two previously identified protein-binding domains, LNS 2 and LNS 6, which bind to several endogenous partner proteins (see Introduction). The previously described NLGN binding site on LNS 6 is solvent accessible in the stabilized conformation found in the α-NRXN_2-6 structure. To model the complex between α-NRXN and NLGN-1, we superimposed the structure of α-NRXN_2-6 onto the structures of the β-NRXN-1 molecules bound to NLGN-1 (Arac et al., 2007). The overlaid NLGN molecule interfaces with the α-NRXN_2-6 molecule without significant steric obstruction, indicating that NLGN could bind to α-NRXN_2-6 in the crystallized conformation (Figure 4A). The LNS 6 domain of α-NRXN_2-6 shows high structural conservation at the binding interface to the NLGN molecule, as seen in the β-NRXN:NLGN complex structure. Despite the absence of bound Ca2+, the predicted Ca2+-coordinating residues in LNS 6 maintain the same conformational arrangement as observed in Ca2+-bound β-NRXNs either in complex with NLGN or in their free form (Figure 4B&C). The Fourier difference maps for α-NRXN_2-6 clearly show solvent density in place of the Ca2+ in LNS 6 and not the other LNS domains (Figure 4C). While LNS 6 presents the primary binding site for NLGN (Boucard et al., 2005), the overlay shows the bound NLGN to be proximal to the LNS 4 domain of α-NRXN with no significant steric obstruction between the α-NRXN and NLGN molecules. In the overlay, the interfacing surface of NLGN contains a flexible loop that includes the site for splice insert A. The insert introduces an additional 20 residues and would likely influence any interactions potentially mediated by this surface (Supplemental Figure S3). Similar results were produced in a comparison with the structure of NLGN-4 bound to β-NRXN (Fabrichny et al., 2007).
Figure 4. Overlay of α-NRXN_2-6 with β-NRXN bound to NLGN-1.
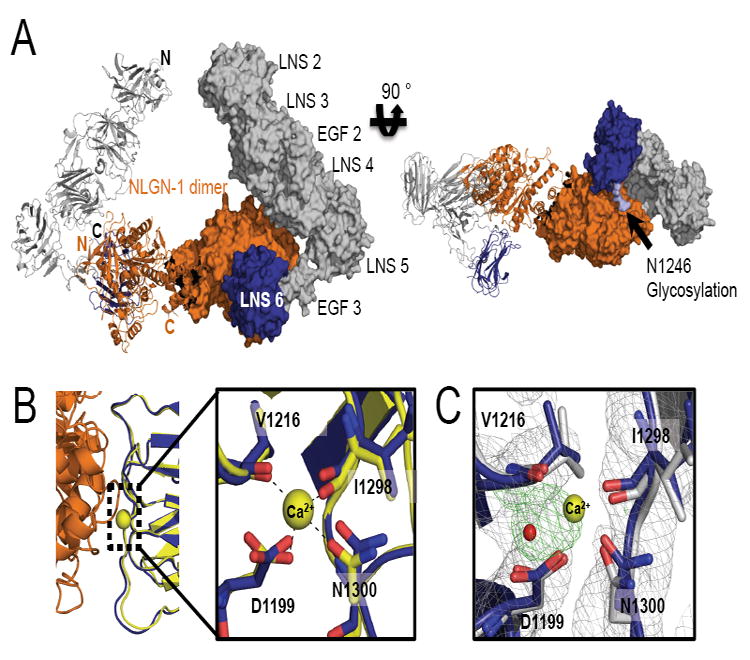
(A) A model of α-NRXN_2-6 in complex with the NLGN-1 dimer obtained by overlaying the LNS 6 domain in α-NRXN_2-6 onto the β-NRXN-1 molecules in the β-NRXN-1:NLGN-1 complex (PDB:3biw). The dimeric complex is shown as a ribbon representation for the left half and a surface representation for the right half. N- and C-termini are indicated on the left half in black for α-NRXN_2-6 and orange for NLGN-1. In the model, α-NRXN_2-6 can accommodate the NLGN molecule without steric hindrance, although with close proximity to LNS 4 (see also Supplemental Figure S3). The NLGN-1 dimer is displayed in orange and α-NRXN_2-6 is shown in gray (LN 2-EGF 3) and blue (LNS 6). On the right panel, a N-linked glycan on LNS 6 (N1246) is shown in light blue.
(B) The NLGN binding interface of α-NRXN_2-6 is conserved, as shown by the structural overlay with the NGLN-1:β-NRXN structure. In particular, the Ca2+-coordinating residues in β-NRXN (yellow) retain their conformation in the α-NRXN_2-6 LNS 6 domain (blue) despite the absence of Ca2+. Residue numbers are those of α-NRXN_2-6.
(C) An overlay of the Ca2+-free α-NRXN_2-6 molecule with Ca2+-bound β-NRXN-1 (PDB:3bod). The Ca2+ and water molecules in the β-NRXN structure are shown as yellow and red spheres, respectively. The 2Fo-Fc maps contoured at 1.2σ are displayed in gray and the Fo-Fc maps contoured at 3.0σ in green. In the absence of Ca2+, the LNS 6 domain in α-NRXN clearly coordinates an ion, which is likely to be sodium. Residue numbers are those of α-NRXN_2-6.
In the structure, the Ca2+ binding site on LNS 2, previously shown to regulate binding to α-dystroglycan (Sugita et al., 2001), is fully solvent accessible. However, absence of domains LNS 1 and EGF 1 in the α-NRXN_2-6 structure precludes a complete analysis of the exposed interfaces of LNS 2.
Binding affinity measurements of the α-NRXN:NLGN-1 complex
α- and β-NRXNs share the same binding domain for NLGNs (Boucard et al., 2005) and both demonstrate synaptogenic properties in cell culture (Graf et al., 2006; Graf et al., 2004; Kang et al., 2008). Yet their evolutionary conservation suggests that they function as independent molecules at the synapse. To address the question of whether α- and β-NRXNs exhibit distinguishing interactions with NLGN, we studied their individual binding properties by SPR. In these studies, the NLGN-1 dimer was immobilized through a C-terminal Fc domain onto a Protein A-coupled sensor chip and the monomeric NRXNs were injected as free ligands.
Soluble constructs of β-NRXN-1 and subsets of the α-NRXN-1 molecule with sequential deletions of each LNS domain (α-NRXN_2-6, α-NRXN_3-6, α-NRXN_4-6, α-NRXN_5-6) were used to analyze binding affinities with NLGN-1 lacking splice inserts A and B (NLGN-1-ΔAB) (Supplemental Figure S4). β-NRXN-1 (ΔSS#4) binds to NLGN-1ΔAB with ~30 nM KD, comparable with previously published results (Arac et al., 2007; Comoletti et al., 2006). The α-NRXN_5-6, α-NRXN_4-6 and α-NRXN_3-6 constructs all bind with a ~2-3 fold lower affinity (higher KD) and similar kinetics involving rapid bimolecular association and unimolecular dissociation (Figure 5 and Supplemental Figure S5). The α-NRXN_2-6 construct consistently yielded a KD of ~10-30 nM, reversing the trend of a slight decrease in KD with the additional domains. When normalized against the maximum binding response (Rmax), compared with the other truncated α-NRXNs, α-NRXN_2-6 shows the same fast bimolecular association, but displays a different dissociation profile consisting of an initial fast dissociation phase of greater amplitude, followed by a slow secondary dissociation (Supplemental Figure S5). Tight clustering in the calculated KD values across multiple surfaces with various densities of immobilized NLGN-1 attests to reproducibility of the measurements (Figure 5). Additionally, as was previously demonstrated by pull-down studies (Boucard et al., 2005), no binding to NLGN-1 was observed using the α-NRXN_2-5 construct at concentrations approaching 3 μM (Supplemental Figure S5). Although the specific recognition properties of the individual LNS domains remain to be resolved, the comparative equilibrium dissociation constants between the β-NRXN and truncated α-NRXNs demonstrate that both molecules can bind with low nM KD’s (high affinity). The 2-3-fold increase in KD (decrease in affinity) for α-NRXN corresponds to only minimal differences in free energy requirements for binding and therefore supports the model of a flexible hinge between LNS 5 and EGF 3 in α-NRXN-1.
Figure 5. NRXN-NLGN Association at Equilibrium Measured by SPR.
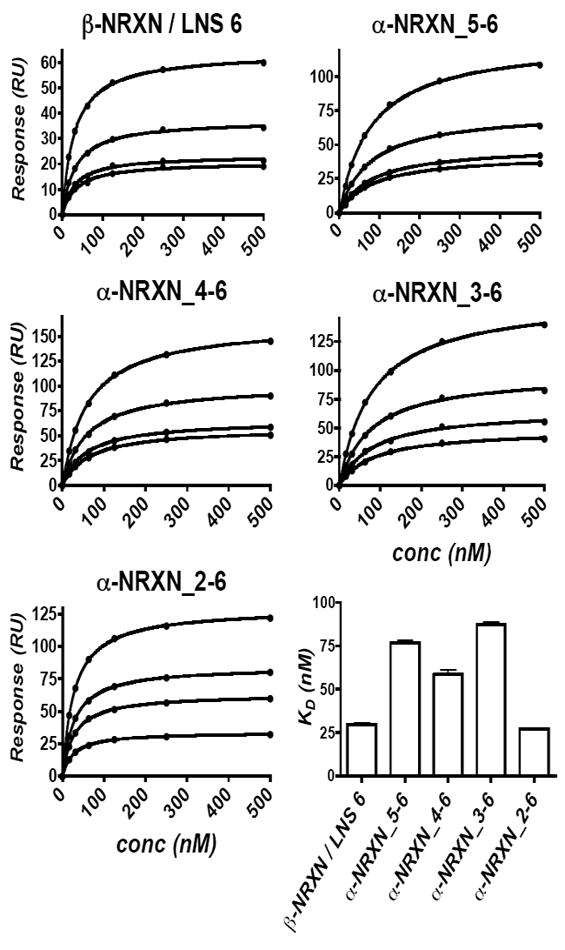
Four truncated α-NRXN constructs and β-NRXN were injected over immobilized NLGN-1. Equilibrium dissociation constants were independently calculated from four surfaces with variable densities of NLGN-1 (the four curves are shown for each construct). The saturation binding curves are similar across the different NLGN surfaces, as shown by the tight error bars in the lower right graph, indicating reproducibility. All five constructs bind to NLGN-1 with low nM affinity, indicating similar accessibility of the NLGN binding site on LNS 6 of the truncated α-NRXN molecules. (see also Supplemental Figure S5)
DISCUSSION
The structural characterization of the α-NRXN-1 extracellular region reveals how the concatenated five LNS and two EGF domains assemble into a L-shaped molecule and provides insights into its function as a receptor for several endogenous partnering proteins in the limited dimensions of a synaptic cleft. A complementary SPR analysis shows that α-NRXN binding to NLGN-1 is mostly triggered by the LNS 6/β-NRXN domain, which is likely to be accessible in solution due to a flexible linker between LNS 5 and EGF 3. The high resolution structure reveals the accessibility to known protein interaction sites, provides details on the atomic interactions leading to the assembled domains and offers more insight on the flexible properties of the protein compared to the previously determined EM and SAXS structures(Comoletti et al., 2010). Overlaying the α-NRXN_2-6 structure on the 2-D EM images of α-NRXN_1-6 demonstrates a similar overall arrangement of the domains despite the distinctive methodologies, and shows possible orientations of the flexible LNS 1 domain in relation to the other domains (Figure 6). Evident in the crystal structure, the extent of inter-domain contacts between LNS 2-5 suggests limited flexibility in their linear arrangement. On the other hand, a lack of stabilizing interactions at the connection between LNS 5 and EGF 3 (P1106) is indicative of innate flexibility at this position in the molecule, suggesting that the extension of the EGF 3-LNS 6 domains observed in our structure is one of the possible conformations affording solvent accessibility. The previously reported EM/SAXS models were specifically discussed in terms of “ensembles of structures” (Comoletti et al., 2010), supporting the notion that the LNS 6 domain can fold in towards the LNS 4 domain, creating either a more compact Y-shaped conformation or an extended L-shaped conformation as observed in the crystal structure. In agreement with the SPR studies, the dynamic arrangement of the two-arms is likely to influence the binding kinetics with partner proteins such as NLGN, with the extended conformation granting an energetically favorable state for association.
Figure 6. 2-D overlay of the α-NRXN_2-6 structure on negative stain EM images.
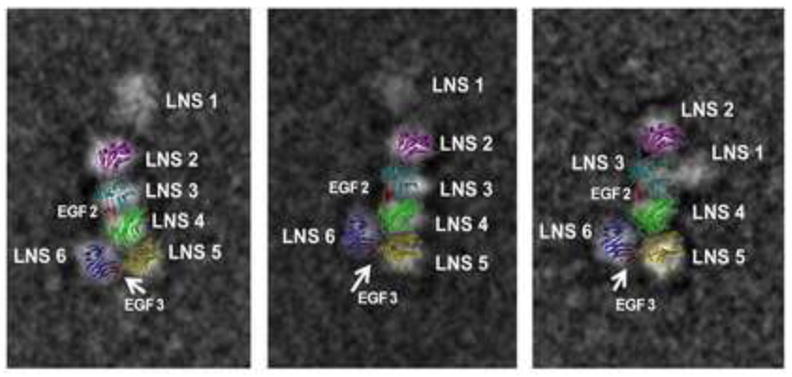
A 2-D image of the α-NRXN_2-6 structure overlaid onto three previously published 2-D negative stain single particle EM images of the α-NRXN_1-6 purified protein (Comoletti et al., 2010). The overall arrangement of the LNS 2-6 domains is conserved and the variable placement of the flexible LNS 1 domain relative to the crystal structure is observed.
Predicted Ca2+-binding sites in the α-NRXN-1 extracellular region
α-NRXN-1 interacts with the post-synaptic NLGNs, LLRTM and α-dystroglycan proteins in a Ca2+-dependent manner through the LNS 2 and/or LNS 6 domains (Arac et al., 2007; Ko et al., 2009; Sugita et al., 2001). In the structure, all of the predicted Ca2+-binding sites are aligned on the outer surface of the molecule with similar inter-site distances. The coordinating atoms at the Ca2+-binding sites in LNS domains 2, 3 and 6 have a conserved spatial arrangement based on a structural overlay with the Ca2+-bound structures of the LNS 2, 4 and 6/β-NRXN domains (Rudenko et al., 1999; Sheckler et al., 2006; Shen et al., 2008). For the LNS 6 domain, the Fourier difference maps show solvent coordination at the previously reported Ca2+-binding site (Koehnke et al., 2008; Shen et al., 2008), and may indicate a higher affinity site for divalent cation coordination compared to the other domains. On the other hand, based on a structural overlay, the inclusion of an insert at SS#3 in LNS 4 near the Ca2+-binding site, may reduce the Ca2+ affinity by altering the conformation of the coordinating L805 carbonyl oxygen (Shen et al., 2008). The loss of measurable Ca2+-binding to the LNS 2 domain due to an insert at SS#2 was previously demonstrated by isothermal titration calorimetry (Sheckler et al., 2006). The predicted Ca2+-binding site in LNS 5 appears intact, however, unlike the LNS 2, 3 and 6 domains that show the conserved spatial distances between the coordinating atoms, the LNS 5 domain has a more open site with increased inter-atomic distances. A definitive analysis of Ca2+-binding of α-NRXN awaits a structure with bound Ca2+.
Splice sites in the α-NRXN_2-6 structure
α-NRXN-1 has five sites of alternative splicing, three of which are found within the α-NRXN_2-6 sequence, SS#2 on LNS 2, SS#3 on LNS 4 and SS#4 on LNS 6. We have crystallized α-NRXN_2-6 containing the 10 amino acid insert at SS#3 (sequence DCIRINCNSS). The structure lacks density for six of the ten residues (R809-S815), probably due to the flexible nature of the segment, but the insert appears to create an extended loop between the β6-β7 sheets that does not significantly alter the conformation of the LNS 4 domain compared with the structure of LNS 4 devoid of SS#3 (Shen et al., 2008). Predicted structural features of the splice insert include a disulfide bond between C807 and C812 and an N-linked glycosylation site at N813. The splice insert lies proximal to the LNS 4 Ca2+-binding site and, together with the LNS 3 loop (K546-V561) that also extends towards the LNS 4 Ca2+-binding site, seems to partially occlude access of potential protein ligands to the Ca2+-binding face.
Our structure does not contain an insert at SS#2 on LNS 2 or SS#4 on LNS 6. A structural comparison between the LNS 6 domain and two structures of β-NRXN-1 containing the splice insert (SS#4) (Koehnke et al., 2010; Shen et al., 2008) shows that, while the two β-NRXN structures propose different conformations of the splice insert, either conformation could exist in the α-NRXN structure presented here.
O-glycosylation of EGF 2 in α-NRXN-1
The α-NRXN_2-6 structure reveals a previously uncharacterized O-linked glycosylation site on the first Ser residue in the EGF 2 domain. The identification of a single O-β-glucose moiety was based on the sequence alignment with a conserved O-glycosylation consensus motif previously identified in EGF-like domains in other cell-surface and secreted proteins, including factor VII, factor IX, protein Z and Notch (Haines and Irvine, 2003; Harris and Spellman, 1993). Two unusual O-linked glycans have been mapped to consensus sequences in EGF-like domains, yielding either an O-glucose or O-fucose post-translational modification (Harris and Spellman, 1993). The Notch receptor is made up of a series of EGF-like repeats in which the O-fucosylation pattern functions to modulate protein interactions that determine signal transduction capabilities (Haines and Irvine, 2003; Okajima et al., 2008). Many of the EGF domains in Notch contain the O-glucose modification, but a specific biological role remains to be determined (Moloney et al., 2000). In α-NRXN_2-6 the EGF 2 domain contains the consensus sequence for the O-glucose modification site. Accordingly, the structure shows a single glucose moiety attached to the EGF 2 domain. Although the preservation of such a moiety in α-NRXN-1 expressed in neurons remains to be determined, we illustrate here the possibility for a new glycosylation pattern of α-NRXN-1 that is likely to influence its function.
Disease-linked mutations in the α-NRXN-1 extracellular region
An increasing number of genetic studies implicate the NRXN-1 gene and its partnering NLGN genes as important determinants for the pathogenesis of several diseases of the central nervous system, including ASD, schizophrenia, Pitt-Hopkins-like syndrome-2, and milder forms of mental retardation (Kim et al., 2008; Rujescu et al., 2009; Sebat et al., 2007; Yan et al., 2008; Zahir et al., 2008; Zweier et al., 2009). Identified genetic abnormalities include copy number variations (Glessner et al., 2009; Rujescu et al., 2009; Sebat et al., 2007; Szatmari et al., 2007), chromosomal alterations (Kim et al., 2008; Yan et al., 2008; Zahir et al., 2008) and a few rare sequence mutations (Feng et al., 2006; Kim et al., 2008; Yan et al., 2008; Zweier et al., 2009). Reported missense mutations in the NRXN-1 gene include four sites in the α-NRXN-1 leader sequence (Kim et al., 2008; Yan et al., 2008), two in the β-NRXN-1 leader sequence (Feng et al., 2006), one in the EGF 2 domain (Kim et al., 2008; Yan et al., 2008), two flanking the EGF 2 domain, in LNS 3 and 4 (Yan et al., 2008) and one in the LNS 5 domain that leads to a premature stop codon (Zweier et al., 2009). The ASD-linked mutations found on the mature protein are clustered around the EGF 2 domain. They include T688I, L731I and E738K (Supplemental Figure S6). Mapping these disease-linked mutations does not reveal a defining role that should be disrupted by the mutations; however, their proximity may point to an important region of the protein for processing and/or functional regulation. A number of rare structural variants have also been identified in the partnering NLGN proteins (Comoletti et al., 2004; Talebizadeh et al., 2006; Yan et al., 2005). One reported NLGN mutation (NLGN-4 G99S), is exposed to a solvent surface of the NLGN dimer and shows proximity to the overlaid α-NRXN molecule such that the mutation may affect the surface or alter its conformation to influence α-NRXN binding (Supplemental Figure S6).
The α-NRXN:NLGN complex
α- and β-NRXNs share the same binding domain for the NLGNs; however, it is not fully understood if these comparable interactions are functionally distinct. Binding properties between the NLGNs and β-NRXNs in their various splice isoforms have been extensively studied using SPR (Arac et al., 2007; Boucard et al., 2005; Comoletti et al., 2003; Comoletti et al., 2006; Koehnke et al., 2010). Information on the α-NRXN binding properties with NLGN has been limited to data from affinity pull-down assays and a single SPR experiment comparing the relative affinities of α-NRXN-1 with NLGN-1 (Reissner et al., 2008). Pull-down experiments concluded that α-NRXN-1, with or without splice insert #4, binds to NLGN-1-3 and is regulated by splice insert B in NLGN-1 and that binding is exclusively mediated by the LNS 6 domain (Boucard et al., 2005). Another pull-down experiment suggested that α-NRXN-1 binding to NLGN-1 containing splice insert B is regulated by the structural constraints imposed by the disulfide bonds in the EGF 3 domain (Reissner et al., 2008). By SPR, the relative binding affinity of α-NRXN-1 compared to β-NRXN-1 with NLGN-1 was reported to be ~2-fold less when α-NRXN lacks SS#4 and ~4-fold less with SS#4 (Boucard et al., 2005).
We suggest that the flexibility of the EGF 3-LNS 6 arm has functional implications on the accessibility of the LNS 6 binding site to receptor proteins, including the NLGNs. By superimposing the α-NRXN_2-6 structure on the β-NRXN:NLGN complexes, it becomes apparent that the EGF 3-LNS 6 arm must be in an extended conformation in order to accommodate the NLGN molecule without steric hindrance. Our SPR data show that the sequential addition of LNS domains 2-5 to LNS 6 results in, at most, a 2-3-fold decrease in affinity for NLGN-1 compared to β-NRXN. These variations in the KD values indicate minimal differences in free energy requirements for binding of the truncated NRXNs. Hence modest conformational adjustments are needed for α-NRXN to accommodate the NLGN molecule, thereby supporting the propensity for α-NRXN to form an extended conformation as revealed in the crystal structure.
In the superimposed crystal structure, the LNS 4, 5 and 6 domains fit snugly around the NLGN molecule without evident hindrance, yet with close proximity to the LNS 4 domain. The position of splice insert A in NLGN-1 and -2 at the proximal interface with LNS 4 prompts the question of whether there is a functional role for splice site A in NLGN binding affinities for the α-NRXNs. A previous study showed that the presence of splice insert A in NLGN-1, in the absence of splice insert B, or in NLGN-2 promotes targeting of either neuroligin to GABAergic synapses (Chih et al., 2006). The insert at splice site A adds 20 amino acid residues to the structure and is heavily populated with both cationic and anionic residues. The proximity of this loop region in the structural overlay suggests that the combination of bulk and charge of the added sequence would impact NLGN binding to the α-NRXN_2-6 in the conformation shown here. Overall, it remains to be determined if the α-NRXN:NLGN complex stabilizes the L-shape configuration of the domains, or if the two arms would preferentially open up to form an obtuse configuration and if there is a role for splicing in establishing selectivity of interaction. In either case the EGF 3 domain separates the LNS 6 domain from the other five LNS domains in order to accommodate the NLGN molecule.
A working model of the α-NRXN:NLGN complex at the synapse
Both the α-NRXN and NLGN molecules have long, semi-flexible stalk domains connecting their extracellular binding domains to the cell membrane. The stalk regions contain a relatively large number of serine and threonine residues that are O-glycosylated (Ushkaryov et al., 1992). The glycosylation pattern confers some hydration to the chain and the combination of oligosaccharides and proline amino acid residues is thought to impart some rigidity to the peptide chain, as demonstrated in NLGN-1 and other cell surface receptors (Comoletti et al., 2007; Merry et al., 2003).
The crystal structure of α-NRXN_2-6 molecule has a maximum length of ~125 Å. The full extracellular region of α-NRXN has an additional N-terminal LNS domain (LNS 1) and EGF domain (EGF 1), which elongates the soluble protein by ~35 Å, but also shows extensive flexibility (Comoletti et al., 2010). Therefore, depending on the orientations of the LNS 1 domain and the stalk domain, the maximum dimension of the full extracellular region of α-NRXN approaches or possibly exceeds the membrane-to-membrane distance of a typical synaptic cleft (~200 Å). However, in a similar case, structural studies on the large cell-adhesion proteins, cadherins, indicate that they may create repulsive forces that increase the distance between pre- and post-synaptic membranes to approximately 250 Å (Craig et al., 2006).
Crystal structures of the complexes between the extracellular domains of NLGN and β-NRXN show a NLGN dimer is formed through a hydrophobic four-helix bundle involving the C-terminal α-helices in each subunit with the two C-termini pointing in one direction and two β-NRXN molecules bound on opposite sides of the long axis of the NLGN dimer with their C-termini pointing opposite to the NLGN C-termini (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007). The opposing directionality of the C-termini of β-NRXN and NLGN is indicative of their tethering to the pre- and post-synaptic membranes, respectively. Overlaying α-NRXN onto the β-NRXN:NLGN complex positions the long LNS 2-5 arm of α-NRXN perpendicular to the membrane (Figure 7). If stabilized in this conformation, the N-terminal domains of α-NRXN would lie in close proximity to the pre-synaptic membrane. α-NRXNs are distinct from β-NRXNs in their capacity to regulate Ca2+-dependent exocytosis at the pre-synapse, a role that is believed to be a function of the α-specific domains (Missler et al., 2003; Zhang et al., 2005). Furthermore, α-NRXNs are receptors for the α-latrotoxin protein and function by recruiting the toxin to the pre-synaptic membrane where it can position itself to stimulate exocytosis (Sudhof, 2001). Therefore, a spatial arrangement of α-NRXN with its N-terminus close to the pre-synaptic membrane could favor the observed regulation of Ca2+-dependent neurotransmitter release.
Figure 7. Model of the α-NRXN:NLGN Complex at the Synapse.
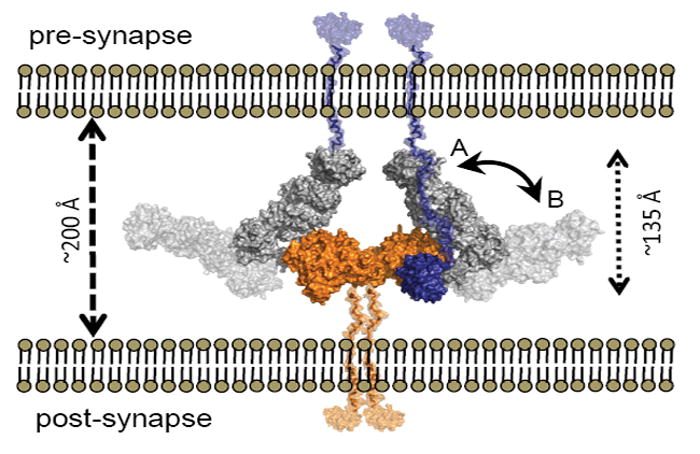
Overlays of α-NRXN_2-6 onto β-NRXN-1 as bound to NLGN-1 or -4 suggests a possible position for the α-NRXN:NLGN complex in the synaptic cleft. The overlaid complex is shown with the C-termini of α-NRXN and NLGN oriented towards the pre- and post-synaptic membranes, respectively. In this conformation the N-terminal domains lay to the pre-synaptic membrane (conformation A, non-transparent surface,). Flexibility at the hinge point between LNS 5 and EGF 3 could allow segmental motion of the α-NRXN_2-6 long arm towards a fully linear conformation of the extracellular region, moving the N-terminal domains in closer proximity to the post-synaptic membrane (conformation B, transparent surface). The relative position of the N-terminal domains is likely to confer selectivity for additional binding partners. The NLGN dimer is colored in orange, the α-NRXN LNS 2-EGF 3 in gray and LNS 6 in blue. Stalk domains for NLGN and α-NRXN_2-6, shown in semi-transparent orange and blue, respectively, were modeled into the figure.
On the other hand, the N-terminal domains of α-NRXN may also function by interacting with proteins on the post-synaptic membrane, as evident from LNS 2 binding to α-dystroglycan (Sugita et al., 2001). The extent of torsional flexibility in the region between LNS 5 and EGF 3, as dictated by the α-NRXN:NLGN complex, may allow for the long arm of α-NRXN to open into a more linear arrangement, lateral to the membrane, thereby allowing the α-NRXN to form additional contacts with post-synaptic receptor proteins (Figure 7). The two identified protein-binding domains, LNS 2 and LNS 6, are separated by ~100 Å, which indicates that with the appropriate alignment, both sites could be occupied at the same time. The potential of α-NRXNs to host multiple partnering proteins suggests a possible scaffold-like functionality that would determine the architectural spacing of the molecular network in the trans-synaptic space. Therefore, future studies to address the possible conformations of the α-NRXN domains when bound to NLGN will be of interest.
EXPERIMENTAL PROCEEDURES
Cloning, expression and protein purification
The original bovine cDNA encoding the secreted, soluble extracellular region of α-NRXN-1 fused with a C-terminal IgG Fc domain to permit affinity chromatography, and inserted in expression vector p-cDNA 3.1, was a gift from Dr. Thomas Sudhof (Ushkaryov et al., 1992). The construct was further modified to introduce a 3Cpro cleavage site upstream from the Fc domain as described previously(Comoletti et al., 2010). All modified α-NRXN constructs contained the original expression vector, signal peptide sequence and Fc sequence. α-NRXN-1 and its various truncated forms were expressed in the mammalian cell line, HEK293 GnT1-, lacking the N-acetyglucosaminyltransferase I gene needed for high-order N-linked glycosylation processing. The soluble recombinant β-NRXN-1 protein used for SPR studies was expressed in bacterial Rosetta Plys cells as an N-terminally 6-His tagged protein as previously described(Fabrichny et al., 2007).
NLGN1ΔAB (residues Q46-S693 without splice inserts A or B) was cloned into p-cDNA 3.1 between the sequences encoding the α-NRXN signal peptide and the C-terminal 3Cpro cleavage site, itself followed by the sequence encoding the Fc domain. This construct was transfected into HEK293 cells for expression in selected stable cell lines.
Crystallization and X-ray Data Collection
α-NRXN-1 containing domains LNS 2 through LNS 6 (α-NRXN_2-6), residues 296-1349 (NP_776829), lacking inserts at SS#2 and #4 but containing the 10-residue splice insert at SS #3 (Supplemental Figure S4) was concentrated to ~3 mg/ml in HEPES buffered saline pH 7.4(HBS). Crystals grew as thin rods by sitting drop vapor diffusion at 14°C in 96-well plates (Innovaplate SD-2, Innovadyne Technologies). Drops of 0.6 μl were set at a 1:1 (v/v) ratio of protein to reservoir buffer (0.1M Bicine pH 9.0, 12% PEG 20,000, 1mM EDTA). Crystals were soaked into the mother liquor complemented with 10-25% glycerol for several minutes before being flash cooled in liquid nitrogen. X-ray diffraction data were collected at a temperature of 100 K from the 11-1 beamline at the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, CA). Diffraction data were processed with XDS(Kabsch, 1993).
The crystals belong to space group C2 and contain one α-NRXN_2-6 molecule per asymmetric unit with a solvent content of 65%.
Extensive attempts to crystallize the protein in the presence of Ca2+ were unsuccessful. Crystals were grown under the Ca2+-free conditions reported here, both with and without the additive EDTA. Soaking the crystals grown without EDTA in a cryo buffer containing 1-10mM Ca2+ did not yield high quality diffraction patterns. The majority of the crystals had a very thin third dimension and were either too fragile to harvest or produced weak, anisotropic diffraction in the 4-8 Å range.
Structure Determination, Refinement and Validation
The structure of α-NRXN_2-6 was determined at 3.0 Å resolution by molecular replacement using the coordinates of previously solved structures for LNS 2 (PDB 2h0b), LNS 4 (PDB 2r16) and β-NRXN (PDB 3bod) as search models for input into PHASER(McCoy et al., 2007). An initial search using the three PDB structures successfully located the LNS 2, 4 and 6 domains. Using the phase information from the previous search, the LNS 3 and 5 domains were located using the coordinates of the LNS 2 and LNS 4 structures, respectively, as the search models in PHASER. Building of LNS 3 and 5 was achieved through subsequent cycles of modeling and refinement. After rigid body refinement of all LNS domains, the EGF-like domains were manually built into the difference Fourier maps using Coot. In the final refinement stages each LNS and EGF domain was used to define seven TLS groups for refinement. All model building and refinement was performed in Coot (Emsley and Cowtan, 2004) and Refmac5 from the CCP4 program suite (CCP4, 1994). A combination of Whatcheck (Hooft et al., 1996) and Molprobity (Chen et al., 2010) were used for structure validation.
The structure was refined to a final Rwork = 0.21 and Rfree = 0.27 and shows 1,005 amino acid residues. It shows no density for residues K551-D552 in LNS 3 and R809-S815 in LNS 4. The latter sequence corresponds to most of SS#3. The structure also includes 33 water molecules, one β-Glc moiety O-linked to S705 in the EGF 2 domain, and part of an N-linked glycan bound to N1246 in LNS 6, fitting the electron density of 2 molecules of GlcNAc and one Man.
Structural Analysis
All analyses of the domains interfaces used the PDBePISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html)(Krissinel and Henrick, 2007). Average rmsds of individual LNS domains were calculated using the secondary structure matching module in COOT(Emsley and Cowtan, 2004). Overlays of the previously solved LNS 2, 4 and 6/β-NRXN domains (PDB: 2h0b, 2r16 and 3bod) onto their respective domains in the α-NRXN_2-6 structure gave average rmsd values between 0.5 and 0.9 Å between Cα atoms. Comparing the five individual LNS domains in the α-NRXN_2-6 structure yielded average rmsds of 1.42-2.3 Å for Cα atoms. Individual Cα rmsds between the LNS domain in α-NRXN_2-6 were calculated using ClustalW(Chenna et al., 2003) and the THESEUS software package(Theobald and Wuttke, 2006). Structural overlays of α-NRXN_2-6 with the β-NRXN:NLGN-1 or β-NRXN:NLGN-4 complexes (PDB: 3biw and 2xb6, respectively), α-NRXN_2-6 with the β-NRXN+SS#4 structures (PDB:2r1b and 3mw2) and the NLGN-2 dimer (PDB:3bl8) with the β-NRXN:NLGN-1 complex were performed using the secondary-structure matching (SSM) module in Coot. All structure figures and electrostatics calculations were generated using PyMol (DeLano Scientific, LLC).
Surface Plasmon Resonance
For details of the NRXN proteins used for the SPR studies see supplemental Figure S4. All NRXN proteins were analyzed for binding against the NLGN-1ΔAB isoform. Affinity binding analysis was performed on a ProteON X36 biosensor with the GLC chip platform (Bio-Rad Laboratories, Inc.) in a running buffer made of HBS, 3mM Ca2+, 0.05% Tween 20, 1mg/mL bovine serum albumin, at 25 °C. For more details see supplemental material.
The GLC chip was equilibrated in HBS buffer at 25 °C for immobilization of NLGN to the mono-layered surface of the sensor chip. Protein A was immobilized on the biosensor chip surface in the vertical direction using amine coupling chemistry(Rich and Myszka, 2001). The surfaces were deactivated with 1M ethanolamine and washed with glycine pH 2.0. NLGN-1ΔAB-Fc was captured over the Protein A surface from filtered medium of cultured HEK293 cells (injected in vertical orientation at 30 μl/min). One channel was dedicated as a control where medium from non-transfected HEK293 cells was injected. Different dilutions of the medium were used to achieve variable amounts of NLGN-1 on the surface, which was necessary to achieve an optimal signal level (RUs) for the various NRXN analytes, which ranged from ~22 kD for β-NRXN to ~140 kD for α-NRXN_1-6.
Affinity binding analysis was performed in a running buffer made of HBS, 3mM Ca2+, 0.05% Tween 20, 1mg/mL BSA at 25 °C. In the experiment shown here each NRXN protein was prepared in a two-fold dilution sequence in running buffer starting at a concentration of 15.6 nM and ending at 1 μM. Additional experiments were performed using a three-fold concentration gradient going from 1.2 nM to 900 nM. For each injection series, one channel was injected with running buffer to use as a double reference(Myszka, 1999). Samples were injected orthogonal to the NLGN surfaces for 60 s, at a flow rate of 50 μl/min, followed by a 600 s dissociation phase. For each injection the NRXN analyte was exposed to 5 independent surfaces with NLGN-1ΔAB and one surface with only Protein A. The surface was regenerated with an 18 s injection of 350mM EDTA, at 100 μl/min after each injection, followed by a 60 s injection of running buffer at 100 μl/min. The lack of binding on control Protein A surfaces upon injection of culture medium from non-transfected HEK293 cells or containing α-NRXN_1-6 with a C-terminal Fc fragment verified the absence of appreciable non-specific binding. The graphs shown here were generated using GraphPad Prism v.4.0b (Graphpad Software, La Jolla, CA).
Supplementary Material
Highlights.
3.0 Å structure of α-NRXN-1, LNS 2-6, reveals asymmetry and a flexible hinge
Domain arrangement allows for simultaneous binding events
A novel O-linked glycosylation site is revealed on the EGF2 domain
α-NRXN binding to NLGN-1 has nM affinity and is governed by LNS 6
Acknowledgments
We are grateful to Drs. Yves Bourne and Pascale Marchot for helpful comments on the manuscript. This work was supported by NIH grants RO1-GM18360-39 and P42ES 10337 to PT and Training Grant GM 07752 to MTM and Autism Speaks #2617 to DC. X-ray data were collected at that the BL11-1 beamline of the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, CA).
List of abbreviations
- NRXN
neurexin
- NLGN
neuroligin
- ASD
autism spectrum disorders
- LNS
laminin neurexin sex hormone-binding globulin
- EGF
epidermal growth factor
- SPR
surface plasmon resonance
Footnotes
Accession codes - Atomic coordinates and structure factors have been deposited with the Protein Data Bank: accession code 3POY
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac D, Boucard AA, Ozkan E, Strop P, Newell E, Sudhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu H, Shim AH, Focia PJ, He X. Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions. Nat Struct Mol Biol. 2008;15:50–56. doi: 10.1038/nsmb1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SB, Amici SA, Ren XQ, McKay SB, Treuil MW, Lindstrom JM, Rao J, Anand R. Presynaptic targeting of alpha4beta 2 nicotinic acetylcholine receptors is regulated by neurexin-1beta. J Biol Chem. 2009;284:23251–23259. doi: 10.1074/jbc.M109.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Comoletti D, De Jaco A, Jennings LL, Flynn RE, Gaietta G, Tsigelny I, Ellisman MH, Taylor P. The Arg451Cys-neuroligin-3 mutation associated with autism reveals a defect in protein processing. J Neurosci. 2004;24:4889–4893. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn R, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Sudhof TC, Taylor P. Characterization of the interaction of a recombinant soluble neuroligin-1 with neurexin-1beta. J Biol Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Grishaev A, Whitten AE, Tsigelny I, Taylor P, Trewhella J. Synaptic arrangement of the neuroligin/beta-neurexin complex revealed by X-ray and neutron scattering. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Miller MT, Jeffries CM, Wilson J, Demeler B, Taylor P, Trewhella J, Nakagawa T. The macromolecular architecture of extracellular domain of alphaNRXN1: domain organization, flexibility, and insights into trans-synaptic disposition. Structure. 2010;18:1044–1053. doi: 10.1016/j.str.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Graf ER, Linhoff MW. How to build a central synapse: clues from cell culture. Trends Neurosci. 2006;29:8–20. doi: 10.1016/j.tins.2005.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletov BA, Krasnoperov V, Hata Y, Petrenko AG, Sudhof TC. High affinity binding of alpha-latrotoxin to recombinant neurexin I alpha. J Biol Chem. 1995;270:23903–23905. doi: 10.1074/jbc.270.41.23903. [DOI] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26:4256–4265. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Harris RJ, Spellman MW. O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology. 1993;3:219–224. doi: 10.1093/glycob/3.3.219. [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- Kang Y, Zhang X, Dobie F, Wu H, Craig AM. Induction of GABAergic postsynaptic differentiation by alpha-neurexins. J Biol Chem. 2008;283:2323–2334. doi: 10.1074/jbc.M703957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Jin X, Trbovic N, Katsamba PS, Brasch J, Ahlsen G, Scheiffele P, Honig B, Palmer AG, 3rd, Shapiro L. Crystal structures of beta-neurexin 1 and beta-neurexin 2 ectodomains and dynamics of splice insertion sequence 4. Structure. 2008;16:410–421. doi: 10.1016/j.str.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Katsamba PS, Ahlsen G, Bahna F, Vendome J, Honig B, Shapiro L, Jin X. Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron. 2010;67:61–74. doi: 10.1016/j.neuron.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Leone P, Comoletti D, Ferracci G, Conrod S, Garcia SU, Taylor P, Bourne Y, Marchot P. Structural insights into the exquisite selectivity of neurexin/neuroligin synaptic interactions. EMBO J. 2010;29:2461–2471. doi: 10.1038/emboj.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry AH, Gilbert RJ, Shore DA, Royle L, Miroshnychenko O, Vuong M, Wormald MR, Harvey DJ, Dwek RA, Classon BJ, et al. O-glycan sialylation and the structure of the stalk-like region of the T cell co-receptor CD8. J Biol Chem. 2003;278:27119–27128. doi: 10.1074/jbc.M213056200. [DOI] [PubMed] [Google Scholar]
- Missler M, Hammer RE, Sudhof TC. Neurexophilin binding to alpha-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. J Biol Chem. 1998;273:34716–34723. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Okajima T, Matsuura A, Matsuda T. Biological functions of glycosyltransferase genes involved in O-fucose glycan synthesis. J Biochem. 2008;144:1–6. doi: 10.1093/jb/mvn016. [DOI] [PubMed] [Google Scholar]
- Reissner C, Klose M, Fairless R, Missler M. Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components. Proc Natl Acad Sci U S A. 2008;105:15124–15129. doi: 10.1073/pnas.0801639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich RL, Myszka DG. BIACORE J: a new platform for routine biomolecular interaction analysis. J Mol Recognit. 2001;14:223–228. doi: 10.1002/jmr.535. [DOI] [PubMed] [Google Scholar]
- Rini JM. Lectin structure. Annu Rev Biophys Biomol Struct. 1995;24:551–577. doi: 10.1146/annurev.bb.24.060195.003003. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J. The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J Biol Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KC, Kuczynska DA, Wu IJ, Murray BH, Sheckler LR, Rudenko G. Regulation of neurexin 1beta tertiary structure and ligand binding through alternative splicing. Structure. 2008;16:422–431. doi: 10.1016/j.str.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci U S A. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu Rev Neurosci. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones IR, Craddock N, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, Marra MA, Friedman JM. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, Sudhof TC. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Rohlmann A, Sargsyan V, Aramuni G, Hammer RE, Sudhof TC, Missler M. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J Neurosci. 2005;25:4330–4342. doi: 10.1523/JNEUROSCI.0497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, Bijlsma EK, Oortveld MA, Ekici AB, Reis A, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


